Abstract
Lactic acid bacteria are found in the gastrointestinal tract of mammals and have received tremendous attention due to their health-promoting properties. We report the development of two dual-color luciferase-producing Lactobacillus (Lb.) plantarum and Lactococcus (Lc.) lactis strains for noninvasive simultaneous tracking in the mouse gastrointestinal tract. We previously described the functional expression of the red luciferase mutant (CBRluc) from Pyrophorus plagiophthalamus in Lb. plantarum NCIMB8826 and Lc. lactis MG1363 (C. Daniel, S. Poiret, V. Dennin, D. Boutillier, and B. Pot, Appl Environ Microbiol 79:1086–1094, 2013, http://dx.doi.org/10.1128/AEM.03221-12). In this study, we determined that CBRluc is a better-performing luciferase for in vivo localization of both lactic acid bacteria after oral administration than the green click beetle luciferase mutant construct developed in this study. We further established the possibility to simultaneously detect red- and green-emitting lactic acid bacteria by dual-wavelength bioluminescence imaging in combination with spectral unmixing. The difference in spectra of light emission by the red and green click beetle luciferase mutants and dual bioluminescence detection allowed in vitro and in vivo quantification of the red and green emitted signals; thus, it allowed us to monitor the dynamics and fate of the two bacterial populations simultaneously. Persistence and viability of both strains simultaneously administered to mice in different ratios was studied in vivo in anesthetized mice and ex vivo in mouse feces. The application of dual-luciferase-labeled bacteria has considerable potential to simultaneously study the interactions and potential competitions of different targeted bacteria and their hosts.
INTRODUCTION
Lactococci and lactobacilli are lactic acid bacteria (LAB) that have been used for thousands of years for the production and preservation of fermented food, such as milk, vegetables, and meat. Some specific strains are commercialized as probiotics and claimed to have health-promoting properties (1). The gastrointestinal tract (GIT) is the most important field of activity of LAB, although distal effects outside the gut have been described (2). Thus, it is important to understand the interactions of the administered bacteria with their host GIT system. LAB are able to survive and adapt to the GIT conditions, as shown by comparative and functional genomic characterization of various human isolates, essentially lactobacilli (for a review, see references 2 and 3). Some LAB, when present in the GIT of mice or humans, express a number of common characteristics that may relate to their intestinal niche adaptation (3, 4). However, direct in vivo tracking of these actions in terms of both spatial and temporal evolution would allow a better understanding of the survival and metabolic activities of these LAB in the gut.
In the past decade, bioluminescence imaging (BLI) has become essential for in vivo noninvasive monitoring of biological processes (4). The technique relies on the detection of photons emitted from cells or tissues in a living organism. Bioluminescence is a biological process that requires an enzyme known as luciferase, an enzyme-specific substrate (e.g., luciferin), oxygen, magnesium, and/or ATP, depending on the luciferase. Luciferases encompass a wide range of enzymes that catalyze light-producing chemical reactions in living organisms. The most used luciferase for in vivo bioluminescence imaging is represented by the Photinus pyralis (firefly) luciferase (Fluc) and the click beetle luciferase from Pyrophorus plagiophthalamus (CBluc). Both use d-luciferin as the substrate, depend on ATP, Mg, and O2, and result in the production of green light. Red click beetle (CBRluc) and firefly luciferase variants with higher emission wavelengths also have been developed and greatly enhance the penetration and sensitivity of BLI in deep tissues (4). We previously showed that CBRluc produced in Lactobacillus (Lb.) plantarum NCIMB8826, one of most studied strains in LAB research, and Lactococcus (Lc.) lactis MG1363, a dairy starter derivative, can be successfully detected in the GIT of anesthetized mice by noninvasive BLI after oral administration (5).
It has been reported that dual-color BLI can be applied in vitro and in vivo using appropriate emission filters for the separation of bioluminescent signals and mathematical corrections for their deconvolution using spectral unmixing algorithms (6–8). Dual-color bioluminescence has been described with red and green luciferases expressed by mammalian cells that can be separated in vivo (9). Very recently, Chang et al. showed that Fluc and CBRluc, which emit distinct light wavelengths, allow simultaneous imaging of two mycobacterial strains during infection (10).
As LAB are studied and given to humans and animals mostly in mixtures with multiple strains (11), we describe the development and application of two dual-color luciferase-expressing Lb. plantarum and Lc. lactis isolates for noninvasive simultaneous monitoring in the GIT of small laboratory animals. We demonstrate that CBRluc is a better-performing luciferase in vivo than the green click beetle luciferase (CBGluc) mutant, which also is described in this study. We also explore the possibility of simultaneously detecting red- and green-emitting LAB by dual-wavelength bioluminescence imaging in combination with spectral unmixing.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids are listed in Table 1. The Lb. plantarum codon-optimized cbgluc gene under the control of Pldh and the Lc. lactis codon-optimized cbgluc gene under the control of Pusp45 (the two DNA fragments were synthetized by Eurogentec [Belgium]) were cloned into pNZ8148 as BglII-XbaI fragments. The two resulting constructs subsequently were introduced into Lc. lactis MG1363 and Lb. plantarum NCIMB8826 by electrotransformation as described elsewhere (12) and named Lc. lactis-CBGluc (Ll-CBGluc) and Lb. plantarum-CBGluc (Lp-CBGluc), respectively. Lc. lactis MG1363 and Lb. plantarum NCIMB8826 containing the empty vector pNZ8148 (named Ll-pNZ8148 and Lp-pNZ8148, respectively), described previously (5), served as controls in all of the in vitro and in vivo experiments. Lp-CBRluc and Ll-CBRluc were described previously (5). Strain stability was tested as described previously (13).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| Lactococcus lactis subsp. cremoris MG1363 | Plasmid free | 12 |
| Lactobacillus plantarum NCIMB8826 | Originally isolated from human saliva | NCIMB |
| Escherichia coli MC1061 | araD139 Δ(ara-leu)7696 lacX74 galV galK hsr-hsm rpsL | Invitrogen |
| Plasmids | ||
| pNZ8148 | Cmr, L. lactis pSH71 replicon | MoBiTech |
| pMEC256 | pNZ8148 carrying CBRluc cDNA optimized for L. plantarum codon fused to the L. plantarum Pldh promoter (lactate dehydrogenase) | 5 |
| pMEC257 | pNZ8148 carrying CBRluc cDNA optimized for L. lactis codon fused to the L. lactis Pusp45 | 5 |
| pMEC271 | pNZ8148 carrying CBGluc cDNA optimized for L. plantarum codon fused to Pldh | This study |
| pMEC269 | pNZ8148 carrying CBGluc cDNA optimized for L. lactis codon fused to Pusp45 | This study |
Cmr, resistance to chloramphenicol.
Escherichia coli was cultured in Luria-Bertani broth at 37°C. Lc. lactis was grown at 30°C in M17 medium (Difco, Becton Dickinson, Sparks, MD) supplemented with 0.5% glucose. Lb. plantarum was grown at 37°C in MRS medium (Difco, Becton Dickinson). Chloramphenicol (Sigma-Aldrich, St. Quentin Fallavier, France) was added to culture media for bacterial selection, when necessary, at a final concentration of 20 μg/ml for E. coli and 10 μg/ml for LAB strains.
In vitro bioluminescence measurement.
The bioluminescence of each recombinant luciferase was quantified as described previously (5). Briefly, recombinant bacteria were grown overnight, harvested by centrifugation, and washed with phosphate-buffered saline (PBS). Fifty microliters of each culture, adjusted to approximately 2.5 × 1011 CFU/ml, was distributed in black microplates (Nunc, Thermo Fisher, NY, USA) and imaged after addition of 50 μl of the Bright-Glo luciferase (Promega, Madison, WI, USA). Luminescence was measured at room temperature on the IVIS Lumina XR imaging system (PerkinElmer, Alameda, CA, USA) using the Living Image software (PerkinElmer). Each individual well was manually selected as a region of interest (ROI). Strains were compared on the basis of the total flux of emitted light, expressed as photons per second (p/s) measured per 1.2 × 1010 CFU of each bacterial culture. Lc. lactis MG1363 and Lb. plantarum NCIMB8826 containing the empty vector pNZ8148 were used to measure the background luminescence.
Preparation of bacteria and administration to mice.
Bacterial strains were grown overnight, harvested by centrifugation, and washed with PBS. Mice received 5 × 1010 CFU in 200 μl gavage buffer (0.2 M NaHCO3 buffer containing 1% glucose, pH 8). Eight-week-old female BALB/c mice were purchased from Charles River (St. Germain sur l'Arbresle, France). Experiments were performed in an accredited establishment (no. A59107; Institut Pasteur de Lille) according to European guidelines (number 86/609/CEE), and animal protocols were approved by the local ethics committee.
In vivo persistence of LAB in the GIT of mice.
Groups of five mice received a unique, daily dose of 5 × 1010 CFU of live recombinant strains (Lp-CBRluc, Ll-CBRluc, Lp-CBGluc, or Ll-CBGluc) or different doses of the two different recombinant strains (one expressing CBRluc and the other CBGluc) administered for four consecutive days by oral gavage. Control mice received Lp-pNZ8148 and Ll-pNZ8148 in the different experiments. Fecal samples were individually collected at different time points and mechanically homogenized in PBS. No chloramphenicol-resistant bacteria were detected in noninoculated mice. Mice were anesthetized with 2% isoflurane and sacrificed by cervical dislocation, and mouse digestive tracts were immediately excised before ex vivo bioluminescence imaging. As described previously, the intestines were injected with air to enhance the bioluminescent signal (5). Experiments were repeated twice.
In vivo bioluminescence imaging.
Bioluminescence imaging was performed using a multimodal IVIS Lumina XR imaging system (PerkinElmer) as described previously (5). Mice were anesthetized with 2% isoflurane, and d-luciferin potassium salt (PerkinElmer) was administered to animals intragastrically 15 min before the oral administration of the bacteria. Two hours later, mice were placed into the specimen chamber of the IVIS imaging system, where a controlled flow of 1.5% isoflurane was administered through a nose cone via a gas anesthesia system (Tem Sega, Lormont, France). A grayscale reference photograph under low illumination was taken as an overlay prior to detection of emitted photons over 1 s to 5 min, depending on signal intensity and using Living Image software (PerkinElmer). A pseudocolor image representing detected light intensity was generated using Living Image software and superimposed over the grayscale reference photograph. For each individual mouse, the ROI corresponding to the mouse digestive tract was selected manually.
In vitro and in vivo spectral unmixing of emission wavelengths. To separate the red bioluminescent signal (CBRluc with an emission peak at 620 nm) from the green signal (CBGluc with an emission peak at 540 nm) in mixed cultures of Lc. lactis and Lb. plantarum, either (i) in vitro in microplates, (ii) in vivo in mice after coadministration of both strains by oral gavage, (iii) ex vivo on dissected GITs, or (iv) directly in feces, images were taken using a set of 20-nm step emission filters (500 series high-spectral-resolution emission filters; PerkinElmer) from 500 nm to 620 nm. Living Image software was used for generating spectrally unmixed images and quantification of each signal.
RESULTS
In vitro comparison of the production of CBGluc and CBRluc in Lc. lactis and Lb. plantarum. The bioluminescent signals produced by the different recombinant Lc. lactis and Lb. plantarum isolates were quantified and compared in vitro on bacterial cultures adjusted to 2.5 × 1011 CFU/ml (Fig. 1). Results show that maximum bioluminescence is obtained with Lp-CBRluc and Lp-CBGluc with mean values of 5.1 × 1010 and 4.1 × 1010 p/s (per 1.2 × 1010 CFU, equivalent to 50 μl of bacterial culture). Approximately 36-fold lower bioluminescent signals were obtained with Lc. lactis strains producing CBGluc and CBRluc. No statistical difference was found between the production of CBGluc and CBRluc for the respective strains. Plasmids 271 and 269 in Lc. lactis and Lb. plantarum, respectively, were remarkably stable, with 100% bioluminescent colonies even after approximately 100 generations (10 subcultures) in nonselective medium.
FIG 1.
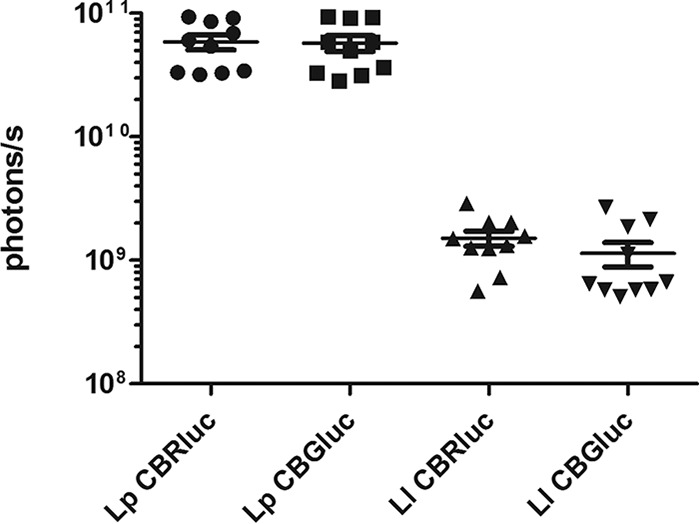
Quantification of bioluminescent signals in cultures of Lp-CBRluc, Lp-CBGluc, Ll-CBRluc, and Ll-CBGluc. The values displayed correspond to the means from 10 independent cultures. Results are given in photons per second for 1.2 × 1010 CFU of each strain. Error bars indicate standard deviations. Lp, Lb. plantarum; Ll, Lc. lactis. Differences between groups were assessed using the Kruskal-Wallis nonparametric test, and no statistical difference between Lp CBRluc and Lp CBGluc or between Ll CBRluc and Ll CBGluc, respectively, was found. There was a statistical difference (P < 0.001) between the two Lb. plantarum and the two Lc. lactis strains. For clarity purposes, the statistics are not shown.
As observed earlier for CBRluc-producing strains (5), there was an excellent correlation between CFU counts and in vitro bioluminescent signals from CBGluc-expressing strains (data not shown). The bioluminescence system allowed the detection of bacterial amounts as low as 5 × 104 CFU/ml for Lp-CBGluc (similar to Lp-CBRluc [5]) and 5 × 105 CFU/ml for Ll-CBGluc (similar to Ll-CBGluc [5]).
Comparison of the persistence of CBGluc- and CBRluc-producing Lc. lactis and Lb. plantarum in the digestive tract of mice after 4 daily oral administrations.
As we had previously shown that differences in bacterial persistence in mice are more pronounced after multiple daily oral administrations (5), groups of mice were given a daily dose of 5 × 1010 CFU of each recombinant strain for four consecutive days. The bioluminescent signal was quantified each day by direct imaging in 5 anesthetized mice (Fig. 2). The same mice were used during the whole experiment. The signal was very strong for the four strains on day 1 and remained at a similar level until the last day of oral administration (day 4). Results show that from day 1 to day 4, Lp-CBRluc emitted the highest bioluminescent signal (mean value of 1.2 × 108 p/s), 6-fold higher than that of Lp-CBGluc, 14-fold higher than that of Ll-CBRluc, and 111-fold higher than that of Ll-CBGluc. Signals of both Lc. lactis strains decreased to background levels at day 5, whereas signals of both Lb. plantarum strains decreased to background levels only at day 7.
FIG 2.
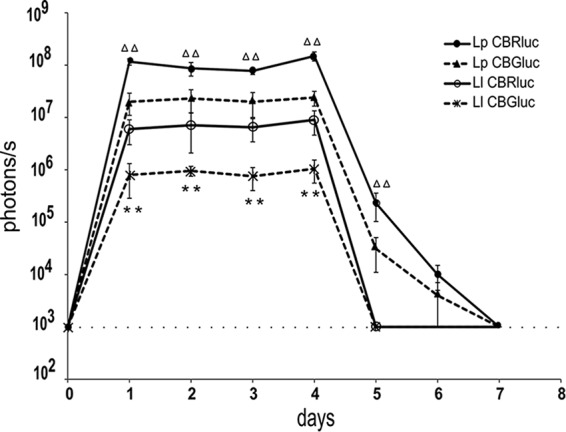
Persistence of Lc. lactis and Lb. plantarum, both producing CBR and CBG, in mice after oral administration. Plots represent the bioluminescence from day 1 to day 7 from each set of five mice, with standard deviations. The background level for the bioluminescent signal is displayed as a dashed line. Differences between groups were assessed using the Kruskal-Wallis test, and those found to be statistically significant are indicated with triangles for comparison between Lp-CBRluc and Lp-CBGluc (two triangles, P < 0.005) and asterisks for comparison between Ll-CBRluc and Ll-CBGluc (P < 0.005).
Simultaneous monitoring of red and green bioluminescent signals by dual-color in vivo imaging.
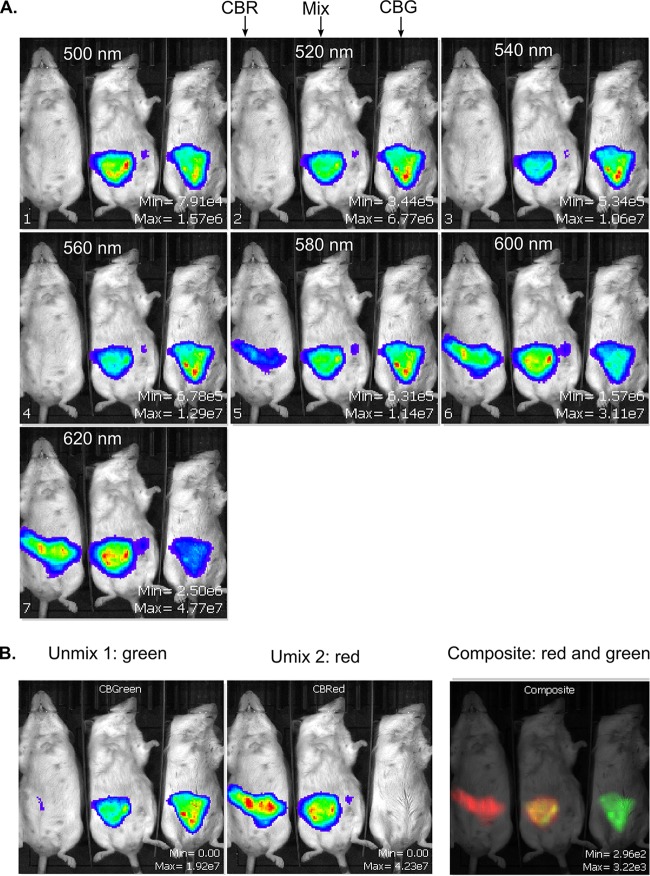
In order to test the CBRluc/CBGluc pair of luciferases for in vivo applications, groups of mice were given an oral dose of Lp-CBRluc, Lp-CBGluc, or a mixture (50/50) of Lb. plantarum expressing CBRluc and expressing CBGluc for 4 days. The bioluminescent signal was measured each day in mice, and a series of images at different wavelengths, ranging from 500 to 620 nm, were acquired (Fig. 3A). Representative images of a spectral unmixing of the red and green signals are shown in Fig. 3. The respective red and green signals are separated and quantified after applying the spectral unmixing algorithm to the acquired image series. The resulting Unmix1 image displays the green signal from Lp-CBGluc, while the resulting Unmix2 image displays the red signal from Lp-CBRluc (Fig. 3B). The red signal was not detected in the green emission light image, and the green signal was not detected in the red emission light image, which confirmed the good spectral unmixing of both signals. Moreover, both signals were detected in mice administered a mixture of Lp-CBRluc and Lp-CBGluc.
FIG 3.
Representative images of spectral unmixing of red and green signals in mice after oral administration of bioluminescent bacteria. (A) Multispectral acquisition of red- and green-emitting Lb. plantarum. Groups of mice were fed with Lp-CBRluc (CBR), Lp-CBGluc (CBG), or a mixture of both strains (50/50; Mix). Images were taken using a set of 20-nm filter step emission filters ranging from 500 nm to 620 nm. (B) Resulting unmixed images used for quantification and recomposition of the two different luciferases in false colors (red for CBR, green for CBG, and yellow for both luciferases).
Simultaneous monitoring of red and green bioluminescent signals after coadministration of Lb. plantarum-CBRluc and Lc. lactis-CBGluc by dual-color in vivo imaging.
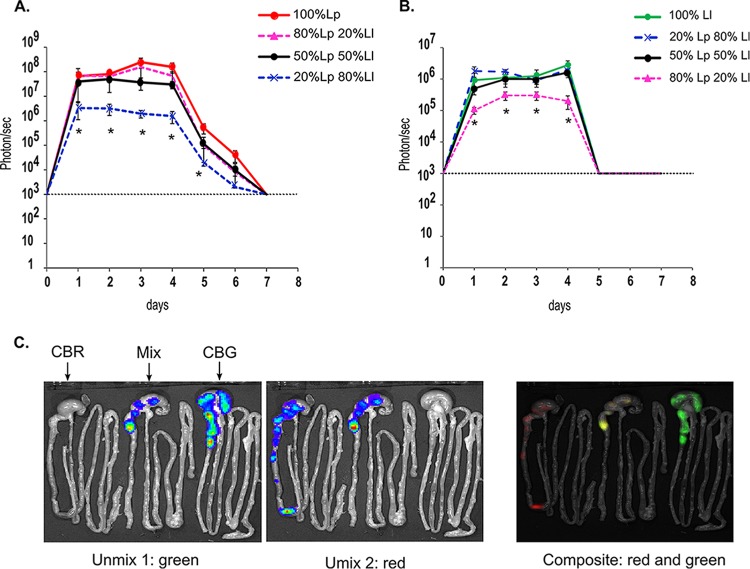
In order to study the persistence of Lb. plantarum and Lc. lactis in mice simultaneously, groups of mice were given an oral daily dose of Lp-CBRluc, Ll-CBGluc, or different mixtures (20/80, 50/50, and 80/20) of Lp-CBRluc and Ll-CBGluc for 4 days. We found that red signals in mice administered a mixture of 50/50 or 80/20 of Lp-CBRluc–Ll-CBGluc are similar to red signals of mice administered 100% Lp-CBRluc (no statistical difference) from day 1 to day 6 (Fig. 4A). The same observation was made for green signals in mice administered only Ll-CBGluc (Fig. 4B). However, red signals in mice administered Lb. plantarum only are statistically higher than red signals in mice administered a mixture of 20% Lb. plantarum and 80% Lc. lactis from day 1 to day 6. Similarly, green signals in mice administered 100% Lc. lactis are statistically higher than green signals in mice administered a mixture of 20% Lc. lactis and 80% Lb. plantarum. The values of red signals from Lb. plantarum given alone (mean value of 1.4 × 108 p/s from day 1 to day 4) are statistically higher than green signals from Lc. lactis given alone (P < 0.001). Signals of Lc. lactis given alone or in mixtures decreased to background levels at day 5, whereas signals of Lb. plantarum given alone or in mixtures decreased to background levels only at day 7.
FIG 4.
Spectral unmixing of red (A) and green (B) signals after oral administration of Lp-CBRluc, Ll-CBGluc, and a mixture of the two strains. Groups of mice were fed for four consecutive days with Lp-CBRluc, Ll-CBGluc, or a mixture of both strains (20/80, 50/50, and 80/20). Plots represent the bioluminescence of each set of five mice, with standard deviations. The background level for the bioluminescent signal is represented by a dashed line. Differences between groups were assessed using the Kruskal-Wallis test, and those found to be statistically significant (P < 0.005) are indicated with an asterisk for comparison between 100% Lp-CBRluc and 20% Lp-CBRluc–80% Ll-CBGluc and between 100% Ll-CBGluc and 80% Lp-CBRluc–20% Ll-CBGluc. (C) Two mice per group were sacrificed from day 4 to day 7, and a representative image of the digestive tract of one mouse per group is shown at day 4 in mice fed with Lp-CBRluc (CBR), a mixture of 50% Lp–50% Ll (Mix), and Ll-CBGluc (CBG).
As observed previously (5), both strains separately administered were localized essentially in the cecum and colon from day 1 to day 4 (Fig. 4C). When both strains are administered simultaneously to mice (data not shown for the 20/80 and 80/20 mixtures), they both also localized in the cecum and colon. On days 5 and 6, Lb. plantarum was localized essentially in the cecum and colon of mice orally administered Lb. plantarum or in mixtures with Lc. lactis, while no green signal was detected.
The persistence of both strains administered separately or in mixtures, as well as the respective red and green bioluminescent signals, also were examined in mouse feces after spectral unmixing (see Fig. S1 in the supplemental material). The Lb. plantarum CBRluc administered alone and in mixtures persisted for 11 days after the last inoculation (day 4), whereas Ll-CBGluc given alone and in mixtures persisted for 4 days after the last inoculation. The bioluminescent signal of Lp-CBRluc given alone and in mixtures was detected 3 days after the last dose, while Ll-CBGluc was undetectable at day 5.
DISCUSSION
As LAB are studied and applied in humans and animals mostly in mixtures with multiple strains (11), there is a need to better understand the fate of the individual strains in the GIT as well as the possible in vivo interactions between the different strains of the mixture. Bioluminescence has been shown to be a useful approach to study the temporal and spatial evolution of live bacteria administered to mice (5). Dual-color luminescence has been described with red and green luciferases in other in vivo systems (8–10) but so far has never been used in Gram-positive LAB.
Here, we describe the development and application of a green-and-red variant of the click beetle luciferase, expressed by strains of Lactobacillus plantarum NCIMB8826 and Lactococcus lactis MG1363 for simultaneous detection of these strains in the GIT of mice. Both luciferases use the same d-luciferin substrate and can be imaged and discriminated simultaneously. We were able to separate the two signals spectrally, allowing the respective strains to be monitored and their respective bioluminescent signals to be quantified.
Our results show that Lb. plantarum expressing CBRluc and CBGluc produced the highest luminescent signal, 30 times brighter than the ones expressed by Lc. lactis. Similar production differences have been observed already for CBRluc in a previous study, even with strong optimized Lc. lactis promoters (5). In vivo, the bioluminescence of CBRluc was greater than the one from CBGluc in both strains, demonstrating that CBRluc is a more sensitive reporter than CBGluc for tracking bacteria in the GIT. Similar observations were made by Chang et al. with CBRluc and Fluc (with an emission spectrum similar to CBGluc's) in the lungs of mice infected with bioluminescent mycobacteria (10). Similar conclusions also have been made by Mezzanotte et al. on imaging the liver and prostate using a red-optimized Fluc with CBGluc (9). The lungs, the liver, the prostate, and the GIT are difficult sites to image optically due to the great tissue depth, and the use of red-emitting luciferases is more adapted to imaging in these tissues, as the longer wavelength enables better penetration through living tissues (4, 8–10).
However, even if the bioluminescence produced in vivo by CBGluc is lower than the one produced by CBRluc, CBGluc is still a good and reliable reporter for LAB in the GIT, as the kinetics of the bioluminescent signals from each respective strain expressing CBGluc and CBRluc were similar during all of the experiments. Both bioluminescent reporters, CBGluc and CBRluc, exhibiting different emission spectra, allowed for dual bioluminescence labeling, permitting us to study the persistence of Lb. plantarum and Lc. lactis in the GIT of mice. We confirmed, based on bioluminescence, that Lb. plantarum persists better than Lc. lactis in the GIT of mice (5, 14). Signals from anesthetized mice and feces showed that the persistence of coadministered Lb. plantarum and Lc. lactis is similar to the persistence of each strain administered separately, illustrating that the strains are not competing with each other, even if they were colocalized in the cecum and colon as described previously (5). The survival and persistence of mixtures of LAB strains have been studied in mice and animals (15–17). However, to our knowledge, the comparison between the persistence of two coadministered LAB and the persistence of each strain administered separately has never been reported.
Our study supports the potential for multiple luciferases to be imaged simultaneously in vivo using spectral unmixing. Further studies are needed to combine both bioluminescence and fluorescence (18), allowing for highly sensitive optical imaging, ranging from single-cell analysis to in vivo whole-body bioluminescence imaging (19–21). The application of dual-luciferase labeled bacteria has significant potential to allow further study of the interactions of various LAB with their mammalian hosts. Our system may be used to perform competition assays between mutant and wild-type strains in the same animal, analyze immunological events in real time, e.g., by labeling a subset of immune cells (22) in conjunction with bioluminescent bacteria, or study the persistence of two LAB which have a different behavior in vivo, and improve our understanding of the effects of a mixture of LAB strains in the GIT.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support of the CNIEL and Syndifrais for this project. We also thank the Institut Fédératif de Recherche IFR142 for cofunding the high-spectral-resolution emission filter.
We are thankful to Pascaline Perrault for her participation in the construction of the recombinant CBGluc strains, as well as the initial animal experiments with spectral unmixing, and to Axel Scotetes for his participation in the final animal experiments.
We thank the BioImaging Center of Lille (Frank Lafont) for the use of the IVIS Lumina XR. We also thank Béatrice David and Guillaume Reveillon from PerkinElmer for their help in setting up the project and in the use of the high-spectral-resolution emission filter set for spectral unmixing.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01042-15.
REFERENCES
- 1.Foligne B, Daniel C, Pot B. 2013. Probiotics from research to market: the possibilities, risks and challenges. Curr Opin Microbiol 16:284–292. doi: 10.1016/j.mib.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Bron PA, van Baarlen P, Kleerebezem M. 2012. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 10:66–78. [DOI] [PubMed] [Google Scholar]
- 3.Douillard FP, de Vos WM. 2014. Functional genomics of lactic acid bacteria: from food to health. Microb Cell Fact 13(Suppl 1):S8. doi: 10.1186/1475-2859-13-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andreu N, Zelmer A, Wiles S. 2011. Noninvasive biophotonic imaging for studies of infectious disease. FEMS Microbiol Rev 35:360–394. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daniel C, Poiret S, Dennin V, Boutillier D, Pot B. 2013. Bioluminescence imaging study of spatial and temporal persistence of Lactobacillus plantarum and Lactococcus lactis in living mice. Appl Environ Microbiol 79:1086–1094. doi: 10.1128/AEM.03221-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. 2006. Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal Chem 78:1520–1527. doi: 10.1021/ac051999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michelini E, Cevenini L, Mezzanotte L, Ablamsky D, Southworth T, Branchini B, Roda A. 2008. Spectral-resolved gene technology for multiplexed bioluminescence and high-content screening. Anal Chem 80:260–267. doi: 10.1021/ac7016579. [DOI] [PubMed] [Google Scholar]
- 8.Foucault ML, Thomas L, Goussard S, Branchini BR, Grillot-Courvalin C. 2010. In vivo bioluminescence imaging for the study of intestinal colonization by Escherichia coli in mice. Appl Environ Microbiol 76:264–274. doi: 10.1128/AEM.01686-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mezzanotte L, Que I, Kaijzel E, Branchini B, Roda A, Lowik C. 2011. Sensitive dual-color in vivo bioluminescence imaging using a new red codon optimized firefly luciferase and a green click beetle luciferase. PLoS One 6:e19277. doi: 10.1371/journal.pone.0019277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang M, Anttonen KP, Cirillo SL, Francis KP, Cirillo JD. 2014. Real-time bioluminescence imaging of mixed mycobacterial infections. PLoS One 9:e108341. doi: 10.1371/journal.pone.0108341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman CM, Gibson GR, Rowland I. 2011. Health benefits of probiotics: are mixtures more effective than single strains? Eur J Nutr 50:1–17. doi: 10.1007/s00394-010-0166-z. [DOI] [PubMed] [Google Scholar]
- 12.Wells JM, Wilson PW, Le Page RW. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol 74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 13.Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl Environ Microbiol 66:4427–4432. doi: 10.1128/AEM.66.10.4427-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grangette C, Muller-Alouf H, Geoffroy M, Goudercourt D, Turneer M, Mercenier A. 2002. Protection against tetanus toxin after intragastric administration of two recombinant lactic acid bacteria: impact of strain viability and in vivo persistence. Vaccine 20:3304–3309. doi: 10.1016/S0264-410X(02)00301-8. [DOI] [PubMed] [Google Scholar]
- 15.Klingberg TD, Budde BB. 2006. The survival and persistence in the human gastrointestinal tract of five potential probiotic lactobacilli consumed as freeze-dried cultures or as probiotic sausage. Int J Food Microbiol 109:157–159. doi: 10.1016/j.ijfoodmicro.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 16.Saxelin M, Lassig A, Karjalainen H, Tynkkynen S, Surakka A, Vapaatalo H, Jarvenpaa S, Korpela R, Mutanen M, Hatakka K. 2010. Persistence of probiotic strains in the gastrointestinal tract when administered as capsules, yoghurt, or cheese. Int J Food Microbiol 144:293–300. doi: 10.1016/j.ijfoodmicro.2010.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Hutt P, Koll P, Stsepetova J, Alvarez B, Mandar R, Krogh-Andersen K, Marcotte H, Hammarstrom L, Mikelsaar M. 2011. Safety and persistence of orally administered human Lactobacillus sp. strains in healthy adults. Benef Microbes 2:79–90. doi: 10.3920/BM2010.0023. [DOI] [PubMed] [Google Scholar]
- 18.Pittet MJ, Weissleder R. 2011. Intravital imaging. Cell 147:983–991. doi: 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mezzanotte L, An N, Mol IM, Lowik CW, Kaijzel EL. 2014. A new multicolor bioluminescence imaging platform to investigate NF-κB activity and apoptosis in human breast cancer cells. PLoS One 9:e85550. doi: 10.1371/journal.pone.0085550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezzanotte L, Blankevoort V, Lowik CW, Kaijzel EL. 2014. A novel luciferase fusion protein for highly sensitive optical imaging: from single-cell analysis to in vivo whole-body bioluminescence imaging. Anal Bioanal Chem 406:5727–5734. doi: 10.1007/s00216-014-7917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito K, Chang YF, Horikawa K, Hatsugai N, Higuchi Y, Hashida M, Yoshida Y, Matsuda T, Arai Y, Nagai T. 2012. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun 3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewandrowski GK, Magee CN, Mounayar M, Tannous BA, Azzi J. 2014. Simultaneous in vivo monitoring of regulatory and effector T lymphocytes using secreted Gaussia luciferase, firefly luciferase, and secreted alkaline phosphatase. Methods Mol Biol 1098:211–227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.