Abstract
Parasitism is now recognized as a major factor impacting the ecology and evolution of plankton, including Daphnia. Parasites that attack the developing embryos of daphniids, known as brood parasites, were first described in the early 1900s but have received relatively little study. Here, we link previous morphological descriptions of the oomycete brood parasite Blastulidium paedophthorum with information on its phylogenetic placement, ecology, and virulence. Based on the morphology and phylogenetic relationship with other members of the Leptomitales, we show that a brood parasite observed in daphniids in the Midwestern United States is B. paedophthorum. We used morphology, DNA sequences, and laboratory infection experiments to show that B. paedophthorum is a multihost parasite that can be transmitted between species and genera. A field survey of six hosts in 15 lakes revealed that B. paedophthorum is common in all six host taxa (present on 38.3% of our host species-lake-sampling date combinations; the maximum infection prevalences were 8.7% of the population and 20% of the asexual adult female population). Although B. paedophthorum was observed in all 15 lakes, presence and infection prevalence varied among lakes. Infection with B. paedophthorum did not reduce host life span but significantly impacted host fecundity. Theory predicts that parasites that affect host fecundity without affecting host life span should have the strongest impact on host population dynamics. Based on its virulence and commonness in natural populations and on the central role of daphniids in freshwater food webs, we predict that B. paedophthorum will influence daphniid ecology and evolution, as well as the larger food web.
INTRODUCTION
While parasites were largely overlooked by plankton biologists for most of the 20th century, it is now clear that parasitism is a major force in the plankton. Approximately 40% of all species are parasitic, so a better understanding of the diversity and function of parasites in aquatic ecosystems is needed if we are to truly understand host diversity, food webs, and nutrient cycling (1). Evidence from recent decades shows that parasites are widespread in freshwater plankton, can influence the ecology and evolution of host populations, and can even impact community structure and ecosystem processes (2). For example, in addition to directly influencing their host populations, parasites can move inedible host resources to higher trophic levels via microbial loops and mycoloops (3, 4).
Daphnia has emerged as an excellent model system for understanding the causes and consequences of parasitism (5, 6), in part because it is genetically tractable and variable, ubiquitous in freshwater habitats, and transparent (making it relatively easy to assess infections). Parasites can influence Daphnia evolution (7–10) and population dynamics (11–13). As expected given their population-level effects and the importance of Daphnia to aquatic ecosystems (14), disease outbreaks can have ecosystem-level impacts (15).
Numerous studies have reported finding a fungal or oomycete parasite attacking the developing embryos of Daphnia. A brood parasite was first reported by Pérez (16), and brood parasites have now been reported from European (see, for example, references 10 and 17–19), North American (17, 20), and Asian populations (21). In some cases, reported infection prevalences have been quite high (e.g., 30 to 40% of the population infected [18, 22]), suggesting that brood parasites might have important ecological and evolutionary consequences. Indeed, infection by a brood parasite has been found to increase susceptibility to a bacterial disease (19) and has been associated with evolutionary changes in host populations (10).
However, whereas brood parasites have been reported routinely since Pérez first described the brood parasite Blastulidium paedophthorum in 1903, little is known about them. Very few studies have attempted to identify brood parasites to species. B. paedophthorum remains the only formally described brood parasite of daphniids, but its precise taxonomic placement is unknown. Originally, Pérez named B. paedophthorum as a new species of Haplosporidia, but it was considered by Chatton (23) to be related to the Chytridiomycetes, because he observed parasite zoospores with apparently a single axial flagellum, a trait absent in Haplosporidia. The most detailed observations come from Manier (24), who determined that the zoospores are biflagellated. This and other ultrastructural features of sporangia led Manier to suggest affinities between B. paedophthorum and the oomycete order Lagenidiales. The biflagellated zoospore and tubular mitochondrial cristae from Manier's study provided convincing association of B. paedophthorum with Oomycetes, but the precise placement within the group has been uncertain, with Karling (25) considering B. paedophthorum allied to Olpidiopsidales and Dick (26) placing B. paedophthorum tentatively near Leptolegniellaceae (Leptomitales), probably because the species therein are holocarpic. To date, it appears most studies observing Daphnia brood parasites identify a distinct fungus-like infection morphologically allied with the Oomycetes, yet no studies have linked information on the phylogenetic placement of a brood parasite with information on its ecology and virulence. This is somewhat surprising, given the commonness of these parasites and their potential to strongly influence the ecology and evolution of Daphnia.
We report here on a study of a brood parasite in 15 lakes in southeastern Michigan. We sought to (i) use parasite morphology and molecular data to resolve the identity and phylogenetic relationships of the brood parasite, (ii) document the frequency of infection in lake populations and determine whether the parasite infects multiple host species, and (iii) quantify the fitness effects of the parasite on host individuals. We conclude that the brood parasite in our study populations is B. paedophthorum, based on morphology and phylogenetic placement. Moreover, we show that B. paedophthorum is a virulent, multihost parasite that might strongly impact the ecology and evolution of daphniids.
MATERIALS AND METHODS
Microscopy.
Photomicrographs were taken using light and dark microscopy at ×25 to ×80 with an Olympus SZX16 dissecting microscope and at ×100 to ×1,000 using differential interference microscopy with an Olympus BX53 microscope; for both of these, we used Olympus CellSens Standard imaging software. Measurements were taken on images taken of B. paedophthorum cells in a Daphnia parvula sample collected from Mill Lake.
Field-collected infected D. dentifera were fixed for transmission electron microscopy (TEM) using 4% glutaraldehyde in Sorensen's phosphate buffer (0.1 M KH2PO4 and 0.1 M Na2HPO4) for 2 h and rinsed and stored at 4°C in the dark until they could be postfixed using 1% OsO4 in Sorensen's phosphate buffer with gentle agitation. After fixation, the D. dentifera samples were again washed in Sorensen's phosphate buffer under agitation, dehydrated in a graded ethanol series, and finally embedded in LR White resin in gelatin capsules heated at 45°C for 3 days. The samples were sectioned through the brood chamber; 70-nm sections were obtained with a PTXL ultramicrotome (RMC; Boeckler Instruments, Tucson, AZ). Ultrathin sections were mounted on 200 Formvar-coated copper mesh grids and stained with 4% uranyl acetate and lead citrate. Sections were visualized using a JEOL 100CX transmission electron microscope (Japan Electron Optics Laboratory, Japan).
DNA sequencing and phylogenetic analysis.
All infected Daphnia individuals used for genetic analysis were identified as infected and stored in 2× CTAB (cetyltrimethylammonium bromide) buffer (27) at −80°C. Individuals were examined under a stereomicroscope at ×25 to ×50 to confirm infection with B. paedophthorum. Infected individuals were then rinsed and moved to a clean slide. DNA was extracted from two whole infected D. dentifera and, for the remaining nine samples analyzed, the parasitized contents of the brood chambers were dissected (Table 1). Cells were extracted using 250 μl of 2× CTAB buffer by homogenization using a Kontes pestle, followed by a standard chloroform-isoamyl alcohol (24:1) purification before an overnight isopropanol DNA precipitation (27). DNA samples were suspended in 50 μl of H2O and stored at −20°C.
TABLE 1.
Locality and species information for material used for DNA sequencinga
| Sample no. | Collection location | Date collected | Species | Brood dissection performed |
|---|---|---|---|---|
| 9, 10 | North Lake | 9/11/2013 | Daphnia dentifera | No |
| 13, 15, 16 | North Lake | 9/11/2013 | Daphnia dentifera | Yes |
| 17, 18, 19 | Pickerel Lake | 9/30/2013 | Daphnia dentifera | Yes |
| 36, 37 | Bishop Lake | 10/10/2013 | Daphnia retrocurva | Yes |
| 43 | Cedar Lake | 10/11/2013 | Daphnia dentifera | Yes |
Brood dissection performed indicates whether the brood contents were removed from the animal's brood chamber before DNA extraction.
PCR was conducted using GoTaq Green master mix using 12.5-μl reactions with 5 μl of extracted DNA. We conducted PCR with general eukaryotic 18S rRNA primers SR1R (28) and NS2 (29) with the following settings: 94°C for 3 min, 45 cycles of 94°C for 1 min, followed by annealing at 54°C for 30 s and extension at 72°C for 1 min, with a final extension of 72°C for 7 min. We also used 18S rRNA oomycete-specific primers (SRSt-1F and SRSt-1R) (30) with the following settings: 94°C for 3 min, 35 cycles of 94°C for 30 s min, followed by annealing at 54°C for 30 s and extension at 72°C for 1.5 min, with a final extension of 72°C for 7 min. The whole internal transcribed spacer (ITS) and partial 28S rRNA regions were amplified from sample 17 using the primers UN-up18S42 and UN-lo28S1220 as described by Robideau et al. (31). To complete the ITS and 28S sequences of sample 17 and to determine the sequence of B. paedophthorum from a D. retrocurva host (sample 37), we designed two internal primers specific to the brood parasite: Brood-ITS-F (5′-TTGGACTTAAATTGCATGGAGA-3′) and Brood-LSU-R (5′-AAAGTCCCGAACAGCAAGAG-3′). The amplification using these primers was the same as for the general eukaryotic primers except using 35 cycles.
PCR products were cleaned using ExoSAP-IT (Promega) and sequenced at the University of Michigan DNA Sequencing Core on an ABI-3730. Sequences were assembled and edited using Sequencher 5.2.4 (Gene Codes). The sequences for samples 17 and 37 have been deposited at GenBank (see the end of Materials and Methods for the accession numbers). We aligned the brood sequences with oomycete 18S and 28S sequences from GenBank using the MAFFT online server with parameters automatically determined (32). The alignments were masked for poorly aligned regions using Gblocks 0.91b (33) with default parameters (except gaps permitted in half of the sequences for the 18S region and gaps permitted in all sequences and 12-bp nonconserved blocks and 8-bp minimum block length for the 28S region). The best-fitting model of evolution was estimated for the combined and separate partitions using jModelTest-2.1.4 (34) using the Akaike information criterion. We selected the general time reversible model plus a proportion of invariable sites and gamma distributed rates for all data sets since this model either was the best model or was not significantly different from the best model, as determined by jModelTest for all partitions. The maximum-likelihood trees for the separate regions and the concatenated regions were estimated using PhyML 3.1 (35) with support estimated using 1,000 bootstrap pseudoreplicates.
Parasite prevalence in natural populations.
We sampled 15 lakes in southeastern Michigan (see Table S1 in the supplemental material). Lakes were sampled every 2 weeks from mid-July through mid-November in 2014. We collected three samples per lake per sampling day from three locations within the deep basin of the lake. At each location, we used a 153-μm-pore-size Wisconsin net to do a vertical tow through the entire water column. We performed three of these at each location, putting one in each of the three sample bottles. Thus, each sample was a pool of tows from three locations. Two samples were preserved in 50 to 90% ethanol and later counted to determine host density. The third was scanned within 24 h of collection. Individuals infected with B. paedophthorum were identified based on the appearance of the developing embryos, which are easily visible through the transparent Daphnia (Fig. 1). We examined all Daphnia and Ceriodaphnia spp. for infection; five species of Daphnia were relatively common (D. dentifera, D. dubia, D. parvula, D. pulicaria, and D. retrocurva), as was Ceriodaphnia dubia. For each lake-day, we determined the infection prevalence in all of the Daphnia and Ceriodaphnia spp. contained in the live sample. If fewer than 20 individuals of a particular host species were present in the sample from a given lake on a given date, we excluded that host species-lake-date combination from our analyses.
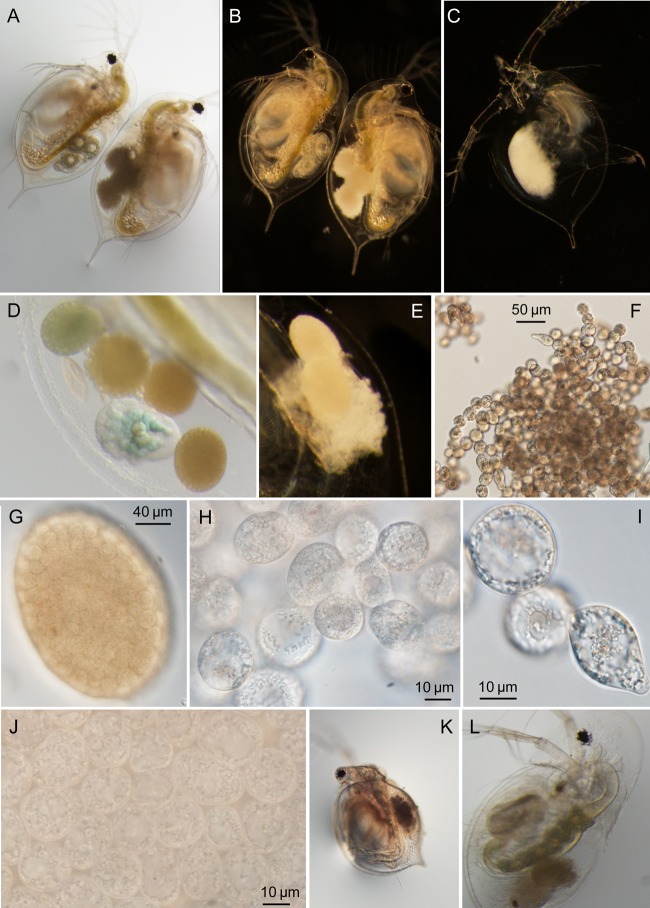
FIG 1.
Morphology of uninfected hosts, infected hosts, and B. paedophthorum cells. (A and B) Individuals of the same D. pulex × pulicaria genotype. The individual on the left is healthy and contains three developing embryos, while the individual on the right has infected embryos. The same two individuals are shown in panel A (bright-field microscopy) and panel B (dark-field microscopy). (C) Molt of D. dentifera containing remnants of infection. (D) Brood chamber of infected D. dentifera containing one developing embryo and four embryos that have been attacked by B. paedophthorum. (Note that this photo is from the same individual and time point as shown in Fig. 7K and L.) (E) Brood chamber of infected D. parvula showing embryos at different stages of infection development. (F to J) B. paedophthorum cells from infected D. parvula at different levels of magnification. Note that in panel G the shape of the original Daphnia embryo is still intact. Panel J shows the same tightly packed cells at a higher magnification. (K) Infected Ceriodaphnia dubia. (L) Infected D. dentifera with full ovaries. The green tissue with yolk globules and lipid droplets along the gut are full ovaries, supporting that infected hosts continue to allocate resources to reproduction (see also the main text).
Here, we present data on the presence of B. paedophthorum and on the maximum infection prevalence in the different host species and lakes. For data on presence/absence of B. paedophthorum, we used lake-host species combinations where we had at least five sampling dates with at least 20 individuals. We then calculated, for each different host and lake, the proportion of days where we saw at least one infected individual. We calculated the maximum infection prevalence as follows: (the number of individuals infected with B. paedophthorum/the total number of individuals in the sample) × 100. We also calculated the maximum percentage of infected asexual adult females as follows: (the number of asexual females infected with B. paedophthorum/the total number of asexual adult females in the sample) × 100. Because B. paedophthorum attacks developing embryos, infections can only be seen in adult females. However, it is possible (although rare) for B. paedophthorum to attack sexually produced offspring in addition to asexual embryos (see Results). Data on the dynamics of infections in these lakes and the relationships between infection prevalence and host density will be presented elsewhere.
Life table measures of parasite virulence.
We used field-collected uninfected and B. paedophthorum-infected D. dentifera and D. retrocurva to determine life tables in order to quantify parasite virulence. We collected D. dentifera and D. retrocurva from two lakes (Table 2). Samples were collected using vertical tows of a 153-μm Wisconsin net in the deep basin of the lake. Adult females were haphazardly isolated from the sample using a side light. Individuals were briefly viewed with a microscope to verify species identification and infection status and then transferred into a 50-ml beaker containing 40 ml of filtered lake water. The sample collected from Cedar Lake did not contain large numbers of infected hosts, limiting our sample size in this lake, especially of infected D. retrocurva (Table 2). Individuals were maintained singly at 20°C and 16:8 light-dark cycles and were given high food (106 cells of the nutritious green alga Ankistrodesmus falcatus daily). Survival and fecundity were recorded every 2 to 3 days, and animals were transferred to clean water weekly. If infected individuals successfully produced offspring, the offspring were transferred to a separate beaker and monitored for infection. Survival was analyzed using a Cox proportional hazards model, implemented using the “survival” package in R (v3.1.2) (36, 37). Fecundity was not normally distributed and was therefore analyzed using a nonparametric permutation test, implemented using the “coin” package in R (38).
TABLE 2.
Fecundity of field-collected uninfected and brood parasite-infected individualsa
| Lake | Host | Yr | Sample size (no. of samples) |
Mean fecundity (offspring/female) |
Z | Pb | ||
|---|---|---|---|---|---|---|---|---|
| U | I | U | I | |||||
| North Lake | D. dentifera | 2013 | 40 | 25 | 29.1 | 0.44 | 5.75 | <0.001 |
| D. dentifera | 2014 | 14 | 13c | 7.2 | 0.46 | 3.22 | <0.001 | |
| D. retrocurva | 2014 | 15 | 15 | 3.53 | 1.33 | 2.05 | 0.04 | |
| Cedar Lake | D. dentifera | 2014 | 10 | 8 | 24.3 | 0.63 | 2.73 | 0.002 |
| D. retrocurva | 2014 | 10 | 4 | 5.4 | 0.00 | 0.656 | 0.61 | |
U, uninfected; I, infected.
Values in boldface are statistically significant.
Two additional individuals were originally set up but were excluded due to handling errors.
Infection progression.
For the life table studies, we noted that some individuals seemed to continually produce clutches but never had viable offspring. To examine this further, we tracked the progression of infection in animals from the life table study to determine whether individuals who remained infected with the parasite continued to produce embryos, or whether they ceased producing embryos. We monitored two D. dentifera from the Cedar Lake life table, as well as one D. retrocurva and one D. dentifera from the North Lake life table in 2014; each individual was monitored for 7 days, except for the North Lake D. retrocurva, which died on day 6.
Experimental infections.
We attempted to infect hosts by placing field-collected B. paedophthorum-infected animals in beakers with uninfected animals from genotypes that had been maintained in the lab for at least 1 year prior to exposure to the parasite. We did this using infected animals collected from two different lakes (Cedar Lake and North Lake), two different species of infected host (D. dentifera and D. retrocurva), and three different species of uninfected host (Ceriodaphnia dubia, D. dentifera, and a D. pulex × pulicaria hybrid). A single infected host (the “donor”) was placed in a 150-ml beaker containing 100 ml of filtered lake water; three uninfected hosts (the “recipients”) were added to this beaker (see Table 2 and the supplemental material for the genotypes used). In three cases, the donor host was placed inside a cage that was constructed by removing the end of a 15-ml centrifuge tube and covering the opening with 500-μm-pore-size Nitex mesh. This cage was placed in the beaker at an angle to allow exchange of water between the beaker and cage.
Each beaker was fed 20,000 cells of A. falcatus/ml daily and maintained at 20°C with a 16:8 light-dark cycle. Each beaker was checked at least twice weekly for 18 days (North Lake) or 22 days (Cedar Lake). When an infected D. dentifera sample was put in with uninfected D. dentifera, the infection trial was only considered successful if two infected individuals were observed at the same time, since it was not possible to identify which individual was the field-collected infected host.
Cultivation attempts.
We attempted to culture the parasite on oatmeal agar containing antibiotics. The medium was prepared by heating 30 g oat flakes in 1 liter of deionized water to boiling and simmering gently for 2 h. The slurry was filtered through cheese cloth and reconstituted to 1 liter. The medium was sterilized by autoclaving at 121°C for 30 min, and streptomycin sulfate and penicillin were added to 100 mg/liter after cooling. On two plates we placed whole, B. paedophthorum-infected D. dentifera; we used an insect pin to poke the host's body after placing it on the plate. In addition, three plates were inoculated by first removing the infected brood from D. dentifera and then transferring the infected brood to a plate using a micropipette. These attempts were unsuccessful and are not discussed further.
Nucleotide sequence accession numbers.
The nucleotide sequences for sample 17 (D. dentifera) have been deposited at GenBank (accession numbers KR869807 [18S] and KR869810 [ITS and 28S]). Identical sequences obtained from sample 37 (D. retrocurva) have been deposited at GenBank (accession numbers KR869808 [18S] and KR869809 [28S]).
RESULTS
Morphology and phylogeny.
Daphnia individuals infected with B. paedophthorum reveal symptoms confined to the brood chamber. Within the brood chamber, the outlines of embryos can be observed, but the contents become opaque and hyaline and are replaced with spherical to ovoid thalli of 22.5-μm (standard deviation [SD], 5.7-μm) width (Fig. 1). Each embryo becomes filled with hundreds of thalli (Fig. 1G and J). Occasionally, broods are mixtures of infected and uninfected embryos (Fig. 1D) as noted by Manier (24). The contents of the thalli have a distinct centrifugal development (Fig. 1H and I), giving them a vacuolated middle and a gross appearance to a blastula, from whence the name Blastulidium derives. Thalli may be in chains, and they may be pear-shaped (Fig. 1F, H, and I). After molting, the molted carapace can still contain signs of infection (Fig. 1C).
Ultrastructural analysis revealed similarities to B. paedophthorum studied previously by Manier (24); because of these similarities, the thalli are considered zoosporangia. The samples we analyzed appeared to be in earlier stages of development, because we seldom observed centrioles and cleavage furrows that delimit individual zoospores. However, similarly to Manier, we observed a vacuolated medullary region containing dense masses unbound by membrane (Fig. 2A and C), spherical to oblong mitochondria (Fig. 2E), a bilayered cell wall (Fig. 2B), and septa between sporangia with a zone of least resistance for cleavage (Fig. 2B). When observed, basal bodies were observed as pairs of tubular structures at a right angle to each other (Fig. 2D). In addition, we observed numerous dense bodies, surrounded by a single membrane, that are similar to the dense corpuscles observed by Manier (24) and are likely organelles homologous to dense body vacuoles (labeled “d” in Fig. 2B and C). These electron-dense organelles may be homologous to similar structures in other oomycetes that are known to contain storage reserves, such as the mycolaminarin carbohydrates (39).
FIG 2.
Ultrastructure of B. paedophthorum infection of Daphnia. (A) Individual thallus showing centrifugal development with a clear medullary region and a cortical region containing organelles. (B) Close-up of septum (s) between two thalli. In the upper thallus, orthogonal basal bodies (bb) can be observed. Several dense body vacuoles (d)—or dense corpuscles according to Manier (24)—can be seen in the lower cell. (C) In developing thalli, the medullary region contains large dense amorphous masses (arrowheads), which are likely components of the dense body vacuole system. (D) Basal bodies were found at orthogonal angles when observed. (E) Mitochondria (m) appear spherical or oblong. Dense body vacuoles (d) are clearly delimited by a single membrane, as well as smaller membrane-bound organelles that may be encystment vesicles (e).
Because our parasite was of uncertain phylogenetic affinities, we began by using universal 18S rRNA primers and amplified DNA from four samples for which the brood chamber contents had been removed, i.e., samples 13, 15, 16, and 43 (Table 1). When these samples were sequenced, all of the fragments showed matches to Oomycota using BLAST to GenBank, but only two sequences gave clean chromatograms. Using this information, we then amplified and sequenced the 18S rRNA gene from samples 9, 10, 17, 18, 19, 36, and 37 (see Table 1) using the oomycete-specific primers SRSt-1F and SRSt-1R. Comparison of these sequences revealed identical sequences over this ∼850-bp region for all samples across these three lakes and two host species. To increase the amount of phylogenetic data, we then amplified and sequenced the ITS and 28S rRNA regions of sample 17 with a combination of oomycete- and brood parasite-specific primers.
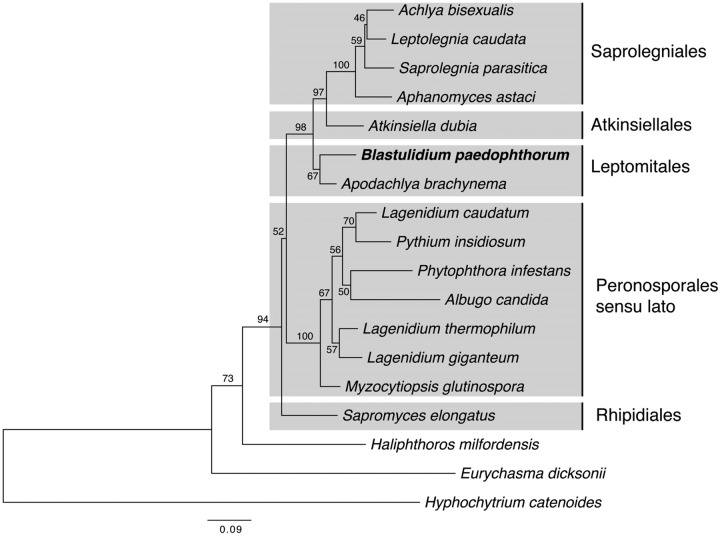
The combined analysis of the 18S and 28S gene regions places B. paedophthorum sister to Apodachlya with limited support (Fig. 3, 67% bootstrap support) but firmly places it with 98% bootstrap support in a basal position among what have been termed the Saprolegnian clades: Saprolegniales, Apodachlya, and Atkinsiella (39). Analysis of 18S and 28S regions, separately, allowed us to include larger numbers of related sequences. Unfortunately, few members of Leptomitales, the order in which Apodachlya is placed, have available DNA sequences. In the 18S phylogeny, B. paedophthorum groups again with Apodachlya and with a Chlamydomyzium sp. isolated from a Japanese rhabditid nematode (40) (see Fig. S1 in the supplemental material). In the 28S gene phylogeny, B. paedophthorum groups among a paraphyletic group of Leptomitales with little support for its precise placement (see Fig. S2 in the supplemental material). Overall, the morphology and phylogenetic placement of our brood parasite strongly suggests that it is Blastulidium paedophthorum and a member of the Leptomitales, as suggested by Dick (26).
FIG 3.
Phylogeny of the brood parasite, Blastulidium paedophthorum, based on a concatenated 18S and 28S rRNA alignment. Phylogeny was estimated by maximum likelihood using PhyML 3.1 with statistical support shown above nodes as bootstrap percentages estimated from 1,000 pseudoreplicated data sets. See Fig. S1 and S2 in the supplemental material for the GenBank accession numbers used.
B. paedophthorum prevalence in natural populations.
B. paedophthorum infections were observed in all six host species and all 15 lakes (Fig. 4). Pictures of infected hosts of three different host species are shown in (Fig. 1A, K, and L). B. paedophthorum was relatively common in all six host species (Fig. 4, left panels), although the lakes showed substantial variation in infection (Fig. 4, right panels). For the different host species, the median percentage of sampling dates on which B. paedophthorum was observed was 25.0 to 49.2%; for the different lakes, the median percentage of dates on which B. paedophthorum was observed was 5.56 to 70.8% (Fig. 4, top row). The maximum infection prevalences reached 4.9 to 8.7% of the entire population and 9.1 to 20% of the asexual adult female population (that is, of the population excluding juveniles, males and sexual females). Of the 522 B. paedophthorum-infected hosts observed during our field survey, only three were sexual females. (Data and code related to the field survey and the life table studies can be found at http://dx.doi.org/10.5281/zenodo.17804.)
FIG 4.
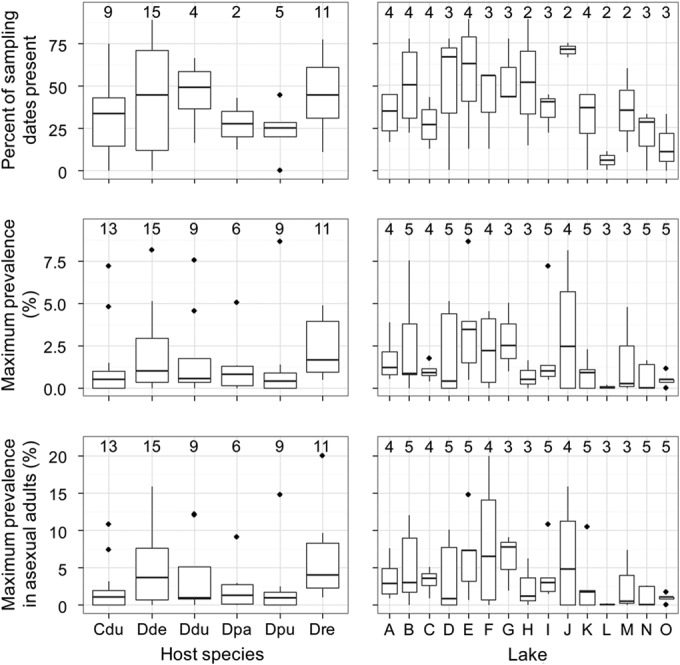
Abundance of B. paedophthorum in 6 host species (left panels) and 15 lake populations (right panels). The top row shows the percentage of sampling dates on which B. paedophthorum was found in a given host (left panel) or lake (right panel). The center row shows the maximum infection prevalence, and the bottom row shows the maximum infection prevalence in asexual adult females. For the panels on the left, the replicates are the lake populations; for the panels on the right, the replicates are the different host species contained within the lake. The number above the box plot indicates the number of samples summarized by the box; for example, the box plot of maximum prevalence of infection in Ceriodaphnia dubia is based on 13 lakes. Cdu, Ceriodaphnia dubia; Dde, Daphnia dentifera; Ddu, D. dubia; Dpa, D. parvula; Dpu, D. pulicaria; Dre, D. retrocurva; A, Appleton; B, Bishop; C, Bruin; D, Cedar; E, Crooked Pinckney; F, Crooked Waterloo; G, Gosling; H, Little Appleton; I, Mill; J, North; K, Pickerel; L, Sullivan; M, Walsh; N, Whitmore; O, Woodland.
B. paedophthorum virulence.
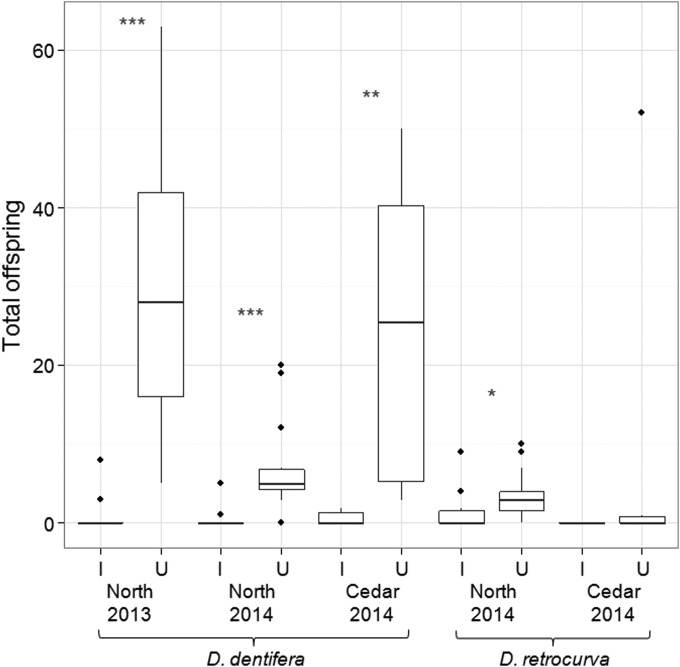
B. paedophthorum infection did not significantly affect host life span (Fig. 5) but did substantially reduce host fecundity (Fig. 6). The mean fecundity of uninfected D. dentifera was much greater than that of B. paedophthorum-infected D. dentifera (Table 2). The effects of infection on fecundity of D. retrocurva were much more modest (and not significant for the Cedar Lake life table). However, this may be due to a combination of low sample size (only four infected D. retrocurva in the Cedar life table) and low reproduction of uninfected hosts (Fig. 6, Table 2); D. retrocurva is difficult to culture in the lab, which may account for this result.
FIG 5.
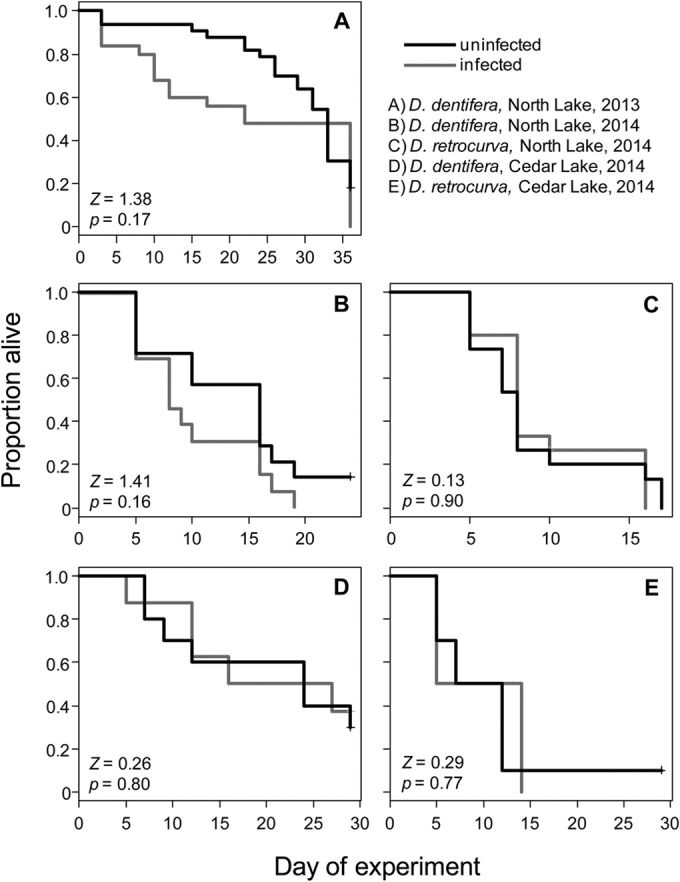
Survivorship of uninfected (black lines) and B. paedophthorum-infected (gray lines) field-collected Daphnia.
FIG 6.
Fecundity of field-collected B. paedophthorum-infected (I) and uninfected (U) Daphnia. Three of the comparisons are of D. dentifera, while two comparisons are of D. retrocurva. Individuals were collected from two lakes (Cedar and North Lakes) in 2 years (2013 and 2014). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Some infected individuals were able to reproduce. In North Lake, two of the infected D. dentifera successfully reproduced in 2013 and 2014 (8 and 15% of the total, respectively), and five (33%) of the infected D. retrocurva reproduced. The average reproduction of these individuals was 5.5 offspring for North Lake D. dentifera in 2013, 3.0 for North Lake D. dentifera, and 4.0 for North Lake D. retrocurva in 2014. No B. paedophthorum-infected individuals of either species collected from Cedar Lake successfully reproduced. No infections were observed in offspring born to infected individuals, suggesting that the parasite is not vertically transmitted.
Infection progression.
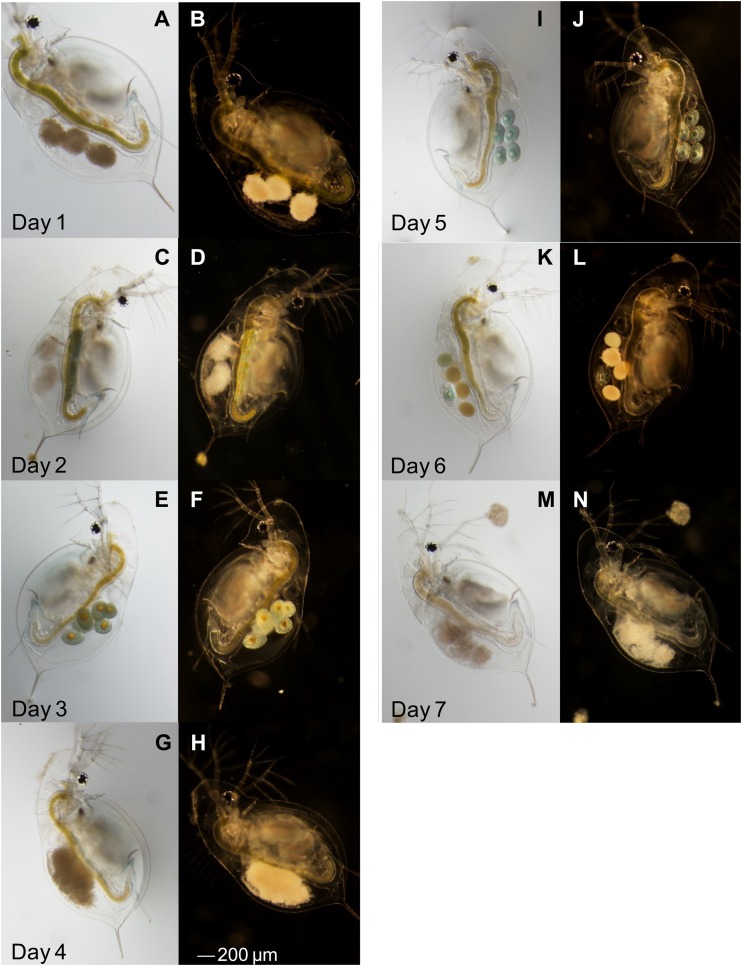
Individuals that did not clear infection continued to produce embryos, which were attacked by the oomycete within 24 h (Fig. 7). This indicates that B. paedophthorum-infected hosts continue to take up resources and allocate them to reproduction.
FIG 7.
Progression of infection in a single Daphnia dentifera individual. On each day, a photo was taken using light and dark microscopy. Images from the same day are shown side by side (e.g., images A and B were both taken on day 1). The D. dentifera had healthy-looking embryos on days 3 and 5 but was visibly infected on the other days.
Experimental infections.
Infections were transmitted in the lab, including between species and genera (Table 3), supporting that this is a multihost parasite (see also the molecular evidence discussed above). In addition, infections were transmitted even when the donor host was contained within a cage, supporting the idea that transmission occurs via a motile transmission stage that can pass through 500-μm-pore-size mesh.
TABLE 3.
Experimental infection species combinations and results
| Infection source | Infected host species | Uninfected host species | Uninfected host genotype(s) | No. of infectionsa |
|---|---|---|---|---|
| North Lake | D. dentifera | D. dentifera | GD 107 | 1/3* |
| D. dentifera | D. dentifera | GD 113 | 0/3 | |
| D. dentifera | D. dentifera | GD 115 | 1/3 | |
| D. dentifera | D. dentifera | Standard | 2/6* | |
| Cedar Lake | D. dentifera | C. dubia | 6A, GC 102, GC 104 | 2/4 |
| D. retrocurva | C. dubia | 6A, GC 102, GC 104 | 0/2 | |
| D. dentifera | D. dentifera | GD 107, GD 113, GD 115 | 3/4 | |
| D. retrocurva | D. dentifera | GD 107, GD 113, GD 115 | 0/2 | |
| D. dentifera | D. dentifera | BD 13, BD 16, WD 50 | 3/4 | |
| D. retrocurva | D. dentifera | BD 13, BD 16, WD 50 | 0/2 | |
| D. dentifera | D. dentifera | Standard | 2/4 | |
| D. retrocurva | D. dentifera | Standard | 0/2 | |
| D. dentifera | D. pulex × pulicaria | Geedey | 1/4 | |
| D. retrocurva | D. pulex × pulicaria | Geedey | 1/2 |
That is, the number of replicates with new infections/the total number of replicates. *, for one GD 107 beaker and two standard beakers the infected host was placed inside a cage, as described in Materials and Methods. New infections were seen in one of the standard beakers.
DISCUSSION
Recently, there has been an explosion in research on Daphnia-parasite interactions, including work that shows that parasites are key drivers of the evolution and population dynamics of Daphnia (2, 5, 41). Although this work has shown that Daphnia-parasite interactions can have important implications for aquatic populations, communities, and ecosystems, it has largely ignored a common type of parasite: brood parasites, which attack developing embryos. Pérez first described the brood parasite B. paedophthorum in 1903 (16), and future work focused on the morphology and development of B. paedophthorum (23, 24, 42, 43). Here, we show that a brood parasite in lakes in southeastern Michigan is B. paedophthorum. More importantly, we link the morphology of B. paedophthorum with its molecular sequence and information on its ecology and virulence. We show that B. paedophthorum is common in natural populations (occurring in all our study lakes and host species), is a multihost parasite, and has strong effects on host fecundity but not life span. Given this, we predict that it may be an important driver of Daphnia ecology and evolution.
Parasites that impact host fecundity but not life span, as is the case with B. paedophthorum, are expected to have the largest impact on host populations and can most easily regulate host populations (44). This is because infected adults consume substantial resources, but these resources do not contribute to the production of more host individuals. As a result, population size should decrease during epidemics. Given the centrality of Daphnia in freshwater food webs, if B. paedophthorum does depress host density (as seems likely, especially given the severity of some epidemics), this might influence ecosystem-level properties such as primary productivity and nutrient cycling.
Here, we showed that the same brood parasite can be found across multiple species and lakes. There are many similarities between the brood parasite in these southeastern Michigan lakes and B. paedophthorum descriptions from the literature, including a large central vacuole in the developing zoosporangia, the presence of dense body vesicles, orthogonal basal bodies, and chains of septate zoosporangia; these similarities confirm its taxonomic assignment to B. paedophthorum. Our phylogeny places B. paedophthorum clearly within the Saprolegnian clades. This assignment is also supported by conserved morphological features: the centrifugal formation of spores (very like Saprolegnia) and the direct (nonvesiculate) liberation of zoospores. B. paedophthorum is currently loosely classified near the Leptolegniellaceae (26) which is now in the order Leptomitales. The precise placement of B. paedophthorum is difficult, because only two of the genera in the order are represented by DNA sequence data (Apodachlya and Leptomitus), and neither is in the Leptolegniaceae. The original observations of Manier (24) were on a very similar parasite in a different daphniid (Simocephalus) on a different continent, which adds even more weight to the widespread, multihost nature of the parasite. Further molecular and developmental studies will be needed to determine whether there are multiple, cryptic species of Blastulidium.
Brood parasites have been reported periodically since Pérez first described B. paedophthorum. However, after Pérez (16, 43), only four studies assigned a species name to the brood parasite under study; all of those studies referred to the parasite being studied as B. paedophthorum (17, 23, 24, 42), based on morphological similarities. Notably, Green reported finding B. paedophthorum in both Europe and North America (in a pond in southwest Michigan). Three other studies referred to the parasite attacking the developing embryos as a member of the Saprolegniaceae; the basis for this assignment was not given by Little and Ebert (18), but the other two studies report hyphae running through and emerging from the developing embryos (22, 45). This morphological difference (hyphae versus the chains of thalli produced by B. paedophthorum), suggests that more than one species of oomycete is a daphniid brood parasite, a finding that was also suggested by Goren and Ben-Ami (21). In a molecular survey of Daphnia parasites, Wolinska et al. (46) reported observing brood parasites; all of their brood parasite sequences are affiliated with the Saprolegniales and clearly distinct from B. paedophthorum and other members of Leptomitales, further supporting the idea that there are multiple oomycetes that attack the developing embryos of Daphnia. Future studies should address the diversity of oomycete brood parasites, the amount of diversity within B. paedophthorum, and the geographic distributions of brood parasites of daphniids. Fortunately, molecular techniques should make addressing these questions relatively straightforward.
Because there has been little work on B. paedophthorum, we do not yet know whether host species or genotypes vary in their susceptibility to or tolerance of infection, nor do we know whether B. paedophthorum varies in infectivity or virulence. However, there is substantial variation in susceptibility of Daphnia to other infectious diseases (see, for example, references 47, 48, and 49), and two earlier studies found significant over- and underinfection of certain genotypes by a brood parasite (10, 18). Thus, it seems likely that there is genetic variation in susceptibility to B. paedophthorum. Indeed, such variation is hinted at by the recovery of some individuals from infections, but not others, and by variation in our infection experiments. In our study, the effects of B. paedophthorum on D. retrocurva fecundity were not as strong as on D. dentifera. However, our sample sizes were small for D. retrocurva. In addition, D. retrocurva can be very difficult to culture, and the relatively low reproduction of the uninfected D. retrocurva suggests that our estimates of effects of B. paedophthorum on fecundity might be conservative. Thus, further work needs to be done to rigorously assess intra- and interspecific variation in susceptibility and tolerance.
B. paedophthorum is likely to influence the ecology of Daphnia via more than just direct virulent effects. First, previous work found that infection with an unidentified brood parasite increased the susceptibility of Daphnia to infection by a bacterial parasite (19). Brood parasites, including B. paedophthorum, co-occur with numerous other parasites (see, for example, references 10, 20, and 21); in our study lakes, we observed infections of B. paedophthorum in populations at the same time as other parasites and occasionally noted individuals who were coinfected with B. paedophthorum and another parasite (e.g., Pasteuria ramosa, White Bacterial Disease, or Caullerya mesnili; M. Duffy, unpublished data). Future studies should address whether infection with B. paedophthorum alters susceptibility to other parasites, and whether dynamics of B. paedophthorum are strongly correlated (positively or negatively) with those of other parasites. Second, Daphnia parasites often influence host susceptibility to fish predation (11, 50). The increased susceptibility to fish predation most likely results from the increased opacity of infected hosts. Although B. paedophthorum-infected hosts are somewhat more opaque than uninfected hosts (Fig. 1), it is a much more modest increase in opacity than is seen with parasites that fill the daphniid body cavity, such as the yeast Metschnikowia bicuspidata and the bacterium Pasteuria ramosa. Thus, we predict that B. paedophthorum might increase the rates of fish predation on infected hosts, but much less so than do parasites that develop in the host hemolymph.
Finally, B. paedophthorum should provide an interesting study system for multihost parasites and examinations of how parasites influence host energetics. Most parasites can infect multiple hosts, and yet such parasites have been studied relatively little, in part due to tractability issues (51). Since several species of the Daphnia host can be cultured, as can Ceriodaphnia, this should provide a tractable study system in which to address important questions related to multihost parasites. In addition, comparisons of B. paedophthorum to other parasites should yield interesting insights into how parasites manipulate host energetics. The sterilizing pathogen Pasteuria ramosa alters energy use by hosts, causing them to shift resources from reproduction to growth, which increases parasite fitness (47, 52, 53). B. paedophthorum is similar to Pasteuria in that it reduces host fecundity but not life span and yet is very different in that its fitness is increased by the host continuing to produce clutches of embryos in which it can then develop. Interestingly, one study of a brood parasite reports that infected Daphnia hosts produced larger embryos than uninfected hosts (45), which supports a potential effect of infection on life history traits. Future studies contrasting the effects of B. paedophthorum and Pasteuria on host energy allocation could yield interesting insights into how parasites influence host life history traits.
Overall, we have shown here that B. paedophthorum, which was first described in the early 1900s in France, is a widespread, virulent, multihost parasite. Based on its effects on host individuals and its abundance in lake populations, we predict that this parasite is likely to have significant impacts on host populations, communities, and ecosystems and that it is likely to provide interesting insights into the ecology and evolution of parasitism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mandy Bromilow, Kailash Dhir, Camden Gowler, Dylan Grippi, Katie Hunsberger, Bella Oleksy, Catherine Searle, and Clara Shaw for assistance in the field. We also thank Katie Hunsberger, Bella Oleksy, and Serena Zhao for assistance in the lab and Gregg Sobocinski, Janine Maddock, and Alicia Pastor for assistance with the TEM. We thank Gordon Beakes for helpful discussions on the taxonomy and ultrastructure of oomycetes and Sandra Goutte, David Hardekopf, and Emile Moacdieh for assistance with the translation of earlier studies on B. paedophthorum.
This study was supported by NSF grants DEB-1305836 to M.A.D. and DEB-1354625 to T.Y.J. A.L.'s participation on the project was supported by the University of Michigan's Undergraduate Research Opportunities Program.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01369-15.
REFERENCES
- 1.Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. 2008. Homage to Linnaeus: how many parasites? How many hosts? Proc Natl Acad Sci U S A 105:11482–11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cáceres CE, Tessier AJ, Duffy MA, Hall SR. 2014. Disease in freshwater zooplankton: what have we learned and where are we going? J Plankton Res 36:326–333. doi: 10.1093/plankt/fbt136. [DOI] [Google Scholar]
- 3.Azam F, Fenchel T, Field JG, Gray JS, Meyerreil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10:257–263. doi: 10.3354/meps010257. [DOI] [Google Scholar]
- 4.Miki T, Takimoto G, Kagami M. 2011. Roles of parasitic fungi in aquatic food webs: a theoretical approach. Freshwater Biol 56:1173–1183. doi: 10.1111/j.1365-2427.2010.02562.x. [DOI] [Google Scholar]
- 5.Ebert D. 2005. Ecology, epidemiology, and evolution of parasitism in Daphnia. National Library of Medicine, National Center for Biotechnology Information, Bethesda, MD. [Google Scholar]
- 6.Ebert D. 2008. Host-parasite coevolution: Insights from the Daphnia-parasite model system. Curr Opin Microbiol 11:290–301. doi: 10.1016/j.mib.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Decaestecker E, Gaba S, Raeymaekers JAM, Stoks R, Van Kerckhoven L, Ebert D, De Meester L. 2007. Host-parasite “Red Queen” dynamics archived in pond sediment. Nature 450:870–873. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 8.Duffy MA, Brassil CE, Hall SR, Tessier AJ, Cáceres CE, Conner JK. 2008. Parasite mediated disruptive selection in a natural Daphnia population. BMC Evol Biol 8:80. doi: 10.1186/1471-2148-8-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy MA, Ochs JH, Penczykowski RM, Civitello DJ, Klausmeier CA, Hall SR. 2012. Ecological context influences epidemic size and parasite-mediated selection. Science 335:1636–1638. doi: 10.1126/science.1215429. [DOI] [PubMed] [Google Scholar]
- 10.Wolinska J, Spaak P. 2009. The cost of being common: evidence from natural Daphnia populations. Evolution 63:1893–1901. doi: 10.1111/j.1558-5646.2009.00663.x. [DOI] [PubMed] [Google Scholar]
- 11.Duffy MA, Hall SR. 2008. Selective predation and rapid evolution can jointly dampen effects of virulent parasites on Daphnia populations. Am Nat 171:499–510. doi: 10.1086/528998. [DOI] [PubMed] [Google Scholar]
- 12.Ebert D, Lipsitch M, Mangin KL. 2000. The effect of parasites on host population density and extinction: experimental epidemiology with Daphnia and six microparasites. Am Nat 156:459–477. doi: 10.1086/303404. [DOI] [PubMed] [Google Scholar]
- 13.Hall SR, Becker CR, Duffy MA, Cáceres CE. 2011. Epidemic size determines population-level effects of parasites. Oecologia 166:833–842. doi: 10.1007/s00442-011-1905-4. [DOI] [PubMed] [Google Scholar]
- 14.Ebert D. 2011. A genome for the environment. Science 331:539–540. doi: 10.1126/science.1202092. [DOI] [PubMed] [Google Scholar]
- 15.Duffy MA. 2007. Selective predation, parasitism, and trophic cascades in a bluegill-Daphnia-parasite system. Oecologia 153:453–460. doi: 10.1007/s00442-007-0742-y. [DOI] [PubMed] [Google Scholar]
- 16.Pérez C. 1903. Sur un organisme nouveau, Blastulidium paedophthorum, parasite des embryons de Daphnies. Comp Rend Séances Soc Biol Filiales 55:715–716. [Google Scholar]
- 17.Green J. 1974. Parasites and epibionts of Cladocera. Trans Zool Soc London 32:417–515. [Google Scholar]
- 18.Little TJ, Ebert D. 1999. Associations between parasitism and host genotype in natural populations of Daphnia (Crustacea: Cladocera). J Anim Ecol 68:134–149. [Google Scholar]
- 19.Tellenbach C, Wolinska J, Spaak P. 2007. Epidemiology of a Daphnia brood parasite and its implications on host life-history traits. Oecologia 154:369–375. [DOI] [PubMed] [Google Scholar]
- 20.Duffy MA, Cáceres CE, Hall SR, Tessier AJ, Ives AR. 2010. Temporal, spatial and between-host comparisons of patterns of parasitism in lake zooplankton. Ecology 91:3322–3331. doi: 10.1890/09-1611.1. [DOI] [PubMed] [Google Scholar]
- 21.Goren L, Ben-Ami F. 2013. Ecological correlates between cladocerans and their endoparasites from permanent and rain pools: patterns in community composition and diversity. Hydrobiologia 701:13–23. doi: 10.1007/s10750-012-1243-5. [DOI] [Google Scholar]
- 22.Bittner K. 2001. Parasitismus bei Daphnia im Bodensee. PhD thesis University of Constance, Constance, Germany. [Google Scholar]
- 23.Chatton E. 1908. Sur la reproduction et les affinités du Blastulidium paedophtorum Ch. Pérez. Comp Rend Séances Soc Biol Filiales 64:34–36. [Google Scholar]
- 24.Manier J-F. 1976. Cycle et ultrastructure de Blastulidium poedophthorum Pérez 1903 (Phycomycète Lagénidiale) parasite des oeufs de Simocephalus vetulus (Mull.) Schoedler (Crustacé, Cladocère). Protistologica 12:225–238. [Google Scholar]
- 25.Karling JS. 1981. Predominantly holocarpic and eucarpic simple biflagellate Phycomycetes. J. Cramer, Vaduz, Liechtenstein. [Google Scholar]
- 26.Dick MW. 2001. Straminipilous fungi: systematics of the peronosporomycetes, including accounts of the marine straminipilous protists, the plasmodiophorids, and similar organisms. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
- 27.James TY, Stenlid J, Olson A, Johannesson H. 2008. Evolutionary significance of imbalanced nuclear ratios within heterokaryons of the basidiomycete fungus Heterobasidion parviporum. Evolution 62:2279–2296. doi: 10.1111/j.1558-5646.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 28.Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal rRNA genes for phylogenetics, p 315–322. In Innis M, Gelfand DH, Sninsky J, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, Inc, San Diego, CA. [Google Scholar]
- 30.Molloy DP, Glockling SL, Siegfried CA, Beakes GW, James TY, Mastitsky SE, Wurdak E, Giamberini L, Gaylo MJ, Nemeth MJ. 2014. Aquastella gen. nov.: a new genus of saprolegniaceous oomycete rotifer parasites related to Aphanomyces, with unique sporangial outgrowths. Fungal Biol 118:544–558. doi: 10.1016/j.funbio.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Robideau GP, de Cock A, Coffey MD, Voglmayr H, Brouwer H, Bala K, Chitty DW, Desaulniers N, Eggertson QA, Gachon CMM, Hu CH, Kupper FC, Rintoul TL, Sarhan E, Verstappen ECP, Zhang YH, Bonants PJM, Ristaino JB, Levesque CA. 2011. DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol Ecol Resources 11:1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 34.Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 35.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 36.Core Team R. 2014. R: a language and environment for statistical computing, v3.1.2. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 37.Therneau TM, Lumley T. 2014. Survival: survival analysis, R package version 2.37-7. The Comprehensive R Archive Network, Institute for Statistics and Mathematics, Vienna, Austria: http://cran.r-project.org/web/packages/survival/index.html. [Google Scholar]
- 38.Hothorn T, Hornik K, van de Wiel MA, Zeileis A. 2014. Coin: conditional inference procedures in a permutation test framework, R package version 1.0-24. The Comprehensive R Archive Network, Institute for Statistics and Mathematics, Vienna, Austria: http://cran.r-project.org/web/packages/coin/index.html. [Google Scholar]
- 39.Beakes GW, Glockling SL, Sekimoto S. 2012. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma 249:3–19. doi: 10.1007/s00709-011-0269-2. [DOI] [PubMed] [Google Scholar]
- 40.Beakes GW, Glockling SL, James TY. 2014. A new oomycete species parasitic in nematodes, Chlamydomyzium dictyuchoides sp. nov: developmental biology and phylogenetic studies. Fungal Biol 118:527–543. doi: 10.1016/j.funbio.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG. 2012. Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proc R Soc B Biol Sci 279:1873–1882. doi: 10.1098/rspb.2011.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jírovec O. 1955. Cizopasníci našich perlooček II. Ceskoslovenská Parasitol II 2:95–98. [Google Scholar]
- 43.Pérez C. 1905. Nouvelles observations sur le Blastulidium paedophthorum. Comp Rend Séances Soc Biol Filiales 58:1027–1029. [Google Scholar]
- 44.Anderson RM, May RM. 1981. The population dynamics of microparasites and their invertebrate hosts. Philos Trans R Soc London B 291:451–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stazi AV, Mantovani A, Fuglieni F, Didelupis GLD. 1994. Observations on fungal infection of the ovary of laboratory cultured Daphnia magna. Bull Environ Contam Toxicol 53:699–703. [DOI] [PubMed] [Google Scholar]
- 46.Wolinska J, Giessler S, Koerner H. 2009. Molecular identification and hidden diversity of novel Daphnia parasites from European lakes. Appl Environ Microbiol 75:7051–7059. doi: 10.1128/AEM.01306-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Auld SKJR, Hall SR, Duffy MA. 2012. Epidemiology of a Daphnia-multiparasite system and its implications for the Red Queen. PLoS One 7:e39564. doi: 10.1371/journal.pone.0039564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carius HJ, Little TJ, Ebert D. 2001. Genetic variation in a host-parasite association: potential for coevolution and frequency-dependent selection. Evolution 55:1136–1145. doi: 10.1554/0014-3820(2001)055[1136:GVIAHP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 49.Decaestecker E, Vergote A, Ebert D, De Meester L. 2003. Evidence for strong host clone-parasite species interactions in the Daphnia microparasite system. Evolution 57:784–792. doi: 10.1554/0014-3820(2003)057[0784:EFSHCS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Duffy MA, Hall SR, Tessier AJ, Huebner M. 2005. Selective predators and their parasitized prey: are epidemics in zooplankton under top-down control? Limnol Oceanogr 50:412–420. doi: 10.4319/lo.2005.50.2.0412. [DOI] [Google Scholar]
- 51.Rigaud T, Perrot-Minnot M-J, Brown MJF. 2010. Parasite and host assemblages: embracing the reality will improve our knowledge of parasite transmission and virulence. Proc R Soc B Biol Sci 277:3693–3702. doi: 10.1098/rspb.2010.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cressler CE, Nelson WA, Day T, McCauley E. 2014. Starvation reveals the cause of infection-induced castration and gigantism. Proc R Soc B Biol Sci 281:20141087. doi: 10.1098/rspb.2014.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebert D, Carius HJ, Little T, Decaestecker E. 2004. The evolution of virulence when parasites cause host castration and gigantism. Am Nat 164:S19–S32. doi: 10.1086/424606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.