Abstract
Anaerobic oxidation of methane (AOM) coupled to nitrite reduction is a novel AOM process that is mediated by denitrifying methanotrophs. To date, enrichments of these denitrifying methanotrophs have been confined to freshwater systems; however, the recent findings of 16S rRNA and pmoA gene sequences in marine sediments suggest a possible occurrence of AOM coupled to nitrite reduction in marine systems. In this research, a marine denitrifying methanotrophic culture was obtained after 20 months of enrichment. Activity testing and quantitative PCR (qPCR) analysis were then conducted and showed that the methane oxidation activity and the number of NC10 bacteria increased correlatively during the enrichment period. 16S rRNA gene sequencing indicated that only bacteria in group A of the NC10 phylum were enriched and responsible for the resulting methane oxidation activity, although a diverse community of NC10 bacteria was harbored in the inoculum. Fluorescence in situ hybridization showed that NC10 bacteria were dominant in the enrichment culture after 20 months. The effect of salinity on the marine denitrifying methanotrophic culture was investigated, and the apparent optimal salinity was 20.5‰, which suggested that halophilic bacterial AOM coupled to nitrite reduction was obtained. Moreover, the apparent substrate affinity coefficients of the halophilic denitrifying methanotrophs were determined to be 9.8 ± 2.2 μM for methane and 8.7 ± 1.5 μM for nitrite.
INTRODUCTION
Anaerobic oxidation of methane (AOM) occurs extensively in natural ecosystems and is a crucial biological sink in the global methane cycle that maintains the balance of greenhouse gas content in the atmosphere (1). To date, five electron acceptors that support AOM, including AOM coupled to sulfate reduction (2), nitrite reduction (3), nitrate reduction (4), iron reduction (5), and manganese reduction (5), have been discovered in natural settings. Moreover, on the basis of bioenergetic calculations, researchers have speculated that several other types of AOM (e.g., AOM coupled to perchlorate reduction, arsenate reduction, and selenate reduction) may exist in nature (6); however, these possible types have not yet been confirmed. In particular, AOM coupled to nitrite reduction has been a predominant research focus in the past several years. AOM coupled to nitrite reduction was also called nitrite-dependent anaerobic methane oxidation (n-damo) in previous reports. It has been demonstrated that AOM coupled to nitrite reduction is mediated by the bacterium “Candidatus Methylomirabilis oxyfera” (denitrifying methanotroph) (7), which is affiliated with the candidate NC10 phylum (8). This candidate division (NC10 phylum) was first defined by classification of environmental sequences retrieved from aquatic microbial formations in flooded caves (9). To date, our knowledge regarding the NC10 phylum has stemmed largely from research into “Ca. Methylomirabilis oxyfera” and “Ca. Methylomirabilis oxyfera”-like bacteria. According to the phylogenetic affiliations of 16S rRNA gene sequences, NC10 bacteria have been previously classified into the following four groups: A, B, C, and D (8). However, only group A members were found to perform AOM coupled to nitrite reduction, and the metabolic capacities of organisms in the other groups remain unknown (8, 10). Consequently, more studies are needed to understand and utilize NC10 phylum bacteria.
In the ocean, AOM coupled to sulfate reduction (equation 1) has been widely studied, and a high abundance of sulfate in the oceanic system makes this process common (1). The process of AOM coupled to sulfate reduction releases only minimal energy (16 kJ mol−1 CH4 under standard conditions) to feed the microorganisms; however, AOM coupled to nitrite reduction (equation 2) is more energetically favorable, because this process releases 928 kJ energy per mole of methane oxidized, which is 57-fold higher than the level released by sulfate reduction. According to the energy analysis presented above, AOM coupled to nitrite reduction is more prone to occur when nitrite is available. Indeed, AOM coupled to nitrite reduction has been observed in several freshwater wetlands (11–13), and both 16S rRNA genes of members of the NC10 phylum and genes encoding particulate methane monooxygenase (pMMO) subunit A (pmoA) of “Ca. Methylomirabilis oxyfera”-like bacteria have been detected in many freshwater systems (11–18).
| (1) |
| (2) |
Recently, NC10 phylum 16S rRNA genes and pmoA genes have also been detected in marine systems (19–22) and saline lakes (23). Moreover, salinity adaption (at 20 g NaCl liter−1) of denitrifying methanotrophs has been observed in a freshwater culture (24). These findings showed that AOM coupled to nitrite reduction may occur in marine systems and that NC10 bacteria may mediate this process, which indicated the potential importance of NC10 bacterium-driven methanotrophy in marine systems. However, no marine denitrifying methanotrophic culture has been procured, and we have virtually no knowledge about these marine NC10 bacteria. Therefore, obtaining a marine denitrifying methanotrophic culture and studying its properties are vital to understanding these denitrifying methanotrophs in marine ecosystems and broadening our knowledge of NC10 phylum bacteria.
In our present work, coastal sediment was selected as an inoculum to enrich possible marine denitrifying methanotrophs in a sequencing batch reactor (SBR). Batch activity testing and quantitative PCR (qPCR) were conducted during the enrichment period. 16S rRNA and pmoA gene sequencing was applied to determine the organisms responsible for the process of AOM coupled to nitrite reduction in the culture, and fluorescence in situ hybridization (FISH) was employed to assess the enrichment of NC10 bacteria. Several basic properties of the obtained denitrifying methanotrophic culture, including its substrates and salinity effects, were also investigated.
MATERIALS AND METHODS
Inoculum and medium.
Coastal sediment was selected as an inoculum and sampled from a mudflat of Xiaogan Island (N29°57′42″, E122°13′50″; sampling depth, 10 to 20 mm) in May 2013. Xiaogan Island is near the East China Sea and was undeveloped at the sampling time.
Natural seawater with additive nitrite and bicarbonate was adopted as an enrichment medium from month 0 to month 16, and an artificial seawater medium was used from month 16 to month 20. Natural seawater was taken from the location where the coastal sediment had been sampled. Subsequently, the collected natural seawater was allowed to settle for 12 h, and then the supernatant was filtered (0.45 μm pore size) and stored at 4°C. Next, 0.1 g liter−1 NaHCO3 and 0.08 g liter−1 NaNO2 were added to the natural seawater medium before usage. The physicochemical characteristics of the natural seawater and the coastal sediment are presented in Table 1. The artificial seawater medium (salinity, ∼20.5‰) contained the following components (per kilogram): 16.12 g NaCl, 2.98 g MgCl2, 0.69 g CaCl2, 0.44 g KCl, 0.12 g NaHCO3, 0.05 g MgSO4, 0.05 g KH2PO4, 0.05 g NaBr, 0.5 ml acidic trace element solution, 0.2 ml alkaline trace element solution, and 0.08 to 0.98 g NaNO2 (as needed). The acidic and alkaline trace element solutions were prepared as previously described (8). In addition, the artificial seawater medium used for activity measurement was free of sulfate and contained the following (per kilogram): 16.12 g NaCl, 3.08 g MgCl2, 0.69 g CaCl2, 0.44 g KCl, 0.12 g NaHCO3, 0.05 g NaBr, and 0 to 0.02 g NaNO2 (as needed). The initial pH in the medium was kept at 7.00 ± 0.01.
TABLE 1.
The physicochemical characteristics of the inoculum (coastal sediment) and the natural seawater utilized in this work
| Sample | Salinity (‰) | SO4− (mM) | NO2− (μM) | NO3− (μM) | NH4+ (μM) |
|---|---|---|---|---|---|
| Natural seawater | 20.5 ± 0.3 | 9.6 ± 0.9 | 2.8 ± 1.4 | 92.4 ± 6.7 | 20.6 ± 1.9 |
| Coastal sedimenta | 21.9 ± 0.2 | 7.5 ± 2.2 | 11.4 ± 1.4 | 16.6 ± 3.7 | 69.1 ± 3.4 |
Concentrations refer to the values for pore water in the coastal sediment.
Enrichment protocol.
Microorganisms were enriched in an SBR (see Fig. S1 in the supplemental material) that consisted of 1.5 liters of inoculum, 0.5 liter of medium, and 0.5 liter of headspace. The reaction mixture in the reactor was mixed by a magnetic stirrer at 400 rpm, and the reaction was run at 25°C and 100 kPa of methane partial pressure. A gas bag (∼40 liter) was linked to the gas outlet of the SBR and was used to collect the exhaust gas.
Every 3 days, 0.4 liter of supernatant was replaced by the same volume of fresh medium after 6 h of biomass settling. During the medium exchange, the gas bag balances the gas pressure in the reactor and prevents air from entering. After the fresh medium was supplied, (99.999%) pure Ar was flushed into the reactor to eliminate the possible leaked oxygen for 10 min, and the Ar in the headspace was then replaced with (99.99%) pure methane. A 2-ml volume of the medium in the reactor was sampled and centrifuged (5 min, 7,440 × g) to measure the nitrite concentration before and after the medium exchange.
Activity measurement.
To monitor the changes in denitrifying methanotrophic activity, the methane oxidation and nitrite reduction rates were measured during the enrichment. Biomass (40 ml) was sampled from the SBR, rinsed with the nitrite-free artificial seawater medium five times, and then transferred at equal volumes into four 76-ml serum bottles. Two serum bottles were randomly selected as an experimental group, and the other two served as a control group. In each bottle, the medium of artificial seawater and the concentrated nitrite solution (100 mM) were added to reach the final volume of 50 ml and the final nitrite concentration of 0.5 mM. Then, the bottles were flushed with (99.999%) pure Ar for 10 min to obtain anoxic conditions, and 2.6 ml of (99.99%) pure methane was injected into only the headspace of the experimental group. The initial methane partial pressure in the experiment group was approximately 10 kPa, whereas that in the control group was 0 kPa (no addition of methane); all other conditions were identical. The serum bottles were incubated on a shaking table at 150 rpm and 25°C. After 2.0 h of preincubation, 0.5 ml of liquid was sampled to measure the nitrite and nitrate concentrations, and 20 μl of gas was extracted to analyze the methane concentration in the headspace in triplicate every 12 h. The nitrite reduction for denitrifying methanotrophs was estimated by calculating the nitrite reduction rate in the experiment group minus the rate in the control group, whereas the methane oxidation rate for denitrifying methanotrophs was directly calculated from the decrease in the methane concentration in the experimental group.
Tests of effects of substrates and salinity.
To assess the effects of nitrite, methane, and salinity on denitrifying methanotrophic activity, the final culture (after 20 months) was incubated at various nitrite concentrations (0 to 0.25 mM), methane partial pressures (0 to 20 kPa), and salinities (1 to 40‰) with an artificial seawater medium. The individual media and conditions were obtained by modifying the standard medium, and the standard medium was set at 0.2 mM nitrite, 10 kPa of methane, and 20.5‰ of salinity. The salinity gradient was established by proportionately mixing distilled water (0‰) and an artificial seawater concentrated solution (50‰). The artificial seawater concentrated solution was prepared by proportionally increasing the concentrations of components in the standard medium (20.5‰, described in the “Inoculum and medium” section above) to a salinity of 50‰. The effect tests were conducted in 76-ml serum bottles in triplicate as described in the “Activity measurement” section above. Under quasi-stable conditions, the methane concentration in the saline water was estimated by use of the Bunsen solubility coefficient (see the supplemental material for details).
Microbial community analysis.
During the enrichment, the biomass samples (0.25 g) were harvested on a regular basis, and DNA extraction and PCR amplification were performed as previously described (25). PCR products were evaluated by agarose gel (1.0%) electrophoresis and cloned by the use of pMD19-T vector (TaKaRa, Bio Inc., Shiga, Japan) as previously described (26). Clones with successful ligation were checked via a blue-white screening technique, and approximately 30 positive clones from each sample (inoculum and enrichment culture after 16 months) were randomly selected and sequenced by BGI-Shanghai (China).
A phylogenetic analysis of 16S rRNA and pmoA gene sequences was performed using Mega 6.0 (27). The retrieved 16S rRNA and pmoA gene sequences were aligned using the ClustalW algorithm, and phylogenetic trees were constructed using the neighbor-joining statistical method with p-distance as the substitution model. The bootstrap replicates were set at a value of 1,000.
The abundance of the NC10 phylum 16S rRNA genes was quantified by qPCR with primer pair qP1F/qP1R (8). qPCRs (in triplicate) and a standard curve analysis were performed using an iCycler iQ5 thermocycler and a real-time detection system (Bio-Rad, CA, Hercules, USA) as previously described by Ettwig et al. (8). The standard curve was constructed from a series of 10-fold dilutions (from 6.6 × 102 to 6.6 × 109 copies) of plasmid DNA inserted by the use of 16S rRNA genes of NC10 bacteria. The copy numbers of the samples were calculated from the threshold cycle values of samples based on the standard curve. Detailed information about the primers used in this work is provided in Table S1 in the supplemental material.
To obtain FISH micrographs, biomass samples were fixed and hybridized after 0, 9, 16, and 20 months of enrichment. The specific oligonucleotide probe, S-*-DBACT-1027-a-A-18 (red) (3), was used for NC10 phylum bacteria, and DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the total DNA. FISH micrographs were acquired by the use of a two-photon laser confocal microscope (Zeiss, Germany).
Chemical analysis.
Levels of nitrite, nitrate, and volatile suspended solids (VSS) were measured by standard methods (28), and methane was quantified by the use of an Agilent 6890 gas chromatograph (Agilent, USA) equipped with a flame ionization detector (FID) as previously described (24). The pH value was determined using a model FE20 pH meter (Mettler-Toledo, Shanghai, China). The experimental data were fitted and plotted in Origin 8.0 (OriginLab, USA).
Nucleotide sequence accession numbers.
The sequences determined in this work have been deposited in the GenBank database under accession numbers KM888188 to KM888243 (16S rRNA) and KM979290 to KM979341 (pmoA).
RESULTS
Enrichment of marine denitrifying methanotrophic culture.
The activity measurement and qPCR results determined throughout the enrichment procedure are presented in Fig. 1. The denitrifying methanotrophic activity is characterized by methane oxidation and nitrite reduction rates, and the number of denitrifying methanotrophs is represented by the 16S rRNA gene copy number of members of the NC10 phylum. Over the entire enrichment period of 20 months, the number of denitrifying methanotrophs incessantly increased. After 9 months of enrichment, the denitrifying methanotrophic activity appeared and then grew rapidly in the next 11 months. After 20 months, the denitrifying methanotrophic activity increased to 13.5 ± 1.2 μmol CH4 h−1 and 32.7 ± 0.9 μmol NO2− h−1. Assuming that the number of 16S rRNA gene copies, the rate of methane oxidation, and the rate of nitrite reduction increased exponentially from month 9 to month 20, the apparent doubling times were estimated to be 1.29, 1.63, and 1.61 months (equivalent to 38.7, 48.9, and 48.4 days), respectively. In the resulting enrichment culture, the specific cell activity was 0.14 ± 0.02 fmol CH4 day−1 cell−1 (for one copy), and the stoichiometric ratio of methane oxidation to nitrite reduction was 3:7.3 ± 0.7, which is very close to the theoretical value (3:8) (equation 2).
FIG 1.
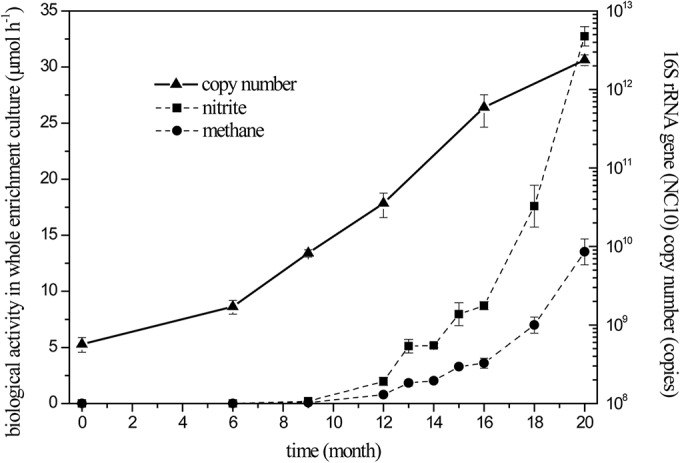
Denitrifying methanotrophic activity and the abundance of NC10 phylum 16S rRNA genes throughout the enrichment procedure. The denitrifying methanotrophic activity is represented by the methane consumption (•) and nitrite reduction (■) rates that were obtained from batch tests. In the batch tests, the initial methane partial pressure and nitrite concentration were 10 kPa and 0.5 mM, respectively. The copy numbers of 16S rRNA genes of NC10 bacteria (▲) were determined using primer pair qP1F/qP1R.
Microbial community of the enrichment culture.
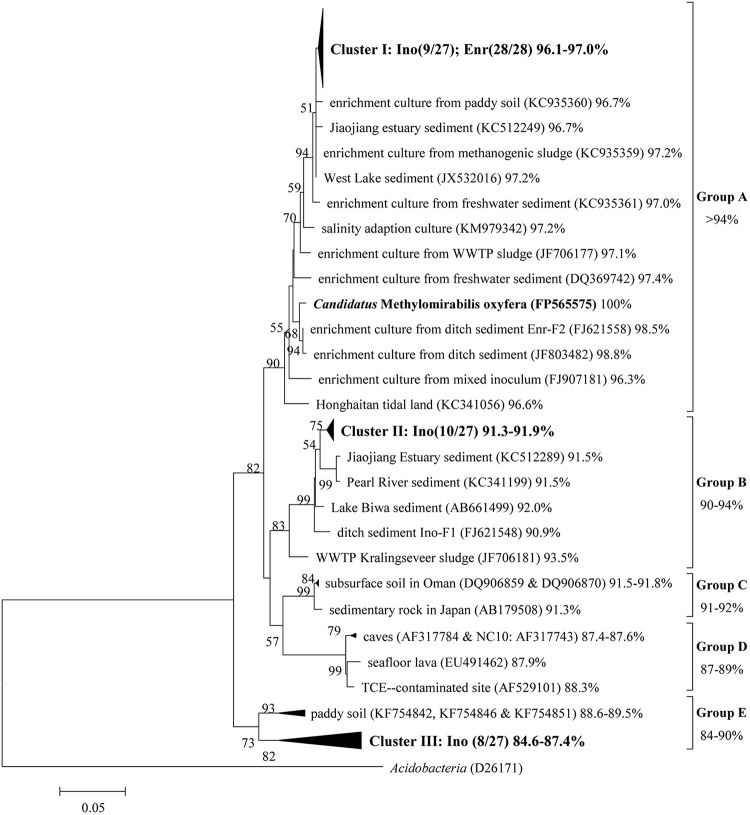
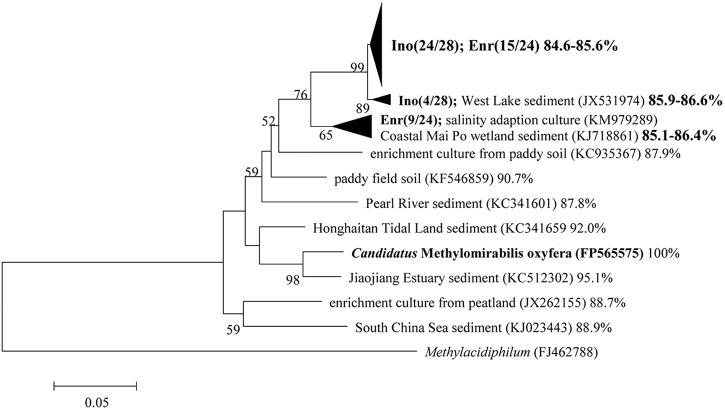
To identify the phylotypes that are responsible for the methane oxidation activity in the marine enrichment culture and to monitor the changes in the microbial community, the inoculum and the enrichment culture were examined after 16 months by cloning the 16S rRNA and pmoA genes. The reconstructed phylogenetic trees determined on the basis of the 16S rRNA and pmoA gene sequences are shown in Fig. 2 and Fig. 3, respectively.
FIG 2.
Phylogenetic tree of the NC10 phylum 16S rRNA gene sequences from inoculum (Ino) and enrichment culture (Enr) after 16 months, with Acidobacteria as the outgroup. The tree was calculated using the neighbor-joining method and the p-distance submodel. The sequence similarities of the 16S rRNA genes of the reference sequences and “Ca. Methylomirabilis oxyfera” are indicated after the sequence indicators in percentages. Groups A, B, C, and D were classified as described by Ettwig et al. (8); group E was newly classified in this work and was distantly related to “Ca. Methylomirabilis oxyfera,” with similarities lower than 90%. The bootstrap value is 1,000, and the scale bar is 5%. WWTP, wastewater treatment plant; TCE, trichloroethylene.
FIG 3.
Phylogenetic tree of the pmoA gene sequences retrieved from inoculum (Ino) and enrichment culture (Enr) after 16 months, with Methylacidiphilum as the outgroup. The tree was calculated with the neighbor-joining method using p-distance correction with the bootstrap value of 1,000. The sequence similarities between the reference sequences and “Ca. Methylomirabilis oxyfera” are indicated after the sequence indicators. Bar, 5%.
The phylogenetic analysis of the 16S rRNA genes indicates that the 16S rRNA gene sequences retrieved in this work and the reference gene sequences selected from the GenBank database could be classified into five phylogenetic groups: groups A, B, C, D, and E (as indicated in Fig. 2). Notably, group E is newly defined and represents a lineage distantly related to “Ca. Methylomirabilis oxyfera” (with similarity lower than 90%), and groups A, B, C, and D were previously classified by Ettwig et al. (8). Group E sequences were previously obtained from samples taken from paddy soil at the depth of 10 to 60 cm. The results showed that the NC10 phylum 16S rRNA gene sequences amplified from the inoculum were present in three clusters (clusters I, II, and III) and scattered into three groups (groups A, B, and E, as shown in Fig. 2); however, only the group A sequences were retrieved in the enrichment culture, which indicated that only the NC10 bacteria clustered in group A were enriched and that the others were eliminated during the enrichment. Similarities between the sequences of the bacteria in clusters I, II, and III retrieved from the inoculum and enrichment culture and that of “Ca. Methylomirabilis oxyfera” were 96.1 to 97.0%, 91.3 to 91.9%, and 84.6 to 87.4%, respectively (as indicated in Fig. 2).
The phylogenetic analysis of pmoA genes (Fig. 3) shows that the pmoA gene sequences obtained from the inoculum and the enrichment culture were 88.0 to 99.8% identical. Moreover, the pmoA gene sequences of the enrichment culture shared relatively lower similarity (84.6 to 86.6%) with that of “Ca. Methylomirabilis oxyfera.”
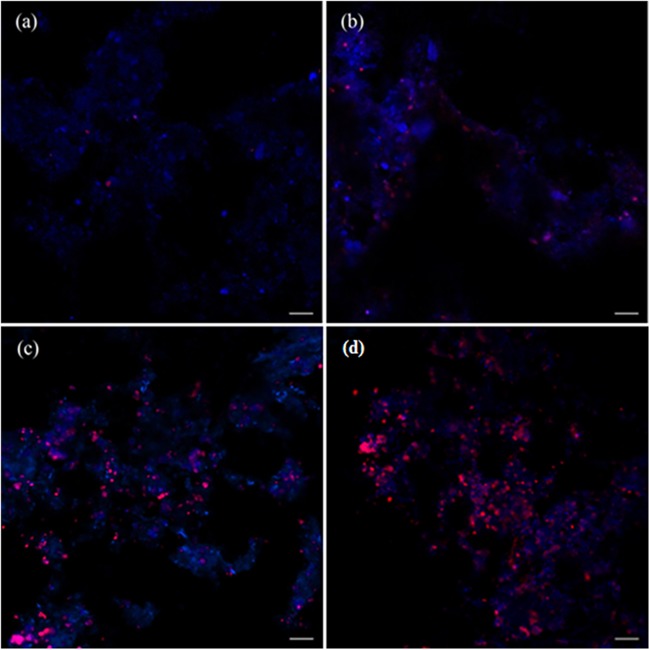
FISH performed with a specific probe, S-*-DBACT-1027-a-A-18 (Fig. 4), revealed that NC10 bacteria were (approximately 70 to 80%) dominant in the enrichment culture after 20 months, and the FISH images showed that the population of NC10 bacteria multiplied continuously during the 20 months of enrichment.
FIG 4.
FISH images of the inoculum (a) and of the enrichment culture after 9 (b), 16 (c), and 20 (d) months. The fluorescence micrographs were taken after hybridization with the NC10 bacterium-specific probe S-*-DBACT-1027-a-A-18 (Cy3, red) and the general DNA stain DAPI (blue). The “Ca. Methylomirabilis oxyfera”-like bacteria appear magenta due to a mixture of the two fluorescence signals (red and blue). Bar, 10 μm.
Effects of substrates and salinity on the culture.
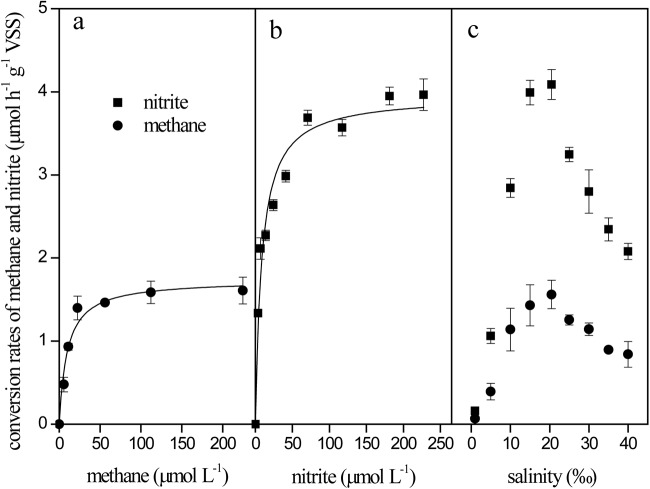
The effects of methane, nitrite, and salinity on the marine enrichment culture were investigated, and the results are shown in Fig. 5. The denitrifying methanotroph-specific activity at various methane concentrations was plotted as shown in Fig. 5a, and the data were fitted by the Monod equation (29). Based on the best fitting in Fig. 5a, the apparent methane affinity coefficient (KCH4) of the denitrifying methanotrophs was estimated to be 9.8 ± 2.2 μM, which is in the range determined for freshwater denitrifying methanotrophic cultures (3 to 92 μM) (29, 30).
FIG 5.
Effects of methane concentration (a), nitrite concentration (b), and salinity (c) on denitrifying methanotroph-specific activity. The denitrifying methanotroph-specific activity was characterized by the methane consumption (•) or nitrite reduction (■) rates. When they were not adjusted, the methane concentration, nitrite concentration, and salinity levels were 0.11 mM, 0.2 mM, and 20.5‰, respectively.
The Monod kinetic equation was also applied to describe the relationship between the denitrifying methanotroph-specific activity and the nitrite concentration (Fig. 5b). The apparent nitrite affinity coefficient was obtained from the best fitting, and the value was 8.7 ± 1.5 μM, which is similar to the nitrite concentration (2.8 to 11.4 μM) at the coast where the inoculum was collected (Table 1).
The salinity effect is of interest with respect to halophiles that have a specific requirement for salts (31), and the salinity effect on the marine enrichment culture was investigated in this work. As shown in Fig. 5c, the denitrifying methanotrophic culture reached the maximum specific activity (1.56 ± 0.17 μmol CH4 h−1 g−1 VSS) at 20.5‰ of salinity (i.e., the salinity of the enrichment media) and decreased at lower or higher salinities, which suggested that a halophilic denitrifying methanotrophic culture was obtained. Denitrifying methanotrophic activity was hardly observed at 1‰ (19.5‰ lower than the optimal salinity) of salinity (near the salinity of the freshwater medium), whereas a previous freshwater culture had the highest activity at this salinity (24). However, the culture was still active at a high salinity level of 40‰ (19.5‰ higher than the optimal salinity), and the denitrifying methanotrophic activity was 0.84 ± 0.16 μmol CH4 h−1 g−1 VSS, approximately half of the maximum activity seen at 20.5‰ salinity. Hence, the results also showed that low salinities caused more serious damage to the marine denitrifying methanotrophic culture than high salinities.
DISCUSSION
Enrichment of marine denitrifying methanotrophs.
To date, numerous freshwater denitrifying methanotrophic cultures have been obtained (3, 8, 10, 32–37), but no marine denitrifying methanotrophic culture has been reported to our knowledge. Although NC10 phylum 16S rRNA and pmoA gene sequences have been retrieved from several marine systems (19–22), these lines of evidence in molecular biology were not sufficient to prove the occurrence of the process of AOM coupled to nitrite reduction in marine systems. On the basis of the existing molecular biological evidence, we can deduce that NC10 bacteria inhabit several marine systems, but we cannot guarantee that these NC10 bacteria perform the process of AOM coupled to nitrite reduction. Very recently, salinity adaption of freshwater denitrifying methanotrophs has been observed at a salinity stress level of 20 g NaCl liter−1 (24), which raised the possibility that the process of AOM coupled to nitrite reduction occurs in marine systems. However, the growth of denitrifying methanotrophs could not be observed in the process of salinity adaption (24); therefore, whether denitrifying methanotrophs are able to grow in saline water remains unknown.
In the present work, a marine denitrifying methanotrophic culture was obtained, and denitrifying methanotrophic activity was observed at 20.5‰ salinity (Fig. 1). The correlated increases in the number of NC10 bacteria and the activity of methane oxidation indicated that NC10 bacteria were responsible for the ever-increasing methane oxidation activity. The specific cell activity of the NC10 bacteria in the final marine denitrifying methanotrophic culture was 0.14 ± 0.02 fmol CH4 day−1 cell−1, which corresponded to the previous measurements (0.09 to 0.34 fmol CH4 day−1 cell−1) (8, 26, 38). Furthermore, the qPCR and FISH results showed that NC10 bacteria could grow in seawater media (20.5‰ of salinity) and that their apparent doubling times (38.7 to 48.9 days) were slightly longer than the doubling times previously observed in freshwater enrichment cultures (14 to 25 days) (3, 29).
Notably, the marine denitrifying methanotrophic culture became inactive at the salinity of freshwater medium (∼1‰), which suggested that these denitrifying methanotrophs in the marine culture had physiological properties distinct from those seen in freshwater cultures (24), although they showed striking resemblances in 16S rRNA and pmoA gene sequences (Fig. 2 and 3).
Potential importance in coastal ecosystems.
AOM coupled to sulfate reduction is a major contributor to the methane oxidation in oceanic systems (1), and the process of AOM coupled to nitrite reduction is a vital sink of methane in freshwater systems (11, 39). However, the contribution of AOM to their transitional zone-coastal ecosystems remains obscure, although several researchers have noted the importance of methane oxidation in coastal ecosystems (40, 41). Moreover, dissolved methane concentrations are dominantly submicromolar in coastal sediments but nanomolar in the water column, which indicates that the methane produced in anoxic underlying sediments was effectively oxidized when transported upward to the water/sediment interfaces (41).
The results of the present work, together with the previous findings of 16S rRNA and pmoA gene sequences in similar areas (19, 21), suggest that the process of AOM coupled to nitrite reduction may be a methane sink in coastal ecosystems. Compared with the data from well-known consortia with respect to AOM coupled to sulfate reduction, denitrifying methanotrophs have the edge in energy yields and methane affinity but are weak in low nitrite concentrations in coastal systems (Table 1). The energy advantage evident in equation 1 and equation 2 helps denitrifying methanotrophs synthesize more biomass and grow relatively quickly. The other advantage is that the halophilic denitrifying methanotrophs have a stronger affinity for methane (KCH4 = 9.8 ± 2.2 μM) than the consortia that perform AOM coupled to sulfate reduction (KCH4 > 1.0 mM) (29). Furthermore, dissolved methane concentrations in coastal sediments are usually <2.0 mM (41), whereas those in the seabed can reach 5 to 15 mM at locations below the sulfate-methane transition zones (SMTZ) (42). The relative low methane concentrations benefit denitrifying methanotrophs in competing for methane against consortia that perform AOM coupled to sulfate reduction. In contrast, the low concentrations of nitrite in coastal sediments, which are approximately 3 orders of magnitude lower than the sulfate concentrations (Table 1), most likely limit bacterial growth. Moreover, marine denitrifying methanotrophs are active in water with a wide range of salinities (from 5 to 40‰; Fig. 5c) and are able to inhabit coastal ecosystems where salinity varies frequently. In short, these findings predict the possible occurrence of the process of AOM coupled to nitrite reduction in coastal ecosystems, but more lines of ecological evidence are needed to confirm it.
Are all NC10 bacteria methanotrophs?
Interestingly, only members of group A of the NC10 phylum were enriched, although three clusters of NC10 bacteria (scattered in groups A, B, and E) were detected in the inoculum. To date, researchers have obtained only group A-containing denitrifying methanotrophic cultures (3, 8, 10, 32–37), and the functions of the members of NC10 phylum groups B, C, D, and E are unknown, although group B members were commonly detected in the inocula. Two possible reasons may explain why members of groups B and E were eliminated during the enrichment process. First, the bacteria from groups B and E may mediate the process of AOM coupled to nitrite reduction, but the existing enrichment media or enrichment conditions were not appropriate for their growth, as discussed in previous literature (10). Second, members of groups B and E could not perform the process of AOM coupled to nitrite reduction or were not even methanotrophs. In this case, they could not be enriched in the existing enrichment media that were prepared to cultivate denitrifying methanotrophs.
Strictly speaking, the denitrifying methanotrophs (or so-called “Ca. Methylomirabilis oxyfera”-like bacteria) consist of members of group A of the NC10 phylum (closely related to “Ca. Methylomirabilis oxyfera,” with 16S rRNA gene sequence similarities of over 94% as shown in Fig. 2) because only group A members have been demonstrated to perform the process of AOM coupled to nitrite reduction. Therefore, the presence of NC10 bacteria does not amount to a guarantee that denitrifying methanotrophic activity will occur, but the presence of group A bacteria (whether in freshwater or in saline water) indicates that they have a high likelihood of exhibiting denitrifying methanotrophic activity. To resolve the issue of whether all NC10 bacteria are methanotrophs, determination of the metabolic capacities of organisms in groups B, C, D, and E in further studies is essential, and the results will be highly significant for improving our understanding of the members of the NC10 phylum and the global methane cycle.
Conclusions.
In the present work, a marine denitrifying methanotrophic culture was obtained from coastal sediments. The phylogenetic analysis of the marine enrichment culture showed that bacteria belonging to group A of the NC10 phylum were responsible for the methane oxidation activity, but the functions of the members of the other groups in the NC10 phylum in coastal sediments remain unknown. Moreover, the findings indicated that the process of halophilic bacterial AOM coupled to nitrite reduction may play a role as a methane sink in coastal ecosystems.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (no. 41276109 and no. 51478415), the Fundamental Research Funds for the Central Universities (no. 2015QNA6012), Zhejiang Provincial Science and Technology Project (2013C33025), and Scientific Research Project of the Department of Education of Zhejiang Province (Y201226062).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00984-15.
REFERENCES
- 1.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 2.Boetius A, Ravenschlag K, Schubert CJ, Rickert D, Widdel F, Gieseke A, Amann R, Jørgensen BB, Witte U, Pfannkuche O. 2000. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 407:623–626. doi: 10.1038/35036572. [DOI] [PubMed] [Google Scholar]
- 3.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Damste JS, Op den Camp HJ, Jetten MS, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 4.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 5.Beal EJ, House CH, Orphan VJ. 2009. Manganese- and iron-dependent marine methane oxidation. Science 325:184–187. doi: 10.1126/science.1169984. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell SL, Laidler JR, Brewer EA, Eberly JO, Sandborgh SC, Colwell FS. 2008. Anaerobic oxidation of methane: mechanisms, bioenergetics, and the ecology of associated microorganisms. Environ Sci Technol 42:6791–6799. doi: 10.1021/es800120b. [DOI] [PubMed] [Google Scholar]
- 7.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 8.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu Rev. Microbiol 57:369–394. doi: 10.1146/annurev.micro.57.030502.090759. [DOI] [PubMed] [Google Scholar]
- 10.He Z, Cai C, Shen L, Lou L, Zheng P, Xu X, Hu B. 2015. Effect of inoculum sources on the enrichment of nitrite-dependent anaerobic methane-oxidizing bacteria. Appl Microbiol Biotechnol 99:939–946. doi: 10.1007/s00253-014-6033-8. [DOI] [PubMed] [Google Scholar]
- 11.Hu BL, Shen LD, Lian X, Zhu Q, Liu S, Huang Q, He ZF, Geng S, Cheng DQ, Lou LP, Xu XY, Zheng P, He YF. 2014. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc Natl Acad Sci U S A 111:4495–4500. doi: 10.1073/pnas.1318393111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen LD, Huang Q, He ZF, Lian X, Liu S, He YF, Lou LP, Xu XY, Zheng P, Hu BL. 2015. Vertical distribution of nitrite-dependent anaerobic methane-oxidising bacteria in natural freshwater wetland soils. Appl Microbiol Biotechnol 99:349–357. doi: 10.1007/s00253-014-6031-x. [DOI] [PubMed] [Google Scholar]
- 13.Shen LD, Liu S, Huang Q, Lian X, He ZF, Geng S, Jin RC, He YF, Lou LP, Xu XY, Zheng P, Hu BL. 2014. Evidence for the cooccurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in a flooded paddy field. Appl Environ Microbiol 80:7611–7619. doi: 10.1128/AEM.02379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deutzmann JS, Schink B. 2011. Anaerobic oxidation of methane in sediments of Lake Constance, an oligotrophic freshwater lake. Appl Environ Microbiol 77:4429–4436. doi: 10.1128/AEM.00340-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kojima H, Tsutsumi M, Ishikawa K, Iwata T, Mussmann M, Fukui M. 2012. Distribution of putative denitrifying methane oxidizing bacteria in sediment of a freshwater lake, Lake Biwa. Syst Appl Microbiol 35:233–238. doi: 10.1016/j.syapm.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MS, Yin C, Op den Camp HJ. 2012. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett 336:79–88. doi: 10.1111/j.1574-6968.2012.02654.x. [DOI] [PubMed] [Google Scholar]
- 17.Shen LD, Liu S, Zhu Q, Li XY, Cai C, Cheng DQ, Lou LP, Xu XY, Zheng P, Hu BL. 2014. Distribution and diversity of nitrite-dependent anaerobic methane-oxidising bacteria in the sediments of the Qiantang River. Microb Ecol 67:341–349. doi: 10.1007/s00248-013-0330-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhu G, Zhou L, Wang Y, Wang S, Guo J, Long XE, Sun X, Jiang B, Hou Q, Jetten MS, Yin C. 2015. Biogeographical distribution of denitrifying anaerobic methane oxidizing bacteria in Chinese wetland ecosystems. Environ Microbiol Rep 7:128–138. doi: 10.1111/1758-2229.12214. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Zhou Z, Gu JD. 2015. Complex community of nitrite-dependent anaerobic methane oxidation bacteria in coastal sediments of the Mai Po wetland by PCR amplification of both 16S rRNA and pmoA genes. Appl Microbiol Biotechnol 99:1463–1473. doi: 10.1007/s00253-014-6051-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Zhou ZC, Gu JD. 2014. Occurrence and diversity of nitrite-dependent anaerobic methane oxidation bacteria in the sediments of the South China Sea revealed by amplification of both 16S rRNA and pmoA genes. Appl Microbiol Biotechnol 98:5685–5696. doi: 10.1007/s00253-014-5733-4. [DOI] [PubMed] [Google Scholar]
- 21.Li-Dong S, Qun Z, Shuai L, Ping D, Jiang-Ning Z, Dong-Qing C, Xiang-Yang X, Ping Z, Bao-Lan H. 2014. Molecular evidence for nitrite-dependent anaerobic methane-oxidising bacteria in the Jiaojiang Estuary of the East Sea (China). Appl Microbiol Biotechnol 98:5029–5038. doi: 10.1007/s00253-014-5556-3. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Jiang X-W, Gu J-D. 2014. Existence of novel phylotypes of nitrite-dependent anaerobic methane-oxidizing bacteria in surface and subsurface sediments of the South China Sea. Geomicrobiology J 32:1–10. [Google Scholar]
- 23.Yang J, Jiang H, Wu G, Hou W, Sun Y, Lai Z, Dong H. 2012. Co-occurrence of nitrite-dependent anaerobic methane oxidizing and anaerobic ammonia oxidizing bacteria in two Qinghai-Tibetan saline lakes. Front Earth Sci 6:383–391. doi: 10.1007/s11707-012-0336-9. [DOI] [Google Scholar]
- 24.He Z, Geng S, Shen L, Lou L, Zheng P, Xu X, Hu B. 2015. The short- and long-term effects of environmental conditions on anaerobic methane oxidation coupled to nitrite reduction. Water Res 68:554–562. doi: 10.1016/j.watres.2014.09.055. [DOI] [PubMed] [Google Scholar]
- 25.Hu BL, Shen LD, Zheng P, Hu AH, Chen TT, Cai C, Liu S, Lou LP. 2012. Distribution and diversity of anaerobic ammonium-oxidizing bacteria in the sediments of the Qiantang River. Environ Microbiol Rep 4:540–547. doi: 10.1111/j.1758-2229.2012.00360.x. [DOI] [PubMed] [Google Scholar]
- 26.Hu B, He Z, Geng S, Cai C, Lou L, Zheng P, Xu X. 2014. Cultivation of nitrite-dependent anaerobic methane-oxidizing bacteria: impact of reactor configuration. Appl Microbiol Biotechnol 98:7983–7991. doi: 10.1007/s00253-014-5835-z. [DOI] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.American Public Health Association (APHA), American Water Works Association (AWWA), and Water Environment Federation (WEF). 2005. Standard methods for the examination of water and wastewater, 21 ed American Public Health Association, Washington, DC. [Google Scholar]
- 29.He Z, Cai C, Geng S, Lou L, Xu X, Zheng P, Hu B. 2013. Modeling a nitrite-dependent anaerobic methane oxidation process: parameters identification and model evaluation. Bioresour Technol 147:315–320. doi: 10.1016/j.biortech.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Winkler MK, Ettwig KF, Vannecke TP, Stultiens K, Bogdan A, Kartal B, Volcke EI. 2015. Modelling simultaneous anaerobic methane and ammonium removal in a granular sludge reactor. Water Res 73:323–331. doi: 10.1016/j.watres.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 31.Madigan MT, Martinko JM, Stahl D, Clark DP. 2010. Brock biology of microorganisms, 13th ed Pearson Education Inc, San Francisco, CA. [Google Scholar]
- 32.Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ Microbiol Rep 1:377–384. doi: 10.1111/j.1758-2229.2009.00083.x. [DOI] [PubMed] [Google Scholar]
- 33.Luesken FA, Sanchez J, van Alen TA, Sanabria J, Op den Camp HJ, Jetten MS, Kartal B. 2011. Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl Environ Microbiol 77:6802–6807. doi: 10.1128/AEM.05539-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TL, Zeeman G, Temmink H, Strous M, Op den Camp HJ, Jetten MS. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol 92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kampman C, Hendrickx TL, Luesken FA, van Alen TA, Op den Camp HJ, Jetten MS, Zeeman G, Buisman CJ, Temmink H. 2012. Enrichment of denitrifying methanotrophic bacteria for application after direct low-temperature anaerobic sewage treatment. J Hazard Mater 227–228:164–171. [DOI] [PubMed] [Google Scholar]
- 36.Zhu B, van Dijk G, Fritz C, Smolders AJ, Pol A, Jetten MS, Ettwig KF. 2012. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl Environ Microbiol 78:8657–8665. doi: 10.1128/AEM.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding ZW, Ding J, Fu L, Zhang F, Zeng RJ. 2014. Simultaneous enrichment of denitrifying methanotrophs and anammox bacteria. Appl Microbiol Biotechnol 98:10211–10221. doi: 10.1007/s00253-014-5936-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhu B, Sanchez J, van Alen TA, Sanabria J, Jetten MS, Ettwig KF, Kartal B. 2011. Combined anaerobic ammonium and methane oxidation for nitrogen and methane removal. Biochem Soc Trans 39:1822–1825. doi: 10.1042/BST20110704. [DOI] [PubMed] [Google Scholar]
- 39.Deutzmann JS, Stief P, Brandes J, Schink B. 2014. Anaerobic methane oxidation coupled to denitrification is the dominant methane sink in a deep lake. Proc Natl Acad Sci U S A 111:18273–18278. doi: 10.1073/pnas.1411617111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeburgh WS. 2007. Oceanic methane biogeochemistry. Chem Rev 107:486–513. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 41.Burgos M, Sierra A, Ortega T, Forja JM. 2015. Anthropogenic effects on greenhouse gas (CH and NO) emissions in the Guadalete River Estuary (SW Spain). Sci Total Environ 503–504:179–189. doi: 10.1016/j.scitotenv.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 42.Sansone FJ, Graham AW, Berelson WM. 2004. Methane along the western Mexican margin. Limnol Oceanogr 49:2242–2255. doi: 10.4319/lo.2004.49.6.2242. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.