Abstract
The biosynthesis of the lantibiotic subtilin is autoinduced in a quorum-sensing mechanism via histidine kinase SpaK. Subtilin-like lantibiotics, such as entianin, ericin S, and subtilin, specifically activated SpaK in a comparable manner, whereas the structurally similar nisin did not provide the signal for SpaK activation at nontoxic concentrations. Surprisingly, nevertheless, nisin if applied together with entianin partly quenched SpaK activation. The N-terminal entianin1–20 fragment (comprising N-terminal amino acids 1 to 20) was sufficient for SpaK activation, although higher concentrations were needed. The N-terminal nisin1–20 fragment also interfered with entianin-mediated activation of SpaK and, remarkably, at extremely high concentrations also activated SpaK. Our data show that the N-terminal entianin1–20 fragment is sufficient for SpaK activation. However, if present, the C-terminal part of the molecule further strongly enhances the activation, possibly by its interference with the cellular membrane. As shown by using lipid II-interfering substances and a lipid II-deficient mutant strain, lipid II is not needed for the sensing mechanism.
INTRODUCTION
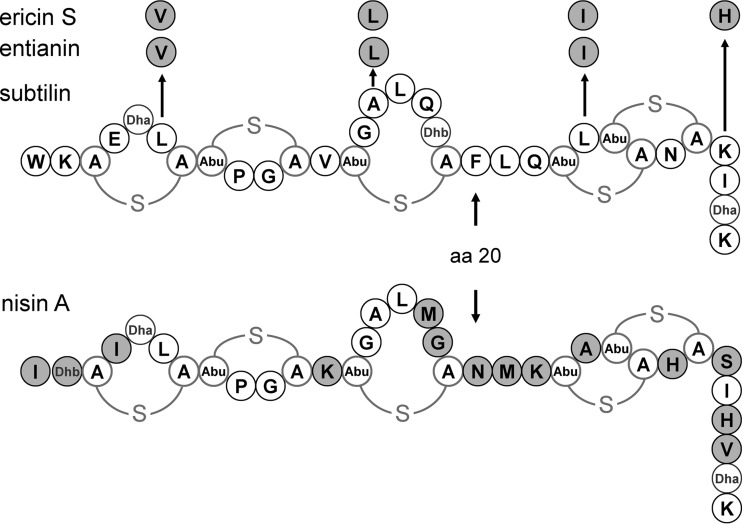
Linear lantibiotics like subtilin from Bacillus subtilis ATCC 6633, ericin S from B. subtilis A1/3, entianin from B. subtilis DSM 15029, nisin from Lactococcus lactis (see Fig. 1), epidermin from Staphylococcus epidermidis, and gallidermin from Staphylococcus gallinarum, form a class of peptide antibiotics (bacteriocins) with high antimicrobial potential (1–6). Members of this special bacteriocin class are ribosomally synthesized and subsequently posttranslationally modified, yielding characteristic lanthionine ring structures from which the names for the lantibiotics are derived (1). The lanthionine structures are derived from dehydration of the amino acid residues serine and threonine, followed by the nucleophilic intramolecular addition of a neighboring cysteine. Both the dehydration and the cyclization are mediated by a multimeric synthetase complex consisting of the enzymes LanB and LanC as well as LanT, important for transport of the lantibiotic molecule out of the cell (7, 8). LanB catalyzes the dehydration reaction (9), and LanC catalyzes cyclization (10).
FIG 1.
Structures of subtilin-like lantibiotics subtilin, entianin, and ericin S, produced by diverse B. subtilis strains, and nisin A, produced by diverse Lactococcus lactis strains. Amino acid residues of ericin S, entianin, and nisin A that differ from those in subtilin are highlighted in gray. Amino acid position 20 is indicated by aa 20. A-S-A, meso-lanthionine; Abu-S-A, 3-methyllanthionine (Abu refers to α-aminobutyric acid); Dha, 2,3-didehydroalanine; Dhb, 2,3-didehydrobutyrine.
Nisin has been shown to bind the cell wall precursor lipid II (11, 12) by forming a unique lipid II-binding motif with the well-conserved N-terminal rings A and B of the nisin and subtilin subfamily of type A lantibiotics (13). Binding to lipid II enables the lantibiotic to permeabilize the cytoplasmic membrane. This pore formation seems to be the primary mode of action for this class of lantibiotics. Structural data revealed that the pyrophosphate moiety of lipid II seems to be the binding target for linear lantibiotics like nisin and subtilin, thus differing from other peptide antibiotics that target lipid II, such as vancomycin, which binds to the dipeptide d-Ala-d-Ala moiety (14, 15), and bacitracin, which prevents the recycling of lipid II upon binding to undecaprenyl pyrophosphate (16, 17). For ramoplanin, which like nisin and subtilin also recognizes the pyrophosphate moiety of lipid II (18, 19), a prevention of nisin-induced membrane poration was demonstrated (11), underlining the competitive nature of the two antibiotics.
Biosynthesis of subtilin is subject to a dual-control mechanism based on two independent regulatory systems (20). A two-component system (TCS) consisting of histidine kinase SpaK and response regulator SpaR (21) is activated by subtilin in a quorum-sensing-like mechanism (20), which has also been described previously for nisin (22, 23). Whereas the TCS NisRK is constitutively expressed (24), the TCS SpaRK is under the control of the major transition state regulator AbrB (20, 25, 26) and the alternative sigma factor H (27, 28). Therefore, in contrast to nisin, subtilin synthesis is growth phase dependent (29). Furthermore, recently published data demonstrated that subtilin synthesis is also retarded in the presence of high glucose concentrations (30).
Biosynthesis of subtilin was followed using a PspaS-lacZ reporter fusion (31) which senses histidine kinase SpaK-mediated autoinduction of subtilin-like lantibiotics such as subtilin, ericin S, and entianin (see Fig. 1). This reporter system allows the quantification of SpaK activation by means of a simple β-galactosidase activity assay. Despite a high similarity between subtilin and nisin, the activation of the TCSs is highly specific for the respective lantibiotic (31), which implies that a unique recognition site must exist within the respective histidine kinases.
In this report, we show that the N-terminal entianin1–20 fragment (comprising the N-terminal amino acids 1 to 20) is sufficient for activation of SpaK histidine kinase and that lipid II is not needed for the autoinduction of subtilin-like lantibiotic biosynthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table 1. B. subtilis wild-type strains B15029 and B6633 were grown in medium A (32, 33) for increased production of the respective lantibiotic and its succinylated version. B168.AUT3 and B168.AUT5 are derivates of strain PDC204 (34) and can be converted from rod-shaped cells to cell wall-deficient L-forms by cultivation on osmotically stabilized medium. For rod-shaped cells, strains were inoculated in nutrient broth medium (Oxoid) or streaked onto nutrient agar (Oxoid), each supplemented with 0.5% xylose. For selection, the antibiotic concentrations used were as follows: erythromycin, 1 μg ml−1; spectinomycin, 100 μg ml−1; kanamycin, 10 μg ml−1. When selection was done with two or more antibiotics simultaneously, the concentrations were reduced to half. The cultivation of L-form cells was performed under nonselective conditions and in the absence of xylose in nutrient broth medium or nutrient agar plates supplemented with 20 mM MgCl2 and 0.5 M sucrose and buffered with 20 mM maleic acid (MSM medium) for osmotic stabilization. Escherichia coli DH5α strains with recombinant plasmids were grown in TB medium (12 g tryptone, 24 g yeast extract, and 4 ml glycerol with the addition of 900 ml H2O plus 100 ml KPP buffer [0.17 M KH2PO4, 0.72 M K2HPO4]), containing 100 μg ml−1 ampicillin.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Descriptiona | Source or referenceb |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| B15029 | Wild type (Ent+) | DSM 15029 |
| B6633 | Wild type (Sub+) | ATCC 6633 |
| B6633.MB1 | ΔspaS amyE::PspaS-lacZ (Specr Cmr Sub−) | 31 |
| PDC204 | W168 trpC2 ΩspoVD::Erm Pxyl-murE (Eryr) | 34 |
| B168.AUT3 | PDC204 pyrD::Kanr PspaRK-spaRK (Eryr Kanr) | This work |
| B168.AUT5 | B168.AUT3 thrC::Specr PspaS-lacZ (Eryr Kanr Specr) | This work |
| E. coli DH5α | recA1 endA1 gyrA96 thi hsdR17(rK− mK+) relA1 supE44 ϕ80ΔlacZΔM15 Δ(lacZYA-argF)U169 | Laboratory stock |
| Plasmids | ||
| pXT | bla thrC′ Spec ′thrC | 35 |
| pPyr-kan | bla pyrD′ Kan ′pyrD | 36 |
| pSB5 | bla amyE′ PspaS-lacZ Cm ′amyE | 20 |
| pAUT1 | pXT PspaS-lacZ | This work |
| pAUT2 | pPyr-kan PspaRK-spaRK | This work |
| Oligonucleotides | ||
| AUT1 | CGATCAATTGAATTCTGGAAGG | This work |
| AUT2 | TAGCAAGCTTTCACGCAGAATTAAAT | This work |
| AUT3 | GCATGGATCCGCATGAAATAAATTCAGGGGTATTG | This work |
| AUT4 | GCTAGAGCTCGAAGATGGATCAGTTCAC | This work |
Ent+, entianin producer; Sub+, subtilin producer; Cmr, chloramphenicol resistant; Specr, spectinomycin resistant; Kanr, kanamycin resistant; Eryr, erythromycin resistant.
DSM, German Resource Centre for Biological Material; ATCC, American Type Culture Collection.
Plasmid construction.
All plasmids and oligonucleotides used in this study are listed in Table 1. The plasmid pAUT1 was constructed after digestion of pXT (35) with MfeI/EcoRI and ligation of the construct PspaS-lacZ that had been PCR amplified from pSB5 with oligonucleotides AUT1 and AUT2 digested with the same enzymes. pAUT2 was constructed after digestion of pPyr-kan (36) with BamHI/SacI and ligation of the construct PspaRK-spaRK that had been PCR amplified from chromosomal DNA of B6633 with oligonucleotides AUT3 and AUT4 digested with the same enzymes. Recombinant plasmids were isolated by an alkaline extraction procedure (37).
Bacillus transformation.
An overnight culture of the preferred B. subtilis strain was inoculated to an optical density at 600 nm (OD600) of 0.1 in 10 ml MNGE medium, which consisted of 9.2 ml 1× MN medium (136 g K2HPO4·3 H2O, 60 g KH2PO4, 10 g Na citrate·2 H2O, add 1 liter), 1 ml 20% glucose, 50 μl 40% K glutamate, 50 ml Fe(III)-ammonium-citrate (2.2 mg ml−1), 100 μl tryptophan (5 mg ml−1), and 30 μl 1 M MgSO4. For strains carrying an insertion in the thrC or pyrD locus, 100 μl threonine (5 mg ml−1) or 100 μl uracil (4 mg ml−1), respectively, was added. The cells were grown to an OD600 of 1.1 to 1.3 at 37°C with an agitation of 200 rpm. For transformation, 400 μl cells was transferred in a small glass test tube containing 1 to 5 μg of linearized plasmid. The culture was further incubated as described above. After 1 h, 100 μl expression mix (500 μl 5% yeast extract, 250 μl 10% Casamino Acids, 250 μl H2O, and 50 μl tryptophan [5 mg ml−1]) was added. The cells were further cultivated for 1 h and thereafter plated on selective medium. The protocol was provided after personal communication with Thorsten Mascher, TU Dresden.
L-form strain construction.
Transformation of PDC204 and derivates was performed as described above by additional supplementation of 0.5% xylose. Transformation of PDC204 with pAUT2 linearized with ScaI resulted in strain B168.AUT3, and transformation of B168.AUT3 with pAUT1 linearized with SpeI resulted in strain B168.AUT5 containing PspaRK-spaRK and PspaS-lacZ fusions (Table 1).
Purification of entianin and subtilin and N-terminal fragments.
Strain B15029 or B6633 was grown in 50 ml medium A at 37°C for 48 h under permanent shaking at 155 rpm. The supernatant was harvested by centrifugation. Afterwards, a modification of previously published purification procedures was used (33, 38, 39). To the supernatant, 0.5 volume of precooled n-butanol was added, and the mixture was stirred for 1 to 2.5 h at 4°C. The emulsion was transferred in chilled polypropylene centrifuge tubes and subsequently centrifuged for 30 min at 4°C at 20,400 × g. The butanol phase was collected, and subsequently 2 volumes of precooled (−20°C) acetone were added. The solution was briefly mixed and stored overnight at −20°C. The next day, the solution was centrifuged for 30 min at 4°C at 20,400 × g and the supernatant was carefully removed. The pellet was dried under vacuum and stored at −20°C until further processing. For the final purification step, the pellet was resuspended in 1 ml high-performance liquid chromatography (HPLC) eluent A (20% acetonitrile [HPLC grade], 0.1% trifluoroacetic acid [TFA]) per 50 ml supernatant. The solution was loaded in portions onto a semipreparative reversed-phase (RP) HPLC column (Gemini, 5 μm, NX-C18, 110-Å, 250- by 10-mm LC column [Phenomenex, Torrance, CA]). Entianin and subtilin and their succinylated variants S-entianin and S-subtilin were separated with a linear gradient of 17% to 29.5% eluent B (99.9% acetonitrile [HPLC grade], 0.1% TFA) within 13 min. The N-terminal fragments entianin1–20 and S-entianin1–20 could be isolated similarly. To isolate entianin1–19 and S-entianin1–19, a linear gradient of 19.5% to 22% eluent B within 28 min was used. The absorbance was monitored at 214 nm and 280 nm. The collected fractions were dried under vacuum and resuspended in 5% acetonitrile for an in vivo test or in 30% acetonitrile–0.1% TFA for identification by mass spectrometry. Entianin, subtilin, and the N-terminal fragments were quantified via their absorption at 214/280 nm, as described previously (30).
Purification and digestion of nisin.
Since purified nisin is not commercially available, we isolated nisin from lyophilized milk powder containing 2.5% nisin (balance, sodium chloride and denatured milk solids) (Sigma). Thirty micrograms of milk powder was dissolved in 1 ml 50 mM lactic acid, pH 3, and filtered through a 0.45-μm membrane filter. The nisin solution was loaded in 500-μl portions onto a semipreparative RP-HPLC column (as described above). The elution buffers consisted of 5% acetonitrile and 0.1% TFA (eluent A) and 99% acetonitrile and 0.1% TFA (eluent B). Nisin was separated with a linear gradient of 15% to 30% eluent B within 40 min. The absorbance was monitored at 214 nm. The collected fractions were handled as described above. Nisin1–20 was obtained by digestion of nisin with α-chymotrypsin as described previously (40) and could be isolated with a linear gradient from 0% to 100% eluent B within 19 min. Since the nisin molecules do not contain a tryptophan, the concentration of nisin and nisin1–20 could be determined via the absorption of the peptide bonds at 214 nm as described previously (30).
Measurement of induction capacity by β-galactosidase assay.
For time kinetic studies, a fresh overnight culture of the subtilin reporter strain B6633.MB1 was inoculated to an optical density of 0.1 at 37°C in TY medium containing 0.3 M NaCl (31). After 30 min, 2 ml of culture was transferred into small test tubes, containing different concentrations of (S-)entianin or (S-)subtilin. After various time points, samples were taken, and the cells were harvested by centrifugation and stored for the β-galactosidase assay at −20°C.
For rapid-induction capacity comparison, a fresh overnight culture of the subtilin reporter strain B6633.MB1 was inoculated to an optical density of 0.1 in TY medium containing 0.3 M NaCl as described above. The cells were grown to an optical density of approximately 1.0 and subsequently transferred in 2-ml portions into small test tubes containing different concentrations of entianin or nisin or the N-terminal fragments. After 1 h, samples were taken and handled as described above. The same procedure was used in the experiments when two substances, e.g., bacitracin (Applichem), ramoplanin (Sigma), or vancomycin (Applichem), were applied simultaneously.
The induction of PspaS-lacZ in lipid II-deficient cells was investigated using strain B168.AUT5. B168.AUT5 was cultivated as rod-shaped cells or in the L-form state, as described above. Investigation of PspaS-lacZ induction in rod-shaped cells of B168.AUT5 was performed just like for reporter strain B6633.MB1. The L-form cells were incubated at 30°C with gentle shaking. L-form cells were induced in the mid-log phase and harvested after 6 h, which corresponds to approximately one generation time.
The activity of the promoter PspaS was monitored by quantification of the β-galactosidase activity using the reporter strain B6633.MB1 (PspaS-lacZ) or strain B168.AUT5 (PspaS-lacZ). Cell pellets were resuspended in working buffer (20 mM β-mercaptoethanol, 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7), and cell lysis was achieved by addition of 0.2 mg ml−1 lysozyme and subsequent incubation at 37°C for 30 min. The β-galactosidase activity was measured as described previously, with normalization to cell density (41).
RESULTS
S-subtilin and S-entianin autoinduce subtilin biosynthesis at high concentrations.
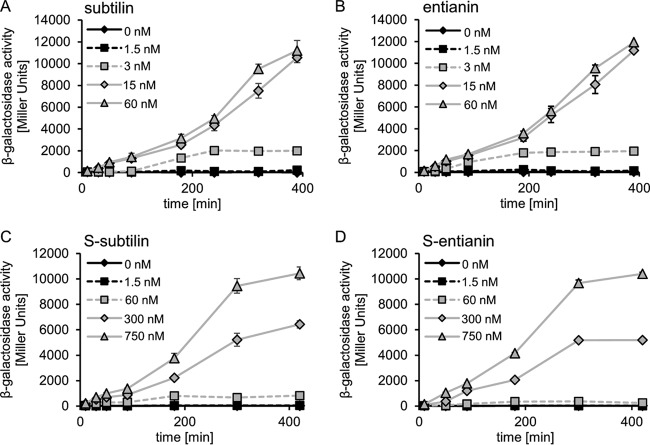
Subtilin-like lantibiotics have only minor and conservative differences in their primary sequences (Fig. 1). Using a ΔspaS PspaS-lacZ reporter strain (B6633.MB1), we have recently shown that subtilin-like lantibiotics have the same efficacy to induce all promoters in the subtilin cluster. Therefore, we consider all subtilin-like lantibiotics as autoinducers of subtilin biosynthesis (3, 31, 42). In contrast to other linear lantibiotics, such as nisin, epidermin, and gallidermin, subtilin-like lantibiotics are succinylated to a major extent at their N-terminal tryptophan residue. The level of succinylation is strain and carbon source dependent (30). To investigate the influence of N-terminal succinylation on autoinduction, we compared the induction capacities of homogeneously purified and quantified subtilin, entianin, and their succinylated versions, S-subtilin and S-entianin. Furthermore, the kinetics of the autoinduction were followed (Fig. 2).
FIG 2.
Induction kinetics of PspaS-lacZ expression by subtilin-like lantibiotics and their succinylated variants. The β-galactosidase activities of the untreated control (black rhombus) and after induction with various concentrations of the lantibiotics were monitored in a ΔspaS PspaS-lacZ reporter strain (B6633.MB1). Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); measurements were carried out in triplicate.
The induction kinetics for subtilin and entianin were similar, with minimal concentrations for autoinduction of 3 nM and a maximum reached at 15 nM (Fig. 2A and B). With concentrations above 15 nM, no further increase of the subtilin- or entianin-mediated induction was observed. With 15 nM and 60 nM, β-galactosidase activity reached its maximum after 400 min. The succinylated variants still activated SpaK and reached maximal PspaS-lacZ expression but at much higher concentrations (Fig. 2C and D). In the case of the succinylated versions, a minimal concentration of 300 nM S-subtilin or S-entianin was needed for detectable β-galactosidase activity, whereas 3 nM subtilin or entianin was sufficient for a comparable activity. For maximal β-galactosidase activity, 750 nM S-subtilin or S-entianin was necessary, whereas 15 nM subtilin or entianin was needed for maximum β-galactosidase activity after 400 min.
Purification of entianin fragments from culture supernatants from B15029.
Supernatants from the entianin-producing strain B15029 were analyzed by RP-HPLC (Fig. 3). In addition to entianin and S-entianin, an N-terminal fragment comprising amino acids 1 to 20 (entianin1–20) and an N-terminal succinylated variant (S-entianin1–20) were also identified and purified to homogeneity. A slight alteration of the elution gradient also showed an N-terminal fragment comprising amino acids 1 to 19 (entianin1–19) and a succinylated N-terminal fragment (S-entianin1–19) to be present in B15029 supernatants (data not shown). The correct masses of entianin ([A + H]+ 3,347.6), entianin1–19 ([A + H]+ 1,921.8), and entianin1–20 ([A + H]+ 2,069.04) as well as their 100.0-Da-larger succinylated variants were verified by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) analysis. Unfortunately, the corresponding C-terminal fragments could not be detected.
FIG 3.
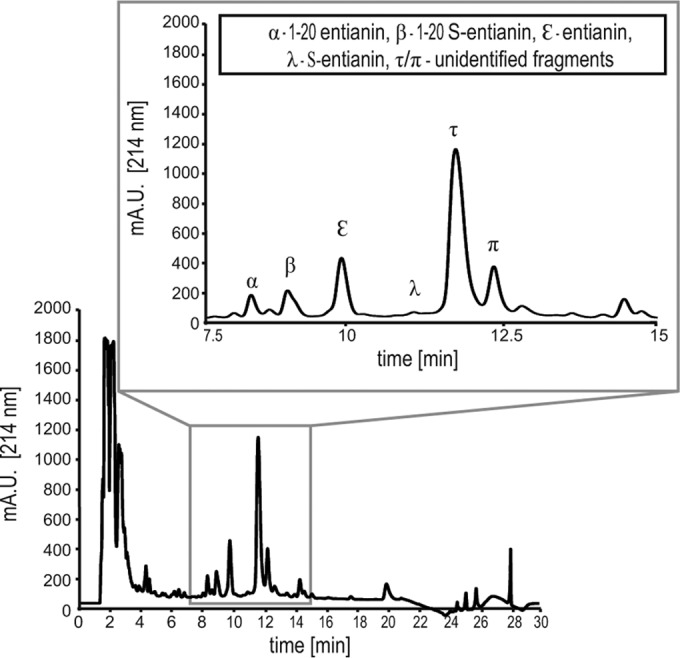
RP-HPLC chromatogram of 100 μl of supernatant from entianin-producing strain B15029 cultivated in 50 ml of medium A for 30 h, recorded at 214 nm. The zoomed area from 7.5 to 15 min shows the entianin-related peaks (α, entianin1–20; β, S-entianin1–20; ε, entianin; λ, S-entianin). Peaks τ and π eluting behind S-entianin could not be unambiguously identified to contribute to entianin fragments by means of the detected mass/charge (m/z) ratio.
Induction mediated by N-terminal entianin fragments.
The N-terminal entianin fragments provided an excellent tool to investigate the possible importance of the C-terminal part for SpaK activation. Since subtilin- and entianin-mediated induction levels were very similar, we focused on the entianin fragments. In these experiments, entianin fragments were added to exponentially growing cells and lacZ induction was followed after 60 min of incubation.
Remarkably, entianin1–20 as well as entianin1–19 fragments activated SpaK. Compared to full-length entianin, in which case 15 nM provided maximum induction of PspaS-lacZ, a 10-fold-higher level of entianin1–20 (150 nM) was needed to reach a comparable β-galactosidase activity (Fig. 4A). The smaller entianin1–19 fragment also activated SpaK; however, a 100-fold-higher level (1.5 μM) was needed to result in a β-galactosidase activity similar to that of entianin (Fig. 4A).
FIG 4.
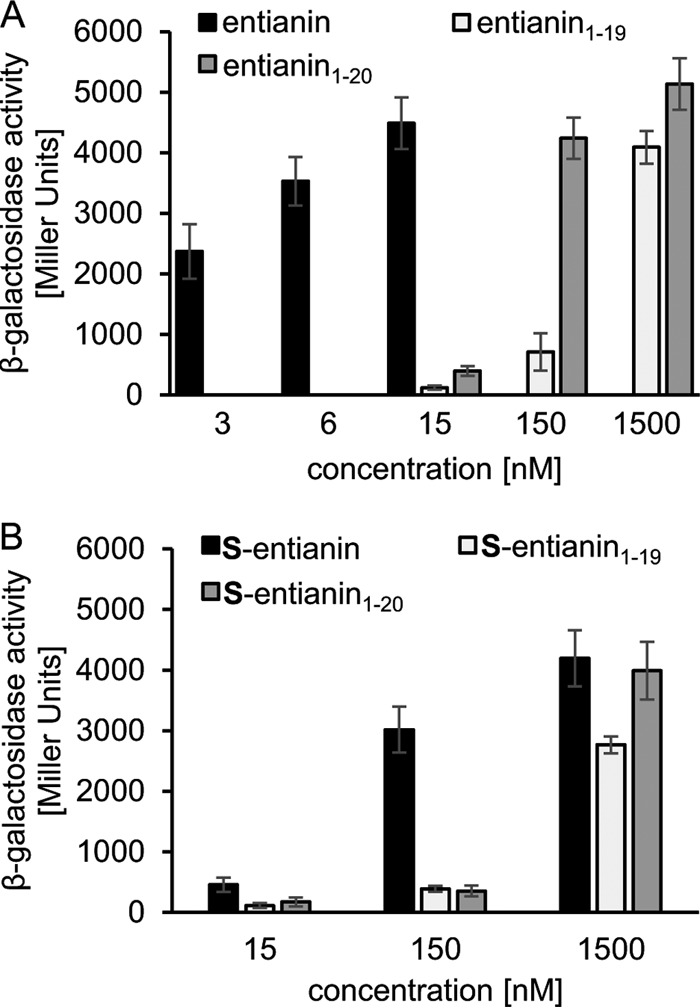
β-Galactosidase activities of PspaS-lacZ inreporter strain B6633.MB1 after 60 min of induction with N-terminal entianin fragments. (A) Induction with entianin (3 to 15 nM) and the corresponding fragments entianin1–19 and entianin1–20. (B) Induction with S-entianin and the corresponding succinylated fragments S-entianin1–19 and S-entianin1–20. Error bars represent the standard deviation between results for samples from at least three separate cultures (n ≥ 3); measurements were carried out in duplicate.
As already observed for S-entianin, maximal values were also reached with the S-entianin1–20 fragment; however, this needed much higher concentrations of 1.5 μM. The S-entianin1–19 fragment also showed a minor SpaK activation at concentrations of 1.5 μM (Fig. 4B).
Our results clearly show that the N-terminal part of entianin is sufficient for SpaK activation; however, with respect to the amount needed, activation is strongly enhanced if the C-terminal part of the lantibiotic is also present.
Specificity of SpaK histidine kinase activation.
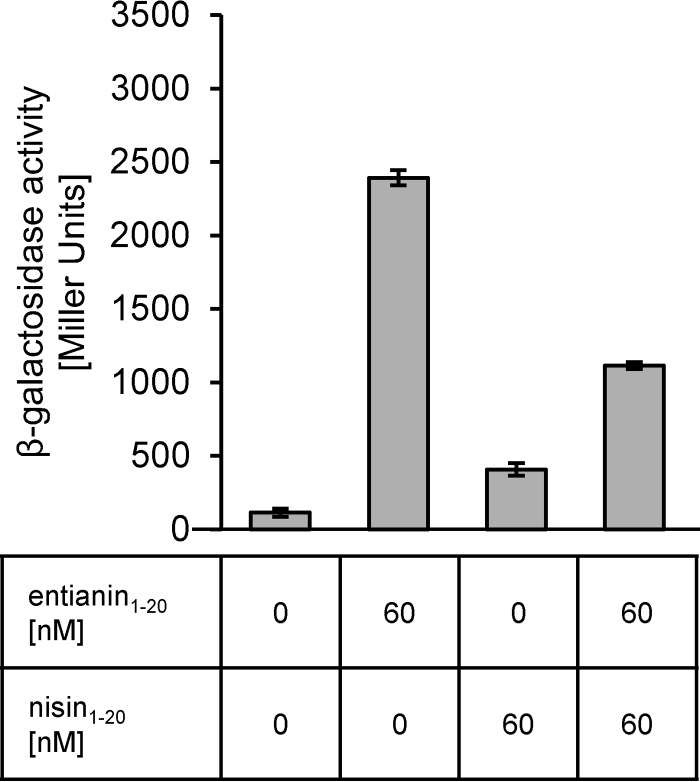
To follow the activation specificity of SpaK by subtilin or entianin, we used the structurally very similar nisin for activation experiments. Even a 2-fold-higher level of nisin (12 nM) compared to entianin (6 nM) did not result in any activation of the β-galactosidase reporter system (Fig. 5). Higher concentrations of nisin, which retard growth due to their toxicity and possibly distort the β-galactosidase assay, were not applied. These data show that the lantibiotic-mediated activation of SpaK seems highly specific for the respective lantibiotic.
FIG 5.
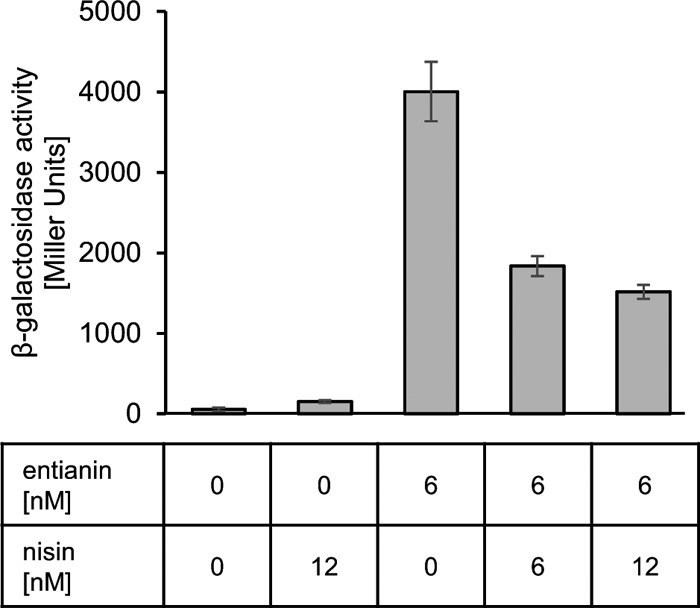
Influence of nisin on entianin-mediated PspaS-lacZ induction. The β-galactosidase activities of PspaS-lacZ in reporter strain B6633.MB1 are shown after 60 min of induction. Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); each measurement was carried out in duplicate.
Surprisingly, the addition of nisin together with entianin diminished entianin-mediated induction of PspaS-lacZ in the reporter strain B6633.MB1. Whereas the addition of 6 nM entianin resulted in a β-galactosidase activity of approximately 4,500 Miller units, the simultaneous addition of nisin decreased entianin-mediated induction significantly (Fig. 5). Even concentrations lower than 1.5 nM nisin caused a slight decrease in entianin-mediated induction of PspaS-lacZ (data not shown). Equimolar concentrations of nisin and entianin reduced entianin-mediated induction to 50%; nisin in 2-fold excess reduced entianin-mediated induction to 40%. Due to their antimicrobial activities, higher concentrations of entianin and nisin became lethal and could not be applied.
To better understand the nisin interference with entianin-mediated SpaK activation, we tested whether nisin also had an influence on SpaK activation mediated by the N-terminal entianin1–20 fragment, and vice versa, we tested whether an N-terminal fragment of nisin, which also comprises the first 20 N-terminal amino acids (nisin1–20), affected entianin-mediated SpaK activation.
Indeed, nisin also interfered with entianin1–20-mediated activation of SpaK to an extent similar to that observed for full-length entianin (Fig. 6A). Using equimolar concentrations of entianin and nisin1–20, no significant difference in β-galactosidase activities was observed. However, in 10-fold excess (60 nM), the nisin1–20 fragment also interfered with SpaK activation by entianin and reduced the entianin-mediated β-galactosidase activity to half, which suggested a direct competition between the two molecules. As nisin1–20 was not toxic and did not inhibit growth (data not shown), higher concentrations could also be tested. Surprisingly, increasing nisin1–20 concentrations to 600 nM did not show a further decrease in entianin1–20-mediated induction of PspaS-lacZ. This is explained by the observation that nisin1–20 alone also caused a moderate increase in β-galactosidase activity at concentrations of 60 nM, which further increased with higher concentrations (Fig. 6B). This shows that nisin1–20 also has some ability to activate the TCS. Taken together, our results suggest that the N-terminal part of entianin is sufficient to provide the specificity for SpaK activation and that the presence of the C-terminal part of entianin strongly enforces the activation.
FIG 6.
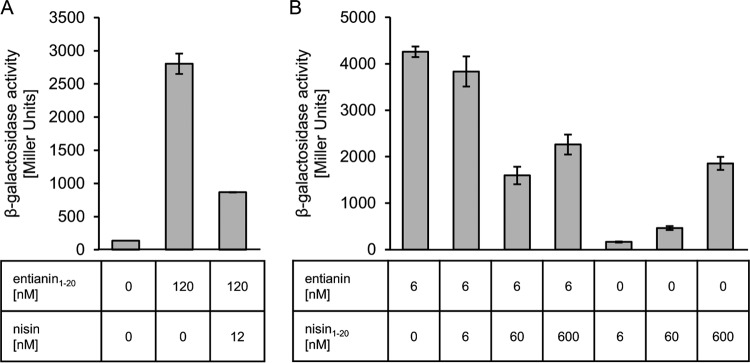
Influence of nisin on entianin1–20-mediated PspaS-lacZ induction (A) and of nisin1–20 on entianin-mediated PspaS-lacZ induction (B). The β-galactosidase activities of PspaS-lacZ in reporter strain B6633.MB1 are shown after 60 min of induction. Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); measurements were carried out in duplicate.
To further validate the importance of the N-terminal part of entianin for SpaK activation and to elucidate the influences of the respective C termini, we examined the competition between entianin1–20 and nisin1–20 in equimolar concentrations. As shown in Fig. 7, equimolar concentrations of nisin1–20 reduced entianin1–20 autoinduction by 50%. This further confirms that the N-terminal part of entianin is crucial for the specific SpaK activation.
FIG 7.
Influence of nisin1–20 on entianin1–20-mediated PspaS-lacZ induction. The β-galactosidase activities of PspaS-lacZ in reporter strain B6633.MB1 are shown after 60 min of induction. Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); measurements were carried out in duplicate.
Lipid II is not needed for SpaK activation.
The current data show that the entianin-mediated activation of SpaK is strongly reduced by adding either nisin or its N-terminal fragment nisin1–20. Therefore, a competition of the two molecules at the sensory binding site of the SpaK histidine kinase seems likely.
As the subtilin-like lantibiotics and nisin interact with lipid II at the pyrophosphate moiety (13) with a high affinity (binding constant = 2 × 107 M−1) (12), we asked if a lipid II interaction is also a prerequisite for the SpaK activation or if subtilin-like lantibiotics activate SpaK independently from lipid II. To test this, the effect on entianin-mediated activation of SpaK was monitored using antimicrobial substances which directly interact with the lipid II molecule or interfere with the recycling intermediates of lipid II.
Therefore, PspaS-lacZ induction was followed after the simultaneous addition of entianin and the cyclic peptide ramoplanin that, like entianin and nisin, binds to the pyrophosphate moiety of lipid II (18, 19). In addition, it has been shown that pretreatment of Micrococcus luteus cells with ramoplanin prevented nisin-induced leakage through pore formation by depletion of lipid II molecules (11). Furthermore, we used the cyclic polypeptide bacitracin, which binds to undecaprenyl pyrophosphate, thereby inhibiting the recycling of the lipid carrier (16, 17), and as a negative control the glycopeptide vancomycin, which specifically interacts with the d-Ala-d-Ala moiety of the pentapeptide chain of lipid II (43, 44).
A 7-fold excess of ramoplanin (44 nM) did not influence the entianin (6 nM)-induced β-galactosidase activity, which shows that the pyrophosphate moiety of lipid II is not important for entianin-mediated SpaK activation (Fig. 8A). This was also the case for a 10-fold excess of vancomycin (Fig. 8B) and a 12,000-fold excess of bacitracin (Fig. 8C).
FIG 8.
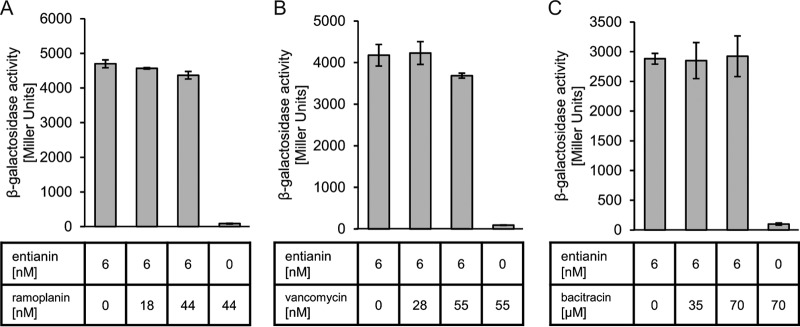
Influence of lipid II-interfering substances on entianin-mediated PspaS-lacZ induction: ramoplanin (A), vancomycin (B), and bacitracin (C). The β-galactosidase activities of PspaS-lacZ in reporter strain B6633.MB1 are shown after 60 min of induction. Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); measurements were carried out in duplicate.
L-form B. subtilis mutant strains show wild-type entianin-mediated activation of SpaK.
Analysis of lipid II-interacting antibiotics did not indicate any importance of lipid II for entianin-mediated activation of SpaK. To finally exclude that the sublethal concentrations used were not sufficient to deplete lipid II, we investigated entianin-mediated activation of SpaK in L-form B. subtilis strain PDC204. In PDC204, the expression of the murE operon, which contains four essential cell wall biosynthesis genes, is expressed using a xylose-inducible promoter (34). Therefore, these strains are conditionally unable to synthesize essential cell wall precursors, including lipid I and lipid II. To analyze subtilin or entianin-mediated activation of SpaK, we introduced the PspaS-lacZ and PspaRK-spaRK constructs into the PDC204 mutant strain, resulting in the L-form reporter strain B168.AUT5. Subtilin-mediated activation of SpaK was monitored in rod-shaped and L-form cells, which were verified by microscopy. Various concentrations of subtilin (0, 3, and 15 nM) were added in the logarithmic growth phase, samples were taken after 6 h, and β-galactosidase activities were measured (Fig. 9). Rod-shaped and L-form cells showed comparable subtilin-mediated PspaS-lacZ induction. This clearly confirmed that lipid II is not needed for subtilin-mediated activation of SpaK.
FIG 9.
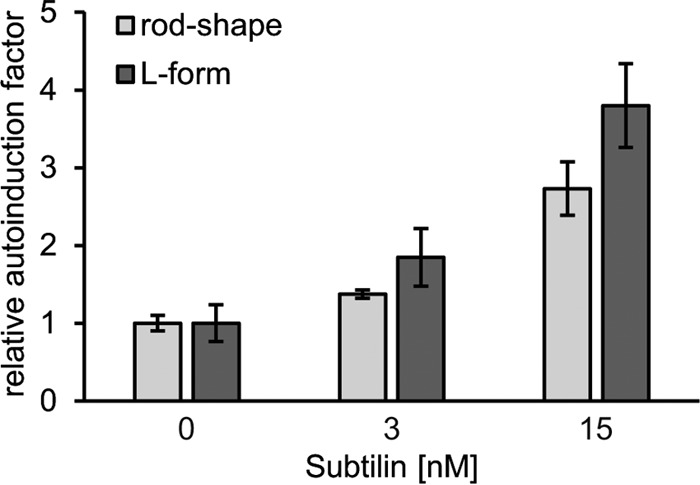
Induction assay of PspaS by subtilin in rod-shaped cells and the corresponding L-form cells of the reporter strain B168.AUT5. Rod-shaped cells and L-form cells were induced in the mid-log phase and harvested after 6 h of induction. The promoter activities were calculated as relative induction factors in comparison to the β-galactosidase activities of the control experiment without subtilin. Error bars represent the standard deviation between results for samples from two separate cultures (n = 2); measurements were carried out in duplicate.
DISCUSSION
The biosynthesis of class I lantibiotics such as subtilin, entianin, nisin, and related peptides is regulated via a two-component regulatory system (TCS) (21, 45). The related lantibiotics themselves act as pheromones and activate their corresponding histidine kinase/response regulator signal transduction system (LanRK) (20, 21, 45, 46). This results in the transcription of the respective lantibiotic operons, including the structural genes, the biosynthetic gene cluster, and the immunity genes.
The histidine kinase SpaK of the subtilin producer B6633 is also capable of sensing the closely related entianin in the same range as subtilin (3). Furthermore, significant activation of SpaK was mediated by the N-terminal fragments entianin1–20 and entianin1–19. This is in accordance with previous investigations, where nisin and a 10-fold excess of nisin1–20 stimulated nisin immunity to similar extents (47). The less efficient activation of SpaK mediated by entianin fragments suggests that the C-terminal part of the molecule supports SpaK activation. For nisin, it has been shown that its C-terminal region is responsible for the interaction of nisin with the target membrane (48). Therefore, we suggest the hypothesis that the C terminus might bring the lantibiotic in close vicinity to the histidine kinase, due to its interaction with the membrane.
Interestingly, in comparison to entianin1–20, the entianin1–19 fragment showed a dramatically reduced induction of PspaS-lacZ. Since both fragments possess the first three lanthionine rings (Fig. 1), the observed differences show that amino acid 20 of subtilin-like lantibiotics is important for the sensing mechanism and enforces the interaction with SpaK. For all succinylated molecules, a reduced PspaS-lacZ induction was observed. Since succinylation provides a negative charge to the N terminus of subtilin-like lantibiotics, the interaction with either the cell membrane or SpaK is obviously impaired. Additionally, the succinyl group could also provide a steric hindrance for an S-entianin–SpaK interaction.
Subtilin-like lantibiotics and nisin have highly similar secondary structures and similar overall charges, although the charges are differently distributed (Fig. 1). Considering the primary structures of the 20 N-terminal amino acids, subtilin-like lantibiotics and nisin differ at 7 to 9 positions which are probably important for SpaK activation. Additionally, the respective histidine kinases SpaK and NisK share only 26% identity with respect to their amino acid sequences (49). These differences are more distinct in the extracellular sensory domains, with a negligible identity of 16%, and in the distribution of polar and nonpolar regions within the sensory domain of the two kinases. The low homology between the two kinases and the differences in the primary structures of the lantibiotics might explain why nisin fails to activate SpaK even in 2-fold excess. Surprisingly, entianin-mediated activation of SpaK could be significantly reduced by equimolar concentrations of nisin, indicating a competition of the two molecules for the SpaK sensory domain. However, complete quenching of the entianin-mediated activation of SpaK was not experimentally possible, since higher nisin concentrations were toxic.
Entianin and nisin are cationic molecules that interact with the negatively charged membrane, and thus the SpaK/NisK sensing mechanism could possibly be based on the interaction of electrical charges. Such a mechanism was found for the PhoQ histidine kinase of Salmonella species and other Gram-negative bacteria (50–52), which sense cationic antimicrobial peptides with an amphipathic structure (53–55). The PhoQ periplasmic sensor domain contains an insertion with an acidic surface that mediates the interaction with the cationic antimicrobial peptides. However, although subtilin and nisin are similarly charged, the NisK sensor domain is acidic (pI of 5) and the SpaK sensor domain is basic (pI of 9) (ExPASy – Compute pI/Mw tool). This makes a charge-dependent sensing similar to the PhoQ system unlikely.
The interference of nisin with entianin-mediated activation of SpaK is probably mediated by the highly similar ring structures of the two molecules, while the difference in amino acid composition between entianin and nisin is possibly responsible for the specific activation.
Our results suggest that the N termini of the two lantibiotics compete for the same binding site at the SpaK histidine kinase due to their similar ring structures. However, although nisin can possibly occupy the binding site of the SpaK histidine kinase, it cannot trigger the autophosphorylation of the histidine kinase in a manner similar to entianin. For transfer of the activation signal, the histidine kinase needs to autophosphorylate using small-molecule phosphor donors and to transfer the respective phosphate to the response regulator (56). At concentrations similar to that of entianin1–20, nisin1–20 obviously cannot activate SpaK but can probably prevent the access of entianin1–20 to its binding pocket. However, at very high concentrations, nisin1–20 fragments can activate SpaK in a less specific manner, which explains that is it not possible to completely quench entianin1–20-mediated activation of SpaK with high nisin1–20 concentrations.
Unfortunately, so far the establishment of an in vitro test system to study SpaK activation remains elusive, since we failed to express and enrich the histidine kinase.
Substances that target lipid II or interfere with its biosynthesis, such as bacitracin, ramoplanin, and vancomycin, were not able to interfere with the entianin-mediated activation of SpaK. This indicated that the interaction of the lantibiotic with lipid II is not necessary for activation of SpaK.
This was also confirmed with lipid II-deficient L-form cells, which could still sense subtilin. Taken together, these results rule out lipid II as a component involved in the activation mechanism of SpaK and argue for a direct interaction of the N terminus of linear lantibiotics with the respective histidine kinase.
ACKNOWLEDGMENTS
This work was supported by DFG (En 134/11-1).
We acknowledge Goethe University and the state of Hesse for providing laboratory space, basic support, and necessary equipment, as well as M. Karas for his support during mass spectrometry analysis. We especially thank J. Errington, R. Daniel, and T. Mascher for providing B. subtilis strains. We further thank S. Düsterhus for technical assistance.
REFERENCES
- 1.Schnell N, Entian KD, Schneider U, Götz F, Zähner H, Kellner R, Jung G. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278. doi: 10.1038/333276a0. [DOI] [PubMed] [Google Scholar]
- 2.Stein T, Borchert S, Conrad B, Feesche J, Hofemeister B, Hofemeister J, Entian KD. 2002. Two different lantibiotic-like peptides originate from the ericin gene cluster of Bacillus subtilis A1/3. J Bacteriol 184:1703–1711. doi: 10.1128/JB.184.6.1703-1711.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuchs SW, Jaskolla TW, Bochmann S, Kötter P, Wichelhaus T, Karas M, Stein T, Entian KD. 2011. Entianin, a novel subtilin-like lantibiotic from Bacillus subtilis subsp. spizizenii DSM 15029T with high antimicrobial activity. Appl Environ Microbiol 77:1698–1707. doi: 10.1128/AEM.01962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H, et al. 2013. Ribosomally synthesized and posttranslationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnell N, Entian KD, Götz F, Hörner T, Kellner R, Jung G. 1989. Structural gene isolation and prepeptide sequence of gallidermin, a new lanthionine containing antibiotic. FEMS Microbiol Lett 49:263–267. [DOI] [PubMed] [Google Scholar]
- 6.Kaletta C, Entian KD. 1989. Nisin, a peptide antibiotic: cloning and sequencing of the nisA gene and posttranslational processing of its peptide product. J Bacteriol 171:1597–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiesau P, Eikmanns U, Gutowski-Eckel Z, Weber S, Hammelmann M, Entian KD. 1997. Evidence for a multimeric subtilin synthetase complex. J Bacteriol 179:1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegers K, Heinzmann S, Entian KD. 1996. Biosynthesis of lantibiotic nisin. Posttranslational modification of its prepeptide occurs at a multimeric membrane-associated lanthionine synthetase complex. J Biol Chem 271:12294–12301. [DOI] [PubMed] [Google Scholar]
- 9.Ortega MA, Hao Y, Zhang Q, Walker MC, van der Donk WA, Nair SK. 2015. Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517:509–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Yu JPJ, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. 2006. Structure and mechanism of the lantibiotic cyclase involved in nisin biosynthesis. Science 311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 11.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 12.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 13.Hsu SD, Breukink E, Tischenko E, Lutters MAG, de Kruijff B, Kaptein R, Bonvin AM, van Nuland NA. 2004. The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi: 10.1038/nsmb830. [DOI] [PubMed] [Google Scholar]
- 14.Sheldrick GM, Jones PG, Kennard O, Williams DH, Smith GA. 1978. Structure of vancomycin and its complex with acetyl-D-alanyl-D-alanine. Nature 271:223–225. doi: 10.1038/271223a0. [DOI] [PubMed] [Google Scholar]
- 15.Molinari H, Pastore A, Lian LY, Hawkes GE, Sales K. 1990. Structure of vancomycin and a vancomycin/D-Ala-D-Ala complex in solution. Biochemistry 29:2271–2277. doi: 10.1021/bi00461a010. [DOI] [PubMed] [Google Scholar]
- 16.Storm DR. 1974. Mechanism of bacitracin action: a specific lipid-peptide interaction. Ann N Y Acad Sci 235:387–398. doi: 10.1111/j.1749-6632.1974.tb43278.x. [DOI] [PubMed] [Google Scholar]
- 17.Stone KJ, Strominger JL. 1971. Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci U S A 68:3223–3227. doi: 10.1073/pnas.68.12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walker S, Chen L, Hu Y, Rew Y, Shin D, Boger DL. 2005. Chemistry and biology of ramoplanin: a lipoglycodepsipeptide with potent antibiotic activity. Chem Rev 105:449–476. doi: 10.1021/cr030106n. [DOI] [PubMed] [Google Scholar]
- 19.Hu Y, Helm JS, Chen L, Ye X, Walker S. 2003. Ramoplanin inhibits bacterial transglycosylases by binding as a dimer to lipid II. J Am Chem Soc 125:8736–8737. doi: 10.1021/ja035217i. [DOI] [PubMed] [Google Scholar]
- 20.Stein T, Borchert S, Kiesau P, Heinzmann S, Kloss S, Klein C, Helfrich M, Entian KD. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol Microbiol 44:403–416. doi: 10.1046/j.1365-2958.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 21.Klein C, Kaletta C, Entian KD. 1993. Biosynthesis of the lantibiotic subtilin is regulated by a histidine kinase/response regulator system. Appl Environ Microbiol 59:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM. 1995. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 23.Kleerebezem M. 2004. Quorum sensing control of lantibiotic production; nisin and subtilin autoregulate their own biosynthesis. Peptides 25:1405–1414. doi: 10.1016/j.peptides.2003.10.021. [DOI] [PubMed] [Google Scholar]
- 24.de Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol 178:3434–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir J, Predich M, Dubnau E, Nair G, Smith I. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J Bacteriol 173:521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauch MA. 1995. Delineation of AbrB-binding sites on the Bacillus subtilis spo0H, kinB, ftsAZ, and pbpE promoters and use of a derived homology to identify a previously unsuspected binding site in the bsuB1 methylase promoter. J Bacteriol 177:6999–7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carter HL, Moran CP. 1986. New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci U S A 83:9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubnau E, Weir J, Nair G, Carter L, Moran C, Smith I. 1988. Bacillus sporulation gene spo0H codes for sigma 30 (sigma H). J Bacteriol 170:1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutowski-Eckel Z, Klein C, Siegers K, Böhm K, Hammelmann M, Entian KD. 1994. Growth phase-dependent regulation and membrane localization of SpaB, a protein involved in biosynthesis of the lantibiotic subtilin. Appl Environ Microbiol 60:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bochmann SM, Spieß T, Kötter P, Entian KD. 2015. Synthesis and succinylation of subtilin-like lantibiotics are strongly influenced by glucose and transition state regulator AbrB. Appl Environ Microbiol 81:614–622. doi: 10.1128/AEM.02579-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burkard M, Entian KD, Stein T. 2007. Development and application of a microtiter plate-based autoinduction bioassay for detection of the lantibiotic subtilin. J Microbiol Methods 70:179–185. doi: 10.1016/j.mimet.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Banerjee S, Hansen JN. 1988. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem 263:9508–9514. [PubMed] [Google Scholar]
- 33.Feeney R, Garibaldi J, Humphreys EM. 1948. Nutritional studies on subtilin formation by Bacillus subtilis. Arch Biochem Biophys 17:435–445. [PubMed] [Google Scholar]
- 34.Domínguez-Cuevas P, Mercier R, Leaver M, Kawai Y, Errington J. 2012. The rod to L-form transition of Bacillus subtilis is limited by a requirement for the protoplast to escape from the cell wall sacculus. Mol Microbiol 83:52–66. doi: 10.1111/j.1365-2958.2011.07920.x. [DOI] [PubMed] [Google Scholar]
- 35.Derré I, Rapoport G, Msadek T. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol Microbiol 38:335–347. doi: 10.1046/j.1365-2958.2000.02124.x. [DOI] [PubMed] [Google Scholar]
- 36.Middleton R, Hofmeister A. 2004. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51:238–245. doi: 10.1016/j.plasmid.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Birnboim HC, Doly J. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu W, Hansen JN. 1992. Enhancement of the chemical and antimicrobial properties of subtilin by site-directed mutagenesis. J Biol Chem 267:25078–25085. [PubMed] [Google Scholar]
- 39.Jansen EF, Hirschmann DJ. 1944. Subtilin, an antibacterial product of Bacillus subtilis: culturing conditions and properties. Arch Biochem 4:297–304. [Google Scholar]
- 40.Chan WC, Bycroft BW, Leyland ML, Lian LY, Roberts GC. 1993. A novel post-translational modification of the peptide antibiotic subtilin: isolation and characterization of a natural variant from Bacillus subtilis A.T.C.C. 6633. Biochem J 291:23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 42.Burkard M, Stein T. 2008. Microtiter plate bioassay to monitor the interference of antibiotics with the lipid II cycle essential for peptidoglycan biosynthesis. J Microbiol Methods 75:70–74. doi: 10.1016/j.mimet.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee AN, Perkins HR. 1966. Compounds formed between nucleotides related to the biosynthesis of bacterial cell wall and vancomycin. Biochem Biophys Res Commun 24:489–494. doi: 10.1016/0006-291X(66)90188-4. [DOI] [PubMed] [Google Scholar]
- 44.Perkins HR. 1969. Specificity of combination between mucopeptide precursors and vancomycin or ristocetin. Biochem J 111:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelke G, Gutowski-Eckel Z, Kiesau P, Siegers K, Hammelmann M, Entian KD. 1994. Regulation of nisin biosynthesis and immunity in Lactococcus lactis 6F3. Appl Environ Microbiol 60:814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmitz S, Hoffmann A, Szekat C, Rudd B, Bierbaum G. 2006. The lantibiotic mersacidin is an autoinducing peptide. Appl Environ Microbiol 72:7270–7277. doi: 10.1128/AEM.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dodd HM, Horn N, Chan WC, Giffard CJ, Bycroft BW, Roberts GC, Gasson MJ. 1996. Molecular analysis of the regulation of nisin immunity. Microbiology 142:2385–2392. doi: 10.1099/00221287-142-9-2385. [DOI] [PubMed] [Google Scholar]
- 48.Breukink E, van Kraaij C, Demel RA, Siezen RJ, Kuipers OP, de Kruijff B. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968–6976. doi: 10.1021/bi970008u. [DOI] [PubMed] [Google Scholar]
- 49.Siezen RJ, Kuipers OP, de Vos WM. 1996. Comparison of lantibiotic gene clusters and encoded proteins. Antonie Van Leeuwenhoek 69:171–184. doi: 10.1007/BF00399422. [DOI] [PubMed] [Google Scholar]
- 50.Fields PI, Groisman EA, Heffron F. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 51.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 52.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, Miller SI. 2014. PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111:1963–1968. doi: 10.1073/pnas.1316901111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol Microbiol 50:219–230. doi: 10.1046/j.1365-2958.2003.03675.x. [DOI] [PubMed] [Google Scholar]
- 54.Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 55.Richards SM, Strandberg KL, Conroy M, Gunn JS. 2012. Cationic antimicrobial peptides serve as activation signals for the Salmonella typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Front Cell Infect Microbiol 2:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]