Abstract
Yersiniabactin (Ybt) is a mixed nonribosomal peptide-polyketide natural product natively produced by the pathogen Yersinia pestis. The compound enables iron scavenging capabilities upon host infection and is biosynthesized by a nonribosomal peptide synthetase featuring a polyketide synthase module. This pathway has been engineered for expression and biosynthesis using Escherichia coli as a heterologous host. In the current work, the biosynthetic process for Ybt formation was improved through the incorporation of a dedicated step to eliminate the need for exogenous salicylate provision. When this improvement was made, the compound was tested in parallel applications that highlight the metal-chelating nature of the compound. In the first application, Ybt was assessed as a rust remover, demonstrating a capacity of ∼40% compared to a commercial removal agent and ∼20% relative to total removal capacity. The second application tested Ybt in removing copper from a variety of nonbiological and biological solution mixtures. Success across a variety of media indicates potential utility in diverse scenarios that include environmental and biomedical settings.
INTRODUCTION
Siderophore production is a strategy utilized by microorganisms to scavenge minerals from the surrounding environment (1). The process is often completed in the context of virulence progression during infection (2–5). Iron sequestration is the primary objective, and several siderophores have been identified from a variety of bacteria and fungi, including enterobactin (Escherichia coli), salmochelin (Salmonella spp.), triacetylfusarinine C (Aspergillus fumigatus), and yersiniabactin (Yersinia pestis) (6–11). The list of representative siderophore compounds provided has a common biosynthetic feature in that all derive from the action of nonribosomal peptide synthetases, with occasional contributions from polyketide synthase enzymes (12). The modular, diverse, and flexible nature of these enzymes offers numerous possibilities for directed manipulation of the biosynthetic pathways for the purpose of new enzymatic insight and molecular variation of the final compound (13–15). In addition, the growing number of successful cases of heterologous production of such compounds provides parallel opportunities to study biosynthesis within a surrogate host (16, 17).
Such is the case for yersiniabactin (Ybt), which was designed for heterologous production within E. coli (18). The biosynthetic process required expression of genes encoding two high-molecular-weight proteins (HMWP1 and HMWP2) to form a mixed nonribosomal peptide synthetase-polyketide synthase complex utilizing three cysteines, a malonyl coenzyme A (malonyl-CoA) unit, and a salicylate starter unit in addition to S-adenosylmethionine (SAM) and NADPH (Fig. 1). Exogenous addition of salicylate allowed successful heterologous biosynthesis, as the remaining substrates and cofactors were native to E. coli metabolism. Success established a production platform independent of handling the native Y. pestis pathogen and capable of extensive engineering given the recombinant features of E. coli.
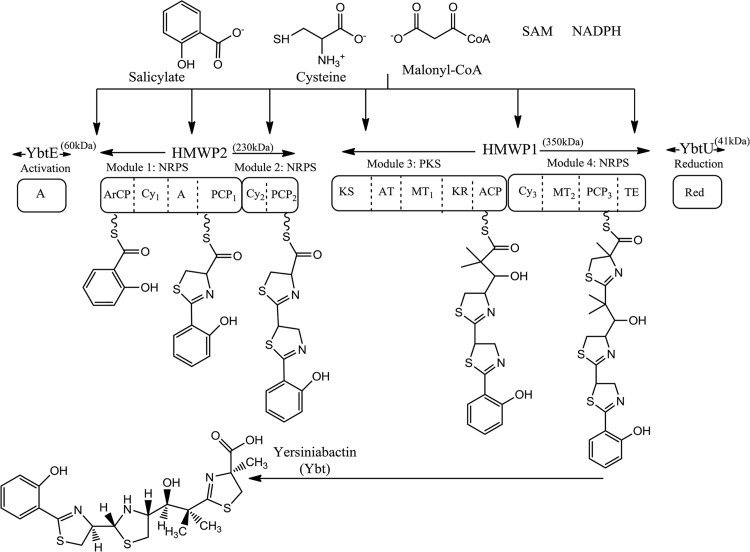
FIG 1.
Modular biosynthesis of yersiniabactin (Ybt). A salicylate starter unit is activated by YbtE, via adenylation (A), prior to transfer to the first nonribosomal peptide synthetase module of HMWP2, which subsequently introduces and cyclizes two cysteine groups. Upon transfer to HMWP1, a polyketide synthase model introduces a malonyl-CoA unit whose carbonyl group is reduced and methylated in NADPH- and SAM-dependent steps via the activity of dedicated ketoreductase (KR) and methyltransferase (MT) domains. The second module of HMWP1 introduces and cyclizes a third cysteine unit. A terminal thioesterase (TE) domain releases the mixed nonribosomal peptide-polyketide chain from HMWP1 prior to a final reduction step by YbtU. ArCP, aryl carrier protein; Cy, cyclization domain; PCP, peptidyl carrier protein; KS, ketosynthase; AT, acyltransferase; Red, reductase.
Recently, the utilities of siderophores have been highlighted across a wide range of scientific settings (19–22). The metal binding properties of siderophores offer useful purposes in settings that include rust removal, corrosion resistance, biosensors, bioremediation, and antimicrobial approaches. In the biomedical community, Ybt was recently recognized for a parallel ability to bind and sequester copper (23, 24). The potential to selectively bind copper also holds promise in treating Wilson's disease, in which an excess of this metal accumulates in tissue (25–27). These and other situations will benefit from the recombinant capabilities that accompany the production of Ybt, and likely future siderophore products, when genetically tractable heterologous hosts such as E. coli are used.
This potential was tested in the current work using the E. coli Ybt production system. Initially, engineering of the metabolic background of E. coli allowed the endogenous production of the salicylate starter unit and, subsequently, the complete intracellular requirements to allow full Ybt biosynthesis. When this new system was used, Ybt production showed a significant boost in production levels, which were directed toward two diverse applications. First, rust removal was assessed and quantified using recombinant Ybt. Next, the Ybt compound was tested in the selective removal of copper from a range of biological and nonbiological fluids, suggesting future use in settings such as heavy metal environmental remediation and physiological imbalance.
MATERIALS AND METHODS
Strains and plasmid construction.
Plasmid cloning was completed using DH5α, whereas Ybt production was completed in strain BAP1 (28). The irp9 gene was obtained from PCR amplification of Yersinia enterocolitica genomic DNA using the primers irp9-F (5′-AATCATATGAAAATCAGTGAATTTCTACAC-3′) and irp9-R (5′-AATCCTCGAGCTACTACACCATTAAATAGGG-3′) (29). The 1.3-kb amplified gene product was cloned into pCDFDuet-1 (EMD Chemicals Inc. [Gibbstown, NJ, USA]) using restriction sites NdeI and XhoI (underlined in the primers above). Plasmids pBP198 (pET21c containing genes encoding HMWP2 and YbtU), pBP205 (pET28a containing genes encoding HMWP1 and YbtE), and pBP200 (pGZ119EH containing the gene encoding YbtT [an auxiliary enzyme expected to play an editing role during biosynthesis]) were described previously (18). Plasmids were transformed into BAP1 using electroporation. Following selection with appropriate antibiotics, the resulting strain-plasmid combinations were stored as 20% glycerol stocks. The negative-control strain was BAP1/pBP198/pBP200/pBP200/pCDFDuet-1-irp9 (in association with “Negative Control Extract” and “XAD-negative” in Fig. 3 and 4, respectively).
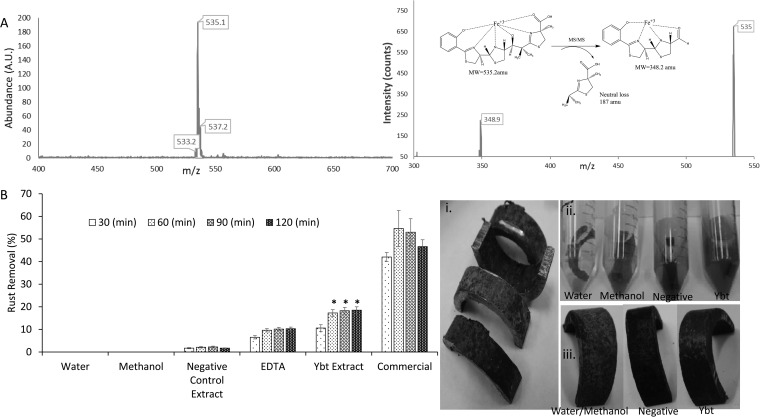
FIG 3.
Ybt iron binding and application. (A) LC/MS analysis of the Ybt-Fe3+ complex (left) and MS/MS analysis of Ybt-Fe3+ fragmentation (right). (B) Removal ability of Ybt-containing extract compared to negative controls (water, methanol, and BAP1/pBP198/pBP200/pCDFDuet-1-irp9) and positive controls (EDTA and commercial product [Rust-Oleum]). Visual indication of rust removal from U-shaped steel tube pieces before (i), during (ii), and after (iii) Ybt treatment is also shown. Asterisks indicate statistical significance (95% confidence) in comparison to corresponding EDTA controls. pBP198 contains genes encoding HMWP2 and YbtU, and pBP205 contains genes encoding HMWP1 and YbtE; strain BAP1/pBP198/pBP205/pBP200/pCDFDuet-1-irp9 is responsible for Ybt production.
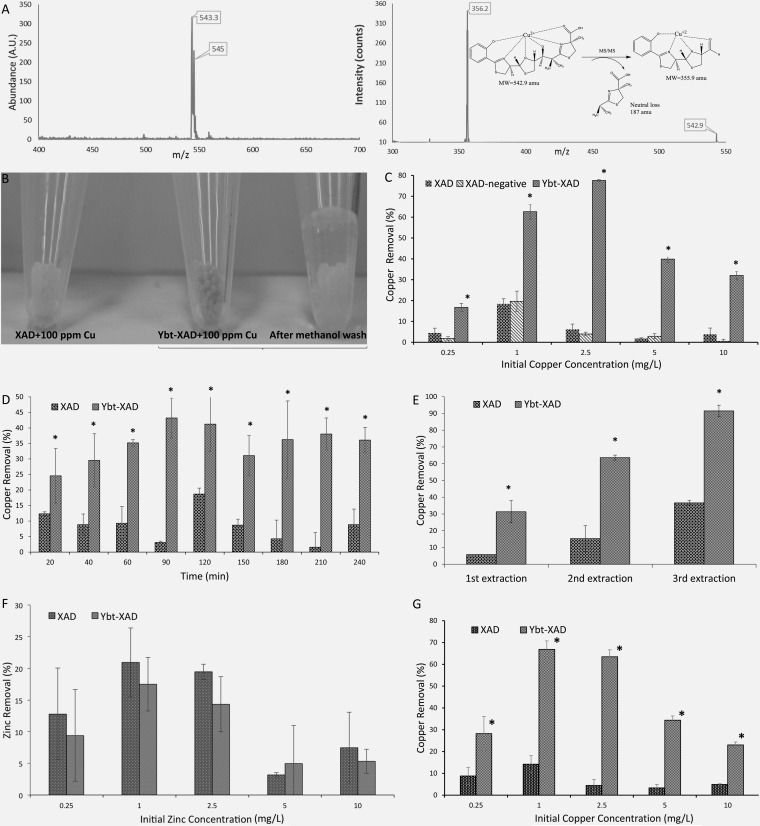
FIG 4.
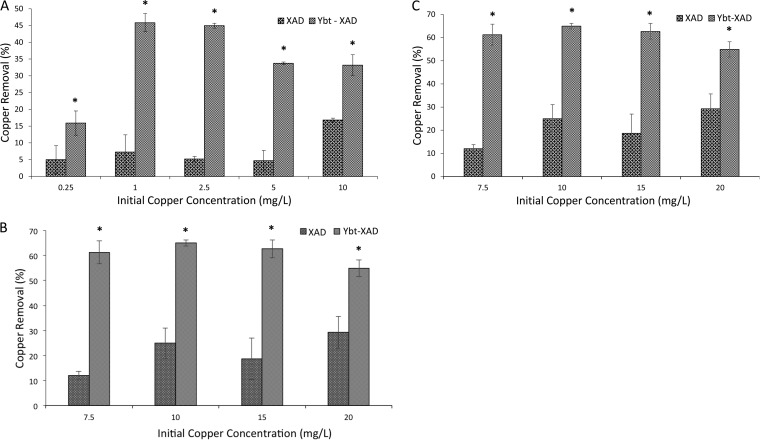
Ybt application to copper removal. (A) LC/MS analysis of Ybt-Cu2+ complex (left) and MS/MS analysis of Ybt-Cu2+ fragmentation. (B) Visual comparison of XAD and Ybt-XAD used to chelate copper. The XAD-bound Ybt and complexed copper are removed upon washing with methanol. (C) Comparison between nonmodified XAD particles, particles modified with extract from negative control BAP1/pBP198/pBP200/pCDFDuet-1-irp9, and Ybt-modified XAD across increasing initial copper concentrations. (D) Copper removal comparison over time. The initial concentration of copper was 10 ppm. (E) Comparison between nonmodified XAD particles and Ybt-modified XAD in a stepwise copper removal process in which fresh beads were added to the same solution to assess copper removal. (F) Zinc tested for Ybt sequestration. (G) Copper removal capability of XAD and Ybt-XAD in the presence of 10 ppm Fe3+. Asterisks indicate statistical significance (95% confidence) in comparison to corresponding negative controls.
Bacterial culture and compound extraction.
Overnight cultures from glycerol stocks were incubated at 37°C with shaking in lysogeny broth (LB) medium and used to inoculate (1% vol/vol) either 25-ml (Ybt production comparisons) or 500-ml (Ybt extraction) cultures of M9 minimal medium (per liter: 12.8 g Na2HPO4 · 7H2O, 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl; pH adjusted to 7.4 with NaOH) supplemented with glycerol (0.5%, by weight) and Casamino Acids (1%, by weight). Postinoculation, cultures were incubated at 22°C with shaking for 5 days with induction initiated at an optical density at 600 nm (OD600) of 0.4 to 0.6 using 100 μM isopropyl β-d-1-thiogalactopyranoside (IPTG). For cultures requiring exogenous supplementation, 1 mM salicylate was added at induction. Culture plasmid selection was maintained with 100 μg/ml ampicillin, 50 μg/ml kanamycin, 20 μg/ml chloramphenicol, and 50 μg/ml spectinomycin, as needed (the same antibiotic levels were used for solid medium transformation selection). Postculture, 1 ml acetone was added per 50 ml of medium and incubated for an additional 30 min prior to centrifugation to collect the resulting supernatant for extraction. For smaller-scale cultures used to compare Ybt production levels, Fe3+ was added to culture supernatants at a final concentration of 5 mM followed by ethyl acetate extraction twice with an equal volume of ethyl acetate each time. Supernatants collected from larger-scale 500-ml cultures were extracted twice with an equal volume of ethyl acetate each time. Extracts from respective cultures were combined, evaporated to dryness under vacuum, and the resulting residue was resuspended in methanol (with the final volume concentrated 100× relative to the original culture volume).
High-performance liquid chromatography (HPLC) and liquid chromatography-mass spectrometry (LC-MS) analysis.
Salicylate quantification was completed using a Zorbax Eclipse XDB-C18 column connected to an Agilent 1120 system equipped with a diode array detector. Solvent A was 0.1% formic acid in water, solvent B was methanol, and samples (20 μl culture supernatant) were analyzed at a flow rate of 1 ml/min using the following gradient: 5 to 50% solvent B for 15 min, 50 to 5% solvent B for 1 min, and 5% solvent B for 4 min. An absorbance wavelength of 304 nm was used, and peak area quantification was conducted compared to a standard calibration curve of pure salicylate (Sigma-Aldrich).
For initial Ybt detection, HPLC was completed using the system described for salicylate analysis. Injected final extract samples (20 μl) were subjected to a gradient of acetonitrile (0 to 2% from 5 to 7 min, 2 to 70% from 7 to 20 min, 70 to 80% from 20 to 22 min, and 80 to 0% from 22 to 27 min) with an initial base buffer of 17 mM formic acid (pH 3.35) using a flow rate of 1 ml/min. The Ybt-Fe3+ product was detected at 385 nm (30). Purified product was achieved using an Agilent 1200 preparatory HPLC system equipped with a Waters C18, 5 μm, 300 Å, 150- by 3.9-mm (inside diameter) column. In this setting, a 1-ml/min flow rate was used together with a 10-to-100% acetonitrile (balance water) gradient over 15 min. Collected fractions containing product were quantified at 385 nm using the known extinction coefficient for Ybt-Fe3+ (ε = 2,884). Purified product was then used to produce a calibration curve for compound quantification via LC-MS analysis using an API 3000 Triple Quad LC-MS with a Turbo ion spray source (PE Sciex) coupled with a Shimadzu Prominence LC system. All MS analyses were conducted in positive-ion mode, and chromatography was performed on a Waters X Terra C18 column (5 μm; 2.1 by 250 mm [inside diameter]). After an injection of 4 μl of final extract, conditions for LC-MS were a 10-to-100% acetonitrile (balance water) gradient over 15 min at a flow rate of 0.2 ml/min.
Rust removal experiments.
A rusted carbon steel tube was machine cut into U-shaped pieces (through the University at Buffalo School of Engineering and Applied Sciences Engineering Machine Shop), and after being cleaned with acetone and rinsed with deionized (DI) water, each piece was immersed in 10 ml Millipore water. Samples were then mixed with 250 μl of methanol or final extracts (∼0.2 mg of Ybt) derived from the larger-scale negative-control or Ybt production cultures. EDTA disodium salt (Fisher Biotech) was tested in parallel at concentrations equimolar to those of Ybt. Samples were then incubated at 30°C with shaking for 2 h.
Rust removal was quantified through a modification of the 1,10-phenanthroline assay to enable detection using a 96-well plate format. In each well, 40 μl of 0.3% (wt/vol) 1,10-phenanthroline, 40 μl of 1% (wt/vol) hydroxylamine hydrochloride, and 40 μl of 0.1 M ammonium acetate were mixed. Ten microliters of diluted samples (to obtain Fe3+ in a range of 1 to 10 ppm) was added to the mixture, and the total volume was adjusted to 250 μl using Millipore water. Absorbance was measured using a microplate reader at a wavelength of 510 nm with concentration measurements determined from a calibration curve of Fe3+ (Sigma-Aldrich).
To assess total rust removal, metal samples recovered after incubation were washed with water and immersed in a solution of 1% (wt/vol) oxalic acid for 30 min to remove the remaining rust on the metal surface. Upon quantification of the amount of Fe3+ removed using the 96-well plate method introduced above, percent rust removal was calculated using the formula [R1/(R1 + R2)] × 100, where R1 is the amount (in milligrams) of Fe3+ in solution after treatment with Ybt extracts or controls and R2 is the amount (in milligrams) of Fe3+ removed with 1% oxalic acid solution.
Ybt-XAD preparation and characterization.
Amberlite XAD-16N beads were activated as recommended by the manufacturer (Sigma-Aldrich). Beads were washed twice with methanol and twice with water before finally being resuspended in Millipore water. Next, 250 μl of Ybt-containing or negative-control final extract was added to 0.5 g of XAD beads, and the final volume was adjusted to 10 ml using Millipore water. Mixtures were incubated at 30°C overnight. Beads were then washed with Millipore water three times and resuspended in Millipore water for further tests. To assess the effectiveness of Ybt binding to XAD beads, Ybt levels were analyzed by LC-MS before and after contact with XAD. XAD beads alone were also used as a negative control (see Fig. 4, 5, and 6) after it was demonstrated that the XAD beads performed similarly to the XAD-negative control (see Fig. 4C).
FIG 5.
Copper removal of unmodified and Ybt-modified XAD beads across different biological fluids, including RPMI 1640 (A), 10% sheep blood in PBS (B), and RPMI 1640 plus 50% FBS (C). Asterisks indicate statistical significance (95% confidence) in comparison to corresponding negative controls.
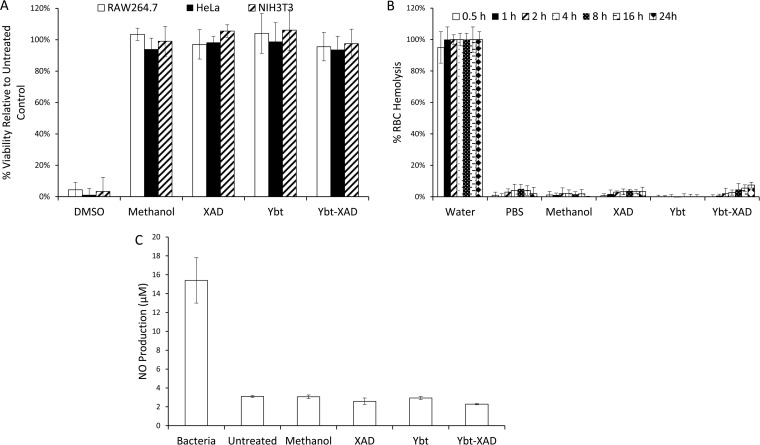
FIG 6.
Biological characterization of Ybt and Ybt-modified XAD. Assays included effects on mammalian cellular viability (A), RBC hemolysis (B), and macrophage immunogenicity (C). Controls included methanol and unmodified XAD.
Copper removal experiments.
Copper and zinc removal assessment was completed using the Zincon assay (31). Briefly, 40-μl samples (diluted if necessary to keep concentrations less than 4 ppm) were added to 200 μl of borate buffer (65.8 mM; pH 9), and the assay was initiated by adding 10 μl of Zincon solution (1.6 mM). After a 5-min incubation at 20°C, absorbance at 610 nm was measured. Standard curves for Zn and Cu were made using pure salts (Sigma-Aldrich) to allow quantification.
To test copper removal, 800 μl of copper solution was added to 40 mg of Ybt-modified XAD beads (or controls) and incubated at 30°C with shaking (for sheep blood and 50% fetal bovine serum [FBS] RPMI samples, this temperature was 22°C). After specific time points, beads were separated by gravity settling and washed with 800 μl of Millipore water twice. The copper content of the original mixture solution and the liquid resulting from the bead-washing steps was measured using the Zincon assay described above. For samples contacted with YBT-modified XAD beads multiple times, freshly prepared beads were used in three successive applications, with the resulting solution's copper concentration being measured at the completion of each stage. The percent copper removal was calculated using the following formula: [(Ci − Cf)/Ci] × 100, where Ci is the copper concentration of the initial solution and Cf is the final concentration of copper, including washes. For samples containing blood or FBS components, an increased copper range of 7.5 to 20 ppm was used (above the 5-ppm level used for nonbiological fluid assays) to account for nonspecific binding of copper to protein and other components of these samples.
Biological interaction assays.
Cytotoxicity resulting from Ybt samples was determined by the 3-(4,5-dimethylthiazol-2-yl)-diphenyltetrazolium bromide (MTT) colorimetric assay. Three distinct cell lines (RAW264.7 [macrophage; provided by Terry Connell, Department of Microbiology and Immunology, University at Buffalo], HeLa [epithelial; provided by Stelios Andreadis, Department of Chemical and Biological Engineering, University of Buffalo], and NIH 3T3 [fibroblast; ATCC]) were cultured in T75 flasks at 37°C and 5% CO2. RAW264.7 cells were maintained in medium prepared as follows: 50 ml of FBS (heat inactivated), 5 ml of 100 mM MEM sodium pyruvate, 5 ml of 1 M HEPES buffer, 5 ml of penicillin-streptomycin solution, and 1.25 g of d-(+)-glucose were added to 500 ml RPMI 1640 and filter sterilized. HeLa and NIH 3T3 cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco BRL, Grand Island, NY) supplemented with 10% (vol/vol) FBS (GIBCO) and 1% (vol/vol) antibiotic-antimycotic (A/A; Gibco). Cells were harvested using mechanical scrapers for RAW264.7 and 0.25% trypsin–1 mM EDTA treatment, trypsin inactivation using an equal volume of DMEM supplemented with 10% FBS, centrifugation (250 × g, 5 min), and resuspension in phosphate-buffered saline (PBS) for HeLa and NIH 3T3 cell lines prior to seeding at 3 × 104 (RAW264.7), 2.9 × 104 (HeLa), and 1.4 × 104 (NIH 3T3) cells/well in tissue culture-treated, sterile, polystyrene 96-well plates in 100 μl medium/well. Postseeding, cells were allowed 24 h for attachment at 37°C and 5% CO2. Final extract was added to 0.03 mg/ml Ybt per well, whereas Ybt-complexed XAD beads (or control bead samples) were added to 50 mg/ml per well (with the solid-phase concentration chosen to equate to the corresponding amount of free Ybt in solution-based assays). A negative control for this experiment (resulting in minimal cytotoxicity) was methanol, whereas dimethyl sulfoxide (DMSO) was used as a positive control. Following 24 h of incubation after sample addition, cells were assayed with MTT solution (5 mg/ml), added at 10% (vol/vol) for 3 h at 37°C and 5% CO2. Medium plus MTT solution was then aspirated and replaced by DMSO to dissolve the formazan reaction products. Following agitated incubation for 1 h, the formazan solution was analyzed by microplate reader at 570 nm, with 630 nm serving as the reference wavelength. Results are presented as a percentage of untreated cells (100% viability). Nitric oxide (NO) production from RAW264.7 cells (a measure of immunogenicity triggered by Ybt sample addition) was determined using a Griess reagent kit (Promega, Madison, WI) according to the manufacturer's instructions. Negative controls for this experiment (resulting in minimal NO levels) were untreated samples and methanol, whereas BAP1 cells (added at a 1:1 ratio to the RAW264.7 cells) were used as a positive control.
To assess potential disruption of red blood cell (RBC) membranes by Ybt samples, a hemolysis assay was modified from a protocol previously described (32). Briefly, a 5% RBC solution was prepared by washing sheep blood (HemoStat Laboratories) with PBS until the supernatant became clear. Next, 100 μl of purified 5% RBC solution was incubated with 900 μl of 0.03-mg/ml crude extract Ybt samples (or controls) or 50-mg/ml Ybt-complexed XAD beads (or controls) in PBS over time at 37°C. A negative control for this experiment (resulting in minimal lysis) was PBS, whereas water was used as a positive control. Triton X-100 (1% solution) was used to construct a standard curve for percent blood lysis by altering the amount of blood added to each sample. For example, for 50 and 100% lysis, 50 and 100 μl of purified 5% RBC solution was mixed with 1% Triton-X (to 1 ml). Samples were centrifuged, and hemolysis was quantified by measuring supernatant at 541 nm and comparing to the percent blood lysis standard curve.
Statistical evaluation.
Unless otherwise indicated, data presented were generated from three independent experiments. Error bars represent standard deviation values. All statistical significance comparisons between groups were performed using a one-way analysis of variance (ANOVA) with Dunnett's (to compare within groups) posttests.
RESULTS AND DISCUSSION
Previously, salicylate was required as an exogenous component of the culture medium during Ybt biosynthesis. In this work, the irp9 gene from Y. enterocolitica was introduced into E. coli (via plasmid) to enable the intracellular provision of this starting substrate (Fig. 2A). The irp9 gene was isolated from a close relative of Y. pestis (Y. enterocolitica) and is dedicated to generating salicylate in the context of Y. enterocolitica Ybt formation (29, 33, 34). As such, this gene was transferred to E. coli as a route to intracellular salicylate formation toward heterologous Ybt biosynthesis. Successful transfer was first confirmed by the accumulation of salicylate in cultures of BAP1/pCDFDuet-1-irp9 (Fig. 2B). The irp9 gene was then tested in the context of Ybt production over time (Fig. 2C) and in comparison to previous production systems, which required salicylate medium supplementation. Improvement in production from 11.5 ± 0.7 to 17.4 ± 0.4 mg/liter was seen between the original system, which is reliant on exogenous salicylate production, and the new strain, which is capable of complete intracellular production of Ybt. The stability of the Ybt-Fe3+ complex was also tested over time in background M9 medium and M9 medium containing the background BAP1 cellular environment (Fig. 2D). In both settings, minimal degradation over time was observed though an initial 1-day drop in product level (∼30%) was observed in medium containing a cellular component, suggesting that some degradation mechanism (such as biochemically derived breakdown or catabolism) may contribute to product loss. Such a concern could be remedied by completing culture conditions with in situ addition of XAD, which could have the dual function of sequestering compound to minimize culture degradation and providing an Ybt-XAD product to then enable metal removal applications as outlined below.
FIG 2.
Heterologous biosynthesis of Ybt. (A) Introduction of the Irp9 enzyme to enable endogenous production of salicylate within E. coli strain BAP1 in support of heterologous Ybt biosynthesis. (B and C) Time course of salicylate produced from BAP1/pCDFDuet-1-irp9 and Ybt produced from BAP1/pBP198/pBP205/pBP200/pCDFDuet-1-irp9. (D) Ybt stability over time as assessed by percent degradation of the compound was analyzed via HPLC upon addition of 64 mg/liter of Ybt-Fe3+ to either M9 medium or M9 medium containing strain BAP1 harboring background plasmids (without inserted biosynthetic genes so as to allow the same antibiotic selection; designated “M9 culture”). The samples were then incubated at 22°C with shaking and IPTG addition. pBP198 contains genes encoding HMWP2, and YbtU and pBP205 contains genes encoding HMWP1 and YbtE.
The intracellular availability of salicylate removes the need for supplementation and consolidates the overall biosynthetic scheme. The introduction of irp9 also ensures that biosynthesis can be derived from standard carbon sources (such as glucose or glycerol). The titers achieved for heterologous salicylate production were substantial, reaching ∼310 mg/liter, which surpassed concentrations used previously (138 mg/liter) during experiments where salicylate was added exogenously (18). Thus, the boost in Ybt production when the irp9 pathway was used suggests a potential bottleneck in exogenous salicylate levels used during previous biosynthetic attempts. The localization of the complete Ybt pathways to E. coli also offers the potential of a simpler and safer screening platform to identify siderophore inhibitors. Namely, emerging approaches and assays designed to screen for small molecule inhibitors of siderophore biosynthesis (35, 36) could incorporate the Ybt system in the experimentally simple E. coli host as opposed to the more complex and significantly more dangerous native Y. pestis source.
The iron binding capabilities of Ybt offer the possibility of widespread applications related to selective metal removal. One such outlet for this activity is in the removal of rust. Rust development proceeds in a layered arrangement and features different types of iron oxide deposits, which are visually distinguishable based upon color differences. Brownish surface layers possess the highest oxygen content, usually FeO(OH) and Fe(OH)3, while the deepest layers consist of black magnetic oxide (Fe2O3) (37, 38). The desire to prevent rust development or remove rust once formed prompted us to test Ybt in a removal setting. Extracts of Ybt-producing cultures were tested against a series of controls (including a common synthetic metal chelator [EDTA] and a commercial product [Rust-Oleum]) to assess the rust removal capabilities of the compound (Fig. 3). As shown, the Ybt extract removed significantly more rust than EDTA, ∼20% of total rust, and ∼40% relative to the commercial control. It is anticipated that further optimization parameters (Ybt concentrations, temperature, and repeated applications) will further enable improved rust removal capabilities. However, the data presented here indicate the raw potential of Ybt in rust removal applications.
The heterologous production process for Ybt removes any need to deal with the native production host (in this case, a priority pathogen) during applications such as rust removal. Furthermore, the compound itself appears to pose little health risk (see below) and is expected to be benign environmentally. However, the compound may target and remove only certain components of a rusted product (such as oxides containing Fe3+) or immediately proximal rust layers, and this likely contributes to the inability of the compound to more closely approach total rust removal. In our experiments, brownish surface layers could be removed using Ybt, but the deeper black oxide layers remained (Fig. 3B). Such a result is not unexpected, since it has been reported that more complete dissolution of iron oxide deposits in siderophore-based desorption efforts involves contributions from small-molecule reductants (39–42).
Extending the metal binding potential of Ybt further, a second target was selected in copper. Ybt has been demonstrated to chelate copper during Y. pestis infection as a means of limiting copper-induced host defense mechanisms (23). Hence, the molecule has evolved to recognize this metal in addition to Fe3+. As such, there exist opportunities to redirect this capability in heterologous production of Ybt. In this work, we initiated experiments that tested copper removal from both nonbiological and biological fluids with the anticipation that successful results would support further assessment in settings such as wastewater treatment or in clinical applications, such as a treatment for Wilson's disease.
In preparation for removal assays, Ybt was first adsorbed to XAD resin beads to establish a solid matrix readily separated from copper-containing fluids. Ybt attachment to XAD was nearly quantitative, as >95% was adsorbed. Conversely, when Ybt-modified XAD beads were incubated with methanol for 3 h, 90% of Ybt was removed from the XAD resin (Fig. 4B). Postattachment, repeated bead washings (three times with water) removed no detectable levels of Ybt, and bead attachment was stable over a 24-h period. The Ybt matrix then constituted a simple means of testing a solid-phase separation process across a range of fluids containing copper.
Ybt-modified XAD demonstrated clear removal capabilities compared to controls (Fig. 4). Increasing concentrations of initial copper and time course analysis resulted in a saturation in removal capability (Fig. 4C and D). However, repeated extractions with Ybt resulted in nearly 100% full solution removal (Fig. 4E). Importantly, Ybt showed no discernible ability to remove zinc (Fig. 4F). Regarding this point, copper and zinc are often found together in environmental and biomedical settings (43–46). In a biomedical context, the potential of Ybt to selectively remove copper would address limitations of current treatments which bind both metals (47). Similarly, copper removal was tested in the presence of iron (Fig. 4G); removal ability remained similar to that seen under conditions without iron inclusion (Fig. 4A).
Copper removal experiments were further conducted in media representative of biological settings, including RPMI mammalian cell growth medium (with and without serum addition) and a solution of sheep blood (Fig. 5). As such, the solutions utilized include biological components, such as serum proteins and red blood cells, expected to limit or inhibit the binding properties of the Ybt samples. Background binding of the XAD control was observed, which is possibly explained by the nonspecific adhesion of proteins and other medium components that have bound copper. In each case, however, the copper removal capability of the Ybt-modified XAD beads was maintained at levels significantly above those of controls. The data provide further support for copper removal in biomedical applications. In such situations, there is also the prospect of using free Ybt in an in vivo setting; however, the risk of simultaneous Ybt-Fe3+ formation and utilization by opportunistic pathogens may prompt an alternative format in which blood metal content is removed using Ybt-XAD beads ex vivo.
A final test of biological compatibility was conducted (Fig. 6). Here, the Ybt samples were tested for detrimental biological effects. Namely, samples were assessed for (i) negative impact upon mammalian cellular viability (using three distinct cell lines), (ii) unwanted immunogenicity (via NO production from macrophage cells), and (iii) RBC hemolysis (a potential concern given the iron-binding properties of Ybt). In each case, the Ybt samples exhibited minimal concerns, further supporting the potential use for metal scavenging applications within biological settings and the likely benign environmental impact of the compound. The data also suggest minimal adverse outcomes as a result of recombinant production from E. coli and remnants from this heterologous host (such as lipopolysaccharide) that may have unwanted cellular effects.
ACKNOWLEDGMENTS
This work was supported from SUNY-Buffalo start-up funds.
We thank David Pawlowski (CUBRC) for providing Y. enterocolitica genomic DNA and Andrew Gulick and Eric Drake (Department of Structural Biology, University at Buffalo) for assistance with preparatory HPLC.
REFERENCES
- 1.Braun V, Killmann H. 1999. Bacterial solutions to the iron-supply problem. Trends Biochem Sci 24:104–109. doi: 10.1016/S0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 2.Miethke M, Marahiel MA. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev 71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratledge C, Dover LG. 2000. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol 54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 4.Caza M, Kronstad JW. 2013. Shared and distinct mechanisms of iron acquisition by bacterial and fungal pathogens of humans. Front Cell Infect Microbiol 3:80. doi: 10.3389/fcimb.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haas H, Eisendle M, Turgeon BG. 2008. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol 46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- 6.Garenaux A, Caza M, Dozois CM. 2011. The ins and outs of siderophore mediated iron uptake by extra-intestinal pathogenic Escherichia coli. Vet Microbiol 153:89–98. doi: 10.1016/j.vetmic.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 7.Muller SI, Valdebenito M, Hantke K. 2009. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals 22:691–695. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- 8.Drechsel H, Stephan H, Lotz R, Haag H, Zahner H, Hantke K, Jung G. 1995. Structural elucidation of yersiniabactin, a siderophore from highly virulent Yersinia strains. Liebigs Ann 10:1727–1733. [Google Scholar]
- 9.Haag H, Hantke K, Drechsel H, Stojiljkovic I, Jung G, Zahner H. 1993. Purification of yersiniabactin: a siderophore and possible virulence factor of Yersinia enterocolitica. J Gen Microbiol 139:2159–2165. doi: 10.1099/00221287-139-9-2159. [DOI] [PubMed] [Google Scholar]
- 10.Perry RD, Balbo PB, Jones HA, Fetherston JD, DeMoll E. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181–1190. doi: 10.1099/13500872-145-5-1181. [DOI] [PubMed] [Google Scholar]
- 11.Wallner A, Blatzer M, Schrettl M, Sarg B, Lindner H, Haas H. 2009. Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus. Appl Environ Microbiol 75:4194–4196. doi: 10.1128/AEM.00479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crosa JH, Walsh CT. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cane DE, Walsh CT, Khosla C. 1998. Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 14.Du L, Sanchez C, Shen B. 2001. Hybrid peptide-polyketide natural products: biosynthesis and prospects toward engineering novel molecules. Metab Eng 3:78–95. doi: 10.1006/mben.2000.0171. [DOI] [PubMed] [Google Scholar]
- 15.Menzella HG, Reeves CD. 2007. Combinatorial biosynthesis for drug development. Curr Opin Microbiol 10:238–245. doi: 10.1016/j.mib.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Boghigian BA, Armando J, Pfeifer BA. 2011. Methods and options for the heterologous production of complex natural products. Nat Prod Rep 28:125–151. doi: 10.1039/C0NP00037J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ongley SE, Bian X, Neilan BA, Muller R. 2013. Recent advances in the heterologous expression of microbial natural product biosynthetic pathways. Nat Prod Rep 30:1121–1138. doi: 10.1039/c3np70034h. [DOI] [PubMed] [Google Scholar]
- 18.Pfeifer BA, Wang CC, Walsh CT, Khosla C. 2003. Biosynthesis of Yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl Environ Microbiol 69:6698–6702. doi: 10.1128/AEM.69.11.6698-6702.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Carvalho CC, Marques MP, Fernandes P. 2011. Recent achievements on siderophore production and application. Recent Pat Biotechnol 5:183–198. doi: 10.2174/187220811797579114. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed E, Holmstrom SJ. 2014. Siderophores in environmental research: roles and applications. Microb Biotechnol 7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schalk IJ, Hannauer M, Braud A. 2011. New roles for bacterial siderophores in metal transport and tolerance. Environ Microbiol 13:2844–2854. doi: 10.1111/j.1462-2920.2011.02556.x. [DOI] [PubMed] [Google Scholar]
- 22.Braud A, Hannauer M, Mislin GL, Schalk IJ. 2009. The Pseudomonas aeruginosa pyochelin-iron uptake pathway and its metal specificity. J Bacteriol 191:3517–3525. doi: 10.1128/JB.00010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaturvedi KS, Hung CS, Crowley JR, Stapleton AE, Henderson JP. 2012. The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat Chem Biol 8:731–736. doi: 10.1038/nchembio.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chaturvedi KS, Hung CS, Giblin DE, Urushidani S, Austin AM, Dinauer MC, Henderson JP. 2014. Cupric yersiniabactin is a virulence-associated superoxide dismutase mimic. ACS Chem Biol 9:551–561. doi: 10.1021/cb400658k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds HV, Talekar CR, Bellapart J, Leggett BA, Boots RJ. 2014. Copper removal strategies for Wilson's disease crisis in the ICU. Anaesth Intensive Care 42:253–257. [PubMed] [Google Scholar]
- 26.Gateau C, Delangle P. 2014. Design of intrahepatocyte copper(I) chelators as drug candidates for Wilson's disease. Ann N Y Acad Sci 1315:30–36. doi: 10.1111/nyas.12379. [DOI] [PubMed] [Google Scholar]
- 27.Brewer GJ. 2012. Metals in the causation and treatment of Wilson's disease and Alzheimer's disease, and copper lowering therapy in medicine. Inorg Chim Acta 393:135–141. doi: 10.1016/j.ica.2012.06.014. [DOI] [Google Scholar]
- 28.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE, Khosla C. 2001. Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291:1790–1792. doi: 10.1126/science.1058092. [DOI] [PubMed] [Google Scholar]
- 29.Pelludat C, Brem D, Heesemann J. 2003. Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J Bacteriol 185:5648–5653. doi: 10.1128/JB.185.18.5648-5653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones AM, Lindow SE, Wildermuth MC. 2007. Salicylic acid, yersiniabactin, and pyoverdin production by the model phytopathogen Pseudomonas syringae pv. tomato DC3000: synthesis, regulation, and impact on tomato and Arabidopsis host plants. J Bacteriol 189:6773–6786. doi: 10.1128/JB.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabel CE, Neureuther JM, Siemann S. 2010. A spectrophotometric method for the determination of zinc, copper, and cobalt ions in metalloproteins using Zincon. Anal Biochem 397:218–226. doi: 10.1016/j.ab.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Jones CH, Ravikrishnan A, Chen M, Reddinger R, Kamal Ahmadi M, Rane S, Hakansson AP, Pfeifer BA. 2014. Hybrid biosynthetic gene therapy vector development and dual engineering capacity. Proc Natl Acad Sci U S A 111:12360–12365. doi: 10.1073/pnas.1411355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerbarh O, Chirgadze DY, Blundell TL, Abell C. 2006. Crystal structures of Yersinia enterocolitica salicylate synthase and its complex with the reaction products salicylate and pyruvate. J Mol Biol 357:524–534. doi: 10.1016/j.jmb.2005.12.078. [DOI] [PubMed] [Google Scholar]
- 34.Kerbarh O, Ciulli A, Howard NI, Abell C. 2005. Salicylate biosynthesis: overexpression, purification, and characterization of Irp9, a bifunctional salicylate synthase from Yersinia enterocolitica. J Bacteriol 187:5061–5066. doi: 10.1128/JB.187.15.5061-5066.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neres J, Wilson DJ, Celia L, Beck BJ, Aldrich CC. 2008. Aryl acid adenylating enzymes involved in siderophore biosynthesis: fluorescence polarization assay, ligand specificity, and discovery of non-nucleoside inhibitors via high-throughput screening. Biochemistry 47:11735–11749. doi: 10.1021/bi801625b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wurst JM, Drake EJ, Theriault JR, Jewett IT, VerPlank L, Perez JR, Dandapani S, Palmer M, Moskowitz SM, Schreiber SL, Munoz B, Gulick AM. 2014. Identification of inhibitors of PvdQ, an enzyme involved in the synthesis of the siderophore pyoverdine. ACS Chem Biol 9:1536–1544. doi: 10.1021/cb5001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oh SJ, Cook DC, Townsend HE. 1998. Characterization of iron oxides commonly formed as corrosion products on steel. Hyperfine Interact 112:59–65. doi: 10.1023/A:1011076308501. [DOI] [Google Scholar]
- 38.Argo J. 1981. A qualitative test for iron corrosion products. Stud Conserv 26:140–142. [Google Scholar]
- 39.Cervini-Silva J, Sposito G. 2002. Steady-state dissolution kinetics of aluminum-goethite in the presence of desferrioxamine-B and oxalate ligands. Environ Sci Technol 36:337–342. doi: 10.1021/es010901n. [DOI] [PubMed] [Google Scholar]
- 40.Reichard PU, Kraemer SM, Frazier SW, Kretzschmar R. 2005. Goethite dissolution in the presence of phytosiderophores: rates, mechanisms, and the synergistic effect of oxalate. Plant Soil 276:115–132. doi: 10.1007/s11104-005-3504-9. [DOI] [Google Scholar]
- 41.Dhungana S, Anthony CR, Hersman LE. 2007. Effect of exogenous reductant on growth and iron mobilization from ferrihydrite by the Pseudomonas mendocina ymp strain. Appl Environ Microbiol 73:3428–3430. doi: 10.1128/AEM.02586-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dehner CA, Awaya JD, Maurice PA, DuBois JL. 2010. Roles of siderophores, oxalate, and ascorbate in mobilization of iron from hematite by the aerobic bacterium Pseudomonas mendocina. Appl Environ Microbiol 76:2041–2048. doi: 10.1128/AEM.02349-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flora SJ, Pachauri V. 2010. Chelation in metal intoxication. Int J Environ Res Public Health 7:2745–2788. doi: 10.3390/ijerph7072745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agarwal S, Ferreira AE, Santos SMC, Reis MTA, Ismael MRC, Correia MJN, Carvalho JMR. 2010. Separation and recovery of copper from zinc leach liquor by solvent extraction using Acorga M5640. Int J Miner Process 97:85–91. doi: 10.1016/j.minpro.2010.08.009. [DOI] [Google Scholar]
- 45.Zhu Z, Zhang W, Pranolo Y, Cheng CY. 2012. Separation and recovery of copper, nickel, cobalt and zinc in chloride solutions by synergistic solvent extraction. Hydrometallurgy 127:1–7. [Google Scholar]
- 46.Zhang XJ, Li XG, Cao HB, Zhang Y. 2010. Separation of copper, iron (III), zinc and nickel from nitrate solution by solvent extraction using LK-C2. Sep Purif Technol 70:306–313. doi: 10.1016/j.seppur.2009.10.012. [DOI] [Google Scholar]
- 47.Seelig MS. 1982. Auto-immune complications of d-penicillamine–a possible result of zinc and magnesium depletion and of pyridoxine inactivation. J Am Coll Nutr 1:207–214. doi: 10.1080/07315724.1982.10718989. [DOI] [PubMed] [Google Scholar]