Abstract
Carbohydrate availability shifts when bacteria attach to a surface and form biofilm. When salivary planktonic bacteria form an oral biofilm, a variety of polysaccharides and glycoproteins are the primary carbon sources; however, simple sugar availabilities are limited due to low diffusion from saliva to biofilm. We hypothesized that bacterial glycoside hydrolase (GH) activities would be higher in a biofilm than in saliva in order to maintain metabolism in a low-sugar, high-glycoprotein environment. Salivary bacteria from 13 healthy individuals were used to grow in vitro biofilm using two separate media, one with sucrose and the other limiting carbon sources to a complex carbohydrate. All six GHs measured were higher in vitro when grown in the medium with complex carbohydrate as the sole carbon source. We then collected saliva and overnight dental plaque samples from the same individuals and measured ex vivo activities for the same six enzymes to determine how oral microbial utilization of glycoconjugates shifts between the planktonic phase in saliva and the biofilm phase in overnight dental plaque. Overall higher GH activities were observed in plaque samples, in agreement with in vitro observation. A similar pattern was observed in GH activity profiles between in vitro and ex vivo data. 16S rRNA gene analysis showed that plaque samples had a higher abundance of microorganisms with larger number of GH gene sequences. These results suggest differences in sugar catabolism between the oral bacteria located in the biofilm and those in saliva.
INTRODUCTION
The oral cavity is a dynamic environment. For instance, both salivary flow and the level of exogenous dietary carbon sources increase during ingestion of food, whereas both are at a minimum during sleep. Therefore, the oral microbiota may be influenced by alterations in the host environment, especially by changes in nutrient supply, local pH, and redox potential (1). One notable example is that the microbial community becomes more acidogenic in the presence of sugar (2). Endogenous salivary glycoconjugates become the most significant carbon source for oral microorganisms when an exogenous dietary carbon source is scarce. Under those circumstances, carbohydrate-active enzymes (CAZymes) play an important role in the adaptation of microorganisms (3–6). These CAZymes include glycosyltransferases and glycoside hydrolases (GHs). The type and availability of carbon sources also influence the development of biofilm at stages throughout the biofilm formation as microorganisms utilize the available carbon sources as nutrients, as building blocks for extracellular polysaccharide production, and as the point of attachment (7). We speculated that the breakdown of complex, endogenous carbohydrates by GH enzymes is one mechanism whereby oral microbes adapt to different carbon sources between the salivary planktonic phase, where exogenous simple sugars are generally more available, and the oral biofilm phase, where, in the absence of exogenous simple sugars, several alternative, more complex endogenous salivary glycoconjugates are the principal carbon source (8–11).
Our hypotheses for the present study were that (i) GH activities are higher under conditions where the primary carbon sources are limited to complex glycan conjugates and (ii) levels of GH enzymes are higher in dental plaque than in saliva due to the shift in microbial population between saliva and plaque.
Here we report the GH levels under conditions where carbon source availabilities include simple sugars (saliva) and those where they are limited to complex glycoconjugates (dental plaque). Our first experiment was to test if carbohydrate availability influences GH activities in in vitro oral biofilm, followed by ex vivo experiments in which GH activities were measured in saliva and overnight dental plaque. In both in vitro and ex vivo experiments, the samples were separated into cell-free supernatant and cellular pellets by centrifuge in order to monitor the enzyme activities that are secreted and those on cell surfaces.
MATERIALS AND METHODS
In vitro biofilm growth.
Gum base-stimulated saliva samples were collected from 13 healthy volunteers (7 females and 6 males, 24 to 45 years of age) at 9 a.m. Subjects were in good general health based on medical history and not being treated for medical or dental conditions or taking antibiotics or other medications which might influence salivary secretion. Subjects refrained from eating, drinking, and oral hygiene for at least 2 h prior to the collection. Samples were collected on ice and immediately treated with phenylmethanesulfonylfluoride (PMSF; final concentration, 1 mM; Sigma-Aldrich, St. Louis, MO). Samples were then centrifuged at 12,000 × g for 30 min at 4°C. The pellet was suspended in phosphate-buffered saline (pH 7.4) to a final optical density at 600 nm (OD600) of 0.600. The pellet suspension was divided and inoculated for biofilm growth into two different media in parallel. Hydroxyapatite disks (Clarkson Chromatography Products, South Williamsport, PA) were coated for 1 h at room temperature in 24-well plates (BD Bioscience, Franklin Lakes, NJ) with pooled supernatant to form salivary pellicles. Pellet suspensions from each individual were then inoculated separately onto disks and grown for 72 h at 37°C in either peptone-yeast base medium containing 5% sucrose (sucrose medium) or 1.25% hog gastric mucin in phosphate-buffered saline (mucin medium) (12). The sucrose medium has been shown to be capable of growing a more diversified community with a microbial profile closer to that of the salivary microbiota when inoculated with pooled human saliva. On the other hand, mucin medium would represent the environment where the carbon source is limited to complex glycoconjugates. The media were refreshed every 24 h. The spent medium was centrifuged at 12,000 × g for 30 min at 4°C, after which the supernatant obtained was filter sterilized for assay of GH activity. After 72 h growth, the biofilm was collected and weighed. It was then centrifuged to remove residual medium and resuspended in 50 mM N-[tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid (TES) buffer, pH 7.4, to a final concentration of 5.0 mg pellet ml−1.
Ex vivo sample collection.
Saliva collection and processing from the same volunteers were done as described above, up to the point of centrifugation. The supernatant and the pellet were weighed, followed by resuspension of the pellet in 50 mM TES buffer to a final concentration of 5.0 mg pellet ml−1. Both supernatant and pellet suspension were subjected for further analyses.
Overnight-grown plaque samples were collected from the same 13 volunteers on a separate day after the volunteers had abstained from normal oral hygiene and consumption of food or drinks for the previous 12 h. A Hayman-style microspatula (Thermo Fisher Scientific Inc., Waltham, MA) was used to scrape the tooth surface to collect supragingival and marginal gingival plaque samples from buccal surfaces of the maxillary and mandibular anterior 10 teeth, i.e., second premolar to second premolar. The collected plaque was weighed immediately and centrifuged at 12,000 × g for 30 min at 4°C. The pellet and plaque fluid were then separated, weighed, and treated as described above for saliva samples.
Total protein analysis and turbidity of plaque suspension.
Total protein was measured using Bradford reagent (Bio-Rad, Hercules, CA). Turbidity was measured at 600 nm.
GH activities.
GH activities were measured separately for each individual using black 96-well microtiter plates (BD Bioscience). Following 1 h incubation at 37°C, sample reactants (pellets and supernatants from in vitro biofilm, saliva, and plaque in TES buffer) were sonicated, vortexed, and diluted 10-fold with TES buffer. This dilution factor was determined based on preliminary experiment to determine the zero-order region. Aliquots (50 μl) of diluted reactants were added to 20 μl of 4-methylumbelliferone (4MU) substrate solution in 100 mM citric acid-Na2HPO4 buffer (pH 5.4) to a final concentration of 4 μM. Each well contained one of the following 4MU-linked glycosides as the substrate: 4MU-α-d-NAc-neuraminic acid, 4MU-α-l-fucoside, 4MU-β-d-glucoside, 4MU-β-d-galactoside, 4MU-NAc-β-d-glucosaminide, or 4MU-NAc-β-d-galactosaminide (Sigma-Aldrich). These substrates were selected based on GH previously detected in oral specimens (13). Plates were then incubated for 1 h at 37°C before the reaction was quenched with 130 μl of 500 mM Na2CO3-NaHCO3 buffer (pH 10.3). Fluorescence was measured using a Victor X3 multilabel reader (excitation/emission wavelengths = 355/460 nm) (PerkinElmer, Waltham, MA). Two negative controls were used, namely, 20 μl citric acid Na2HPO4 buffer (4MU substrate blank) and 50 μl TES buffer (reactant blank). The positive control contained 20 μl of authentic β-glucosidase solution from almonds (Sigma-Aldrich). Known concentrations of 4MU solutions were used to plot a standard curve. The detection limit was 10 nM (signal-to-noise ratio > 3), and the range of linearity was 62.5 nM to 4 μM. All assays were run in triplicate, and data were expressed in terms of unit activity per mg of protein after adjustment of the dilution factors (1 unit was defined as the release of 1 μmol 4MU per min).
qPCR analysis.
The following species were tested: Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, Streptococcus gordonii, Streptococcus mutans, Streptococcus salivarius, and Treponema denticola. 16S rRNA genes were extracted from the cellular pellets using QIAcube (Qiagen Inc., Valencia, CA) and analyzed by quantitative PCR (qPCR) analyses using a CFX-96 real-time system coupled with a C1000 thermal cycler (Bio-Rad) in accordance to the manufacturers' instruction. See Table S2 in the supplemental material for the primer sequences.
Statistical analysis.
All statistical analyses were performed using Microsoft Excel. The statistical significance for GH activities was estimated for each enzyme at a 95% confidence interval using Welch's t test for in vitro results and 2-way analysis of variance (ANOVA) for ex vivo results. The statistical significance for qPCR results was estimated by paired t test at a 95% confidence interval.
RESULTS
In vitro biofilm GH activities.
Overall, higher GH activities were found in spent mucin medium supernatant, except for glucosidase at the 24- and 48-h time points and fucosidase at the 24-h time point (Fig. 1 and Table 1 ). These two enzymes showed continuous increases in their activities for 72 h, whereas the other four GH activities peaked at 48 h. At 72 h, fucosidase, sialidase, and glucosaminidase were found to have higher activities in supernatant than in the cell-bound form (Table 1). Glucosaminidase activity was found to be the highest for both secreted and cell-bound forms among the 6 measured GH, followed by galactosaminidase activity.
FIG 1.
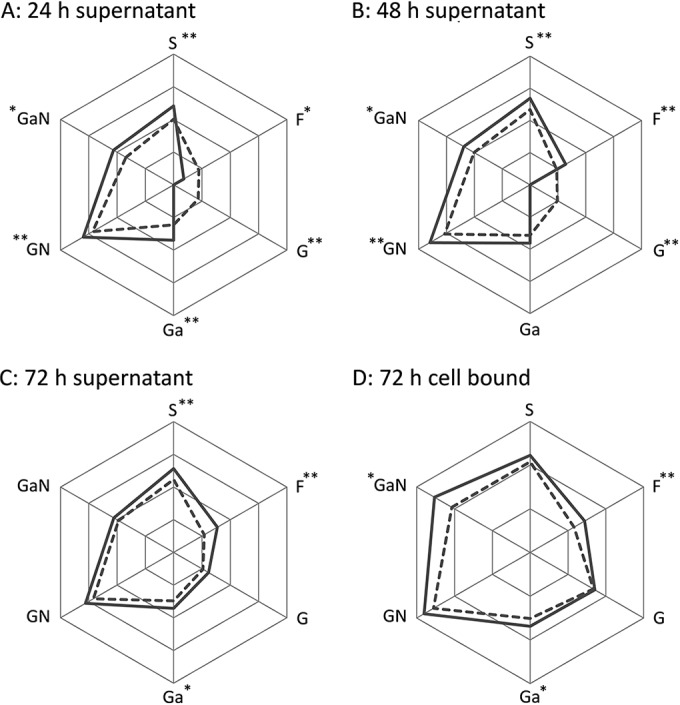
Relative in vitro biofilm GH activities. Overall activities were higher in mucin medium (solid lines) than in sucrose medium (dotted lines) with the exceptions of α-l-fucosidase (F) in 24-h supernatant samples and β-d-glucosidase (G) in 24- and 48-h supernatant samples. The other enzymes are abbreviated as follows: S, β-d-NAc-sialidase; Ga, β-d-galactosidase; GN, β-d-NAc-glucosaminidase; GaN, β-d-NAc-galactosaminidase. *, P < 0.05; **, P < 0.01. The scales are not equal for all enzymes. See Table 1 for the values of enzymatic activities.
TABLE 1.
in vitro GH activities
| Sample and GH | Activity ina: |
Significance (t test) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Mucin medium |
Sucrose medium |
||||||||
| Avg | SD | Min | Max | Avg | SD | Min | Max | ||
| 24-h supernatant | |||||||||
| β-d-N-Ac-sialidase | 258.69 | 58.4 | 192.24 | 365.49 | 101.61 | 19.1 | 66.65 | 123.23 | <0.01 |
| α-l-Fucosidase | 2.29 | 6.5 | ND | 18.32 | 7.74 | 0.8 | 6.53 | 9.03 | <0.05 |
| β-d-Glucosidase | ND | ND | ND | 7.38 | 0.7 | 0.68 | 8.36 | <0.01 | |
| β-d-Galactosidase | 49.62 | 13.8 | 25.62 | 72.19 | 16.70 | 5.1 | 10.93 | 24.79 | <0.01 |
| β-d-N-Ac-glucosaminidase | 1,584.16 | 767.8 | 466.65 | 2,492.35 | 730.74 | 227.5 | 514.25 | 1,139.14 | <0.01 |
| β-d-N-Ac-Galactosaminidase | 133.38 | 96.1 | 51.85 | 330.91 | 48.34 | 22.6 | 28.83 | 93.14 | <0.05 |
| 48-h supernatant | |||||||||
| β-d-N-Ac-sialidase | 489.62 | 127.9 | 264.65 | 604.44 | 218.31 | 61.2 | 129.15 | 314.91 | <0.01 |
| α-l-Fucosidase | 19.15 | 3.7 | 16.35 | 27.10 | 9.05 | 1.4 | 7.08 | 10.79 | <0.01 |
| β-d-Glucosidase | ND | ND | ND | 9.65 | 2.7 | 2.68 | 14.44 | <0.01 | |
| β-d-Galactosidase | 65.14 | 39.1 | 30.42 | 158.66 | 37.09 | 14.3 | 17.79 | 61.65 | 0.078 |
| β-d-N-Ac-glucosaminidase | 3,931.97 | 1,452.5 | 1,574.50 | 5,778.85 | 1,163.54 | 519.7 | 511.46 | 1,904.92 | <0.01 |
| β-d-N-Ac-galactosaminidase | 237.98 | 143.8 | 79.73 | 500.34 | 102.07 | 24.4 | 73.99 | 131.97 | <0.05 |
| 72-h supernatant | |||||||||
| β-d-N-Ac-sialidase | 367.19 | 122.6 | 194.74 | 555.71 | 169.99 | 51.2 | 105.56 | 268.95 | <0.01 |
| α-l-Fucosidase | 35.06 | 3.6 | 33.07 | 43.75 | 12.05 | 3.2 | 8.48 | 18.92 | <0.01 |
| β-d-Glucosidase | 17.12 | 18.4 | ND | 37.49 | 10.75 | 1.1 | 1.10 | 12.06 | 0.344 |
| β-d-Galactosidase | 51.15 | 15.7 | 35.74 | 86.84 | 30.37 | 15.0 | 16.45 | 64.11 | <0.05 |
| β-d-N-Ac-glucosaminidase | 1,307.07 | 838.1 | 339.33 | 2,621.39 | 689.02 | 221.5 | 511.46 | 1,904.92 | 0.063 |
| β-d-N-Ac-galactosaminidase | 130.69 | 79.1 | 44.08 | 288.11 | 88.39 | 22.5 | 50.04 | 114.82 | 0.168 |
| 72-h, cell bound | |||||||||
| β-d-N-Ac-sialidase | 167.82 | 76.9 | 73.52 | 277.80 | 118.39 | 23.0 | 92.06 | 153.00 | 0.103 |
| α-l-Fucosidase | 27.44 | 6.1 | 15.68 | 36.64 | 14.41 | 3.9 | 7.59 | 18.96 | <0.01 |
| β-d-Glucosidase | 50.98 | 10.7 | 10.70 | 58.82 | 46.62 | 21.1 | 21.13 | 86.58 | 0.611 |
| β-d-Galactosidase | 48.87 | 16.6 | 33.10 | 87.03 | 32.39 | 9.8 | 22.07 | 49.02 | <0.05 |
| β-d-N-Ac-glucosaminidase | 636.66 | 442.6 | 511.46 | 1,904.92 | 358.94 | 81.8 | 188.56 | 462.48 | 0.103 |
| β-d-N-Ac-galactosaminidase | 338.31 | 281.2 | 99.91 | 987.08 | 117.83 | 17.9 | 85.86 | 141.46 | <0.05 |
Expressed in units; 1 unit is defined as the release of 1 μmol 4MU per mg protein per minute. ND, not detected.
Salivary GH activities.
Saliva samples, 4.0 ml from each donor, were centrifuged to give cellular and cell-free supernatant. The average total amounts of cellular pellet and supernatant were 141.3 mg and 3,815.1 mg, respectively. The average total protein concentrations were 713.58 μg/ml for the supernatant and 215.40 μg/ml for cellular pellet resuspension, respectively. The average turbidities (OD600) were 0.012 for supernatant and 1.027 for cellular pellet. The removal of cellular material from supernatant was ensured by filter sterilization. Both cellular suspension and cell-free supernatant were assayed for GH activities (Fig. 2).
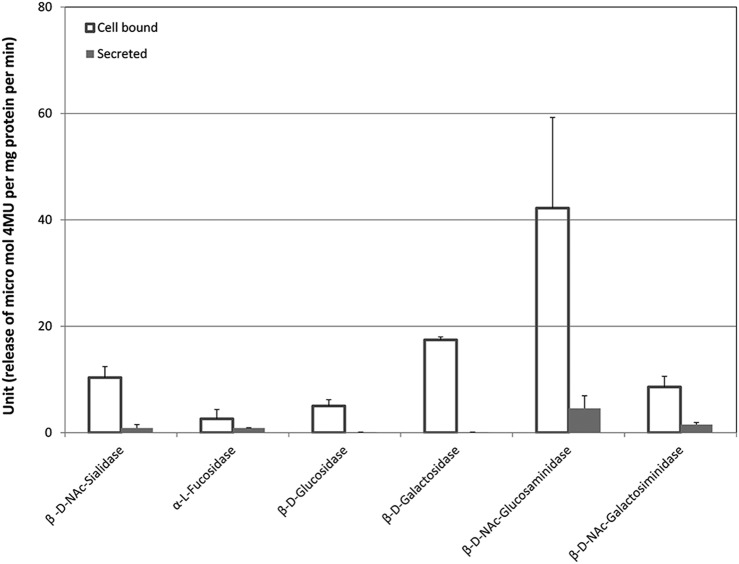
FIG 2.
Salivary GH activities. Open bars represent the activities in cell-bound enzymes, whereas the filled bars represent those in cell-free supernatant. The highest activity was found for cell-bound glucosaminidase. Fucosidase was found to have the overall lowest activity. No secreted glucosidase or galactosidase activities were detected in saliva supernatant. The values are the averages of triplicate measurements from 13 individuals.
The activity of β-glucosaminidase was found to be the highest in both pellet (cell-bound) and supernatant (secreted) in the saliva sample. Secreted fucosidase, hexosaminidase, and sialidase were found in supernatant, while no activity was detected for galactosidase or glucosidase in supernatant. Secreted fucosidase activity level was equivalent with that of cell-bound form.
Overnight plaque GH activities.
Overnight-grown biofilm samples were collected from the same donors and separated into cellular pellet and cell-free supernatant by centrifugation immediately after collection. The total amounts of cellular pellet and supernatant were 70.66 mg and 30.99 mg, respectively. The total protein concentrations were 365.40 μg/ml for the supernatant and 278.13 μg/ml for cellular pellet resuspension. The turbidity values (OD600) were 0.018 for supernatant and 1.078 for cellular pellet. The removal of cellular material from supernatant was ensured by filter sterilization. Both cellular suspension and cell-free supernatant were assayed for glycosidase activities (Fig. 3).
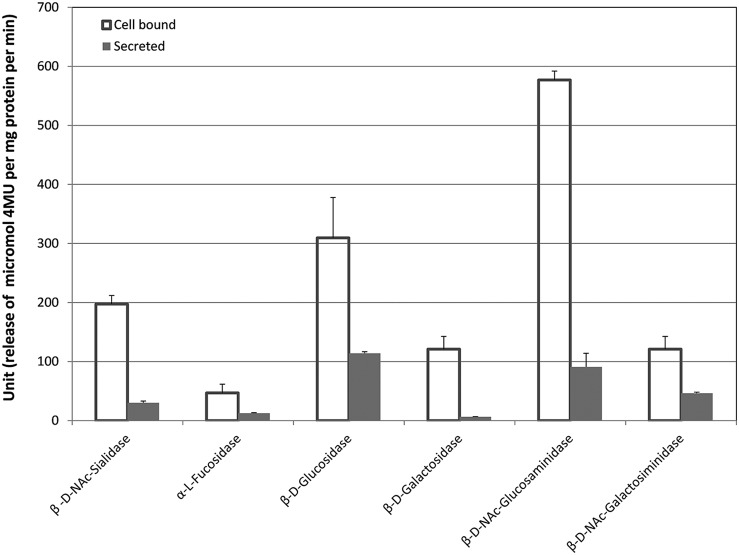
FIG 3.
Biofilm GH activities. Open bars represent the activities in cell-bound enzymes, whereas the filled bars represent those in supernatant. Unlike in saliva samples, glucosidase and galactosidase activities were found in supernatant. Glucosidase showed the highest activity in supernatant. The ratios of activities between secreted and cell-bound forms were higher than in saliva samples, except for fucosidase. The values are the averages of triplicate measurements from 13 individuals.
The overall enzymatic activities for all six enzymes were higher in overnight plaque by a factor of 10 to 50 than those in saliva. All six enzymes were present in both cell-bound and secreted forms, including hexosidases, the secreted form of which was absent in saliva. The ratios of the fucosidase activity of the secreted form to the cell-bound form were lower in overnight plaque than in saliva. In contrast to saliva samples, the secreted forms were observed for galactosidase and glucosidase in overnight plaque. Indeed, the highest activity observed in the secreted form was that of glucosidase, which was more than a third of the activity of its cell-bound counterpart.
Two-way ANOVA showed that the mean activities were statistically different at an α value of 0.05 across four ex vivo sample groups for all enzymes.
qPCR analysis.
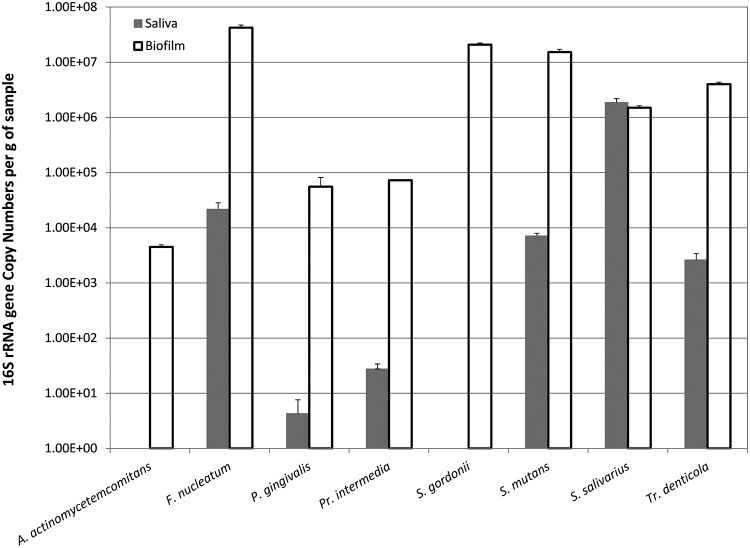
All species showed a 1- to 4-log increase in overnight plaque compared to saliva, except for S. salivarius, which showed slight decrease (Fig. 4). The total numbers of 16S gene copies were 1.90 × 106 for saliva and 8.41 × 107 for plaque, respectively. In saliva samples, the two most abundant species detected, S. salivarius and F. nucleatum, accounted for 98.3 and 1.1% of the 8 measured species, respectively. In plaque samples, the three most abundant species, namely, F. nucleatum, S. gordonii, and S. mutans, accounted for 50.3, 24.8, and 18.2%, respectively. S. gordonii showed the highest increase in numbers, to 105 copies in biofilm. A paired t test showed that the number of DNA copies for all species are significantly different between saliva and biofilm except for S. salivarius.
FIG 4.
qPCR analysis. Solid bars show the number of DNA copies found in the saliva samples, whereas open bars show those in the biofilm samples. There were no measureable copies of DNA detected for A. actinomycetemcomitans and S. gordonii in saliva samples. All measured species showed statistical difference between samples, except for S. salivarius.
DISCUSSION
In vitro biofilm GH activity levels.
Our results supported the first hypothesis, that higher activities of GH were observed in biofilm grown in mucin medium, where the only carbon source available was complex carbohydrate. This is in agreement with previous reports in which single-species streptococci showed upregulation of the GH when fermented in the absence of simple carbon sources (4, 5). The activities of four out of six enzymes peaked at 48 h, in agreement with a previous study where GH of S. gordonii culture were downregulated during exponential growth phase compared to the stationary phase (4). This would suggest that at least some species of oral bacteria are able to shift from a preferred metabolite (glucose) to a more complex carbohydrate (mucin) when conditions require it.
Ex vivo GH activity levels in overnight plaque and saliva.
The results agreed with the second hypothesis, in which overall higher activities of GH occurred in biofilm samples than in saliva samples. While the sample size is small in this study, these results are in line with what has been reported previously for microbiota in saliva and dental plaque: the dental plaque environment favors organisms that are able to metabolize complex organic molecules (14, 15).
Bacteria with the largest numbers of GH genes are found to be most prevalent in dental plaque, and this appears to correlate with higher GH activity. Among the microorganisms measured, S. gordonii and P. gingivalis harbor the largest number of GH coding sequences as well as the most types of GH enzymes. These organisms, along with P. intermedia and T. denticola, have genes for β-N-acetyl-hexosaminidase (GH20) and α-fucosidase (GH29). On the other hand, the most abundant GHs in S. salivarius, which accounted for the majority of salivary species, are α-amylase, lysozyme, and sucrase (see Table S1 in the supplemental material). Our results cannot determine what percentage of the cell-bound GH activities in saliva was derived from bacteria; however, the enzymes in host cells are expressed at low levels and are associated primarily with intracellular metabolism, and hence they are not excreted to a significant extent.
GH activity profiles between secreted and cell-bound forms.
Beside overall enzyme activity levels, the results also showed that secretion patterns of GHs differ under the conditions where carbon source is limited to complex carbohydrates. Our results are in line with previous reports in that (i) all six GHs were present in saliva of healthy adults (16, 17) and (ii) β-d-galactosidase activity was absent in salivary supernatant, while activities of β-d-hexosaminidase and sialidase were present (18). The cell-bound form of glucosaminidase activities was consistently the highest among the measured GH in our samples, both in vitro and ex vivo. β-N-Ac-glucosaminidase and glucosidase have been reported as the predominant GHs in healthy human dental plaque (13). The presence of both cell-bound and secreted forms of sialidase is also in agreement with previous reports (19, 20).
Both fucosidase and sialidase were present in the secreted form in saliva. Although the secreted form of sialidase has been reported, this is the first report on secreted fucosidase to the best of our knowledge. The presence of secreted forms of GH, especially fucosidase and sialidase, can be explained by the modification of terminal glycoside moiety to provide a better scaffold for bacterial adhesion (21, 22). The complete lack of secreted glucosidase and galactosidase activities in saliva suggests that alternative hexose sugar production in response to external glycosides is not significantly induced. Our in vitro experiment showed the onset of glucosidase activity in mucin medium occurred at a later time point than that of the other 5 GH activities. The presence of secreted hexosidases in biofilm is initially counterintuitive, because the sugars produced extracellularly are considered the preferred carbon source for microorganisms and are available for other species in the biofilm. However, since the trigger for production of GH is the energy requirements of the bacteria producing the enzyme, a more global benefit to the biofilm may be an example of kin discrimination (23). Because there is limited diffusion in biofilm, the beneficiaries of such a secreted enzyme could be limited to those in close proximity to the producer, which are more likely to be genetically related to each other (24). It is noteworthy that the secreted form of glucosidase was detected in biofilm grown in nutrient-rich sucrose medium at an earlier time point than that grown in mucin medium in our in vitro experiment. Future studies focusing on how secreted GHs are induced and distributed in biofilm may provide a clue to how oral ecology maintains homeostasis despite constant perturbations in healthy individuals.
Our results showed a reactive shift of glycoside hydrolase activities in microbial communities between planktonic and sessile phases in healthy adults. While further studies are necessary to confirm our findings with a larger number of subjects, the marked increase in activity of several GHs in plaque compared to saliva may serve as a potential biomarker for phenotypic changes associated with dental plaque formation.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xiaoqun Mo, Panagiota Tsatsos, and Lisa Tran at Wm. Wrigley Jr. Co. for technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01180-15.
REFERENCES
- 1.Marsh PD, Devine DA. 2011. How is the development of dental biofilms influenced by the host? J Clin Periodontol 38(Suppl 11):S28–S35. doi: 10.1111/j.1600-051X.2010.01673.x. [DOI] [PubMed] [Google Scholar]
- 2.Marsh PD. 2006. Dental diseases—are these examples of ecological catastrophes? Int J Dent Hyg 4:3–10. doi: 10.1111/j.1601-5037.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Burne RA. 2001. Regulation of the gtfBC and ftf genes of Streptococcus mutans in biofilms in response to pH and carbohydrate. Microbiology 147:2841–2848. [DOI] [PubMed] [Google Scholar]
- 4.Harty DWS, Mayo JA, Cook SL, Jacques NA. 2000. Environmental regulation of glycosidase and peptidase production by Streptococcus gordonii FSS2. Microbiology 146:1923–1931. [DOI] [PubMed] [Google Scholar]
- 5.Homer KA, Whiley RA, Beighton D. 1994. Production of specific glycosidase activities by Streptococcus intermedius strain UNS35 grown in the presence of mucin. J Med Microbiol 41:184–190. doi: 10.1099/00222615-41-3-184. [DOI] [PubMed] [Google Scholar]
- 6.Bradshaw DJ, Homer KA, Marsh PD, Beighton D. 1994. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 140:3407–3412. doi: 10.1099/13500872-140-12-3407. [DOI] [PubMed] [Google Scholar]
- 7.Bowden GH, Li YH. 1997. Nutritional influences on biofilm development. Adv Dent Res 11:81–99. doi: 10.1177/08959374970110012101. [DOI] [PubMed] [Google Scholar]
- 8.Renzi F, Manfredi P, Dol M, Fu J, Vincent S, Cornelis GR. 2015. Glycan-foraging systems reveal the adaptation of Capnocytophaga canimorsus to the dog mouth. mBio 6:e02507-14. doi: 10.1128/mBio.02507-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoodley P, Wefel J, Gieseke A, Debeer D, von Ohle C. 2008. Biofilm plaque and hydrodynamic effects on mass transfer, fluoride delivery and caries. J Am Dent Assoc 139:1182–1190. doi: 10.14219/jada.archive.2008.0333. [DOI] [PubMed] [Google Scholar]
- 10.McLean JS, Ona ON, Majors PD. 2008. Correlated biofilm imaging, transport and metabolism measurements via combined nuclear magnetic resonance and confocal microscopy. ISME J 2:121–131. doi: 10.1038/ismej.2007.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyenal H, Lewandowski Z. 2002. Internal and external mass transfer in biofilms grown at various flow velocities. Biotechnol Prog 18:55–61. doi: 10.1021/bp010129s. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y, He X, Torralba M, Yooseph S, Nelson KE, Lux R, McLean JS, Yu G, Shi W. 2010. Using DGGE profiling to develop a novel culture medium suitable for oral microbial communities. Mol Oral Microbiol 25:357–367. doi: 10.1111/j.2041-1014.2010.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickström C, Svensäter G. 2008. Salivary gel-forming mucin MUC5B—a nutrient for dental plaque bacteria. Oral Microbiol Immunol 23:177–182. doi: 10.1111/j.1399-302X.2007.00407.x. [DOI] [PubMed] [Google Scholar]
- 14.Keijser BJF, Zaura E, Huse SM, van der Vossen JMBM, Schuren FHJ, Montijn RC, ten Cate JM, Crielaard W. 2008. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res 87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 15.Cantarel BL, Lombard V, Henrissat B. 2012. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 7:e28742. doi: 10.1371/journal.pone.0028742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knas M, Choromanska M, Karaszewska K, Dudzik D, Waszkiel D, Borzym-Kluczyk M, Zaniewska A, Zwierz K. 2007. Activity of lysosomal exoglycosidases in saliva of patients with HIV infection. Adv Med Sci 52:186–190. [PubMed] [Google Scholar]
- 17.Waszkiewicz N, Szajda SD, Jankowska A, Waszkiewicz M, Kepka A, Konarzewska B, Szulc A, Snarska J, Zwierz K. 2009. Catabolism of salivary glycoconjugates in acute ethanol intoxication. Med Sci Monit 15:CR413–CR417. [PubMed] [Google Scholar]
- 18.Quinn MO, Miller VE, Dal Nogare AR. 1994. Increased salivary exoglycosidase activity during critical illness. Am J Respir Crit Care Med 150:179–183. doi: 10.1164/ajrccm.150.1.8025747. [DOI] [PubMed] [Google Scholar]
- 19.Corfield T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2:509–521. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- 20.Rogers R, Newbrun E, Tatevossian A. 1979. Neuraminidase activity in human dental plaque fluid. Arch Oral Biol 24:703–705. doi: 10.1016/0003-9969(79)90121-3. [DOI] [PubMed] [Google Scholar]
- 21.Gibbons RJ, Etherden I. 1982. Enzymatic modification of bacterial receptors on saliva-treated hydroxyapatite surfaces. Infect Immun 36:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruhl S, Sandberg AL, Cisar JO. 2004. Salivary receptors for the proline-rich protein-binding and lectin-like adhesins of oral actinomyces and streptococci. J Dent Res 83:505–510. doi: 10.1177/154405910408300614. [DOI] [PubMed] [Google Scholar]
- 23.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat Rev Microbiol 4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 24.Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R, M. Harmsen HJ. 2010. Oral biofilm architecture on natural teeth. PLoS One 5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.