Abstract
Phenotypic heterogeneity describes the occurrence of “nonconformist” cells within an isogenic population. The nonconformists show an expression profile partially different from that of the remainder of the population. Phenotypic heterogeneity affects many aspects of the different bacterial lifestyles, and it is assumed that it increases bacterial fitness and the chances for survival of the whole population or smaller subpopulations in unfavorable environments. Well-known examples for phenotypic heterogeneity have been associated with antibiotic resistance and frequently occurring persister cells. Other examples include heterogeneous behavior within biofilms, DNA uptake and bacterial competence, motility (i.e., the synthesis of additional flagella), onset of spore formation, lysis of phages within a small subpopulation, and others. Interestingly, phenotypic heterogeneity was recently also observed with respect to quorum-sensing (QS)-dependent processes, and the expression of autoinducer (AI) synthase genes and other QS-dependent genes was found to be highly heterogeneous at a single-cell level. This phenomenon was observed in several Gram-negative bacteria affiliated with the genera Vibrio, Dinoroseobacter, Pseudomonas, Sinorhizobium, and Mesorhizobium. A similar observation was made for the Gram-positive bacterium Listeria monocytogenes. Since AI molecules have historically been thought to be the keys to homogeneous behavior within isogenic populations, the observation of heterogeneous expression is quite intriguing and adds a new level of complexity to the QS-dependent regulatory networks. All together, the many examples of phenotypic heterogeneity imply that we may have to partially revise the concept of homogeneous and coordinated gene expression in isogenic bacterial populations.
INTRODUCTION
Bacteria have evolved multiple strategies to cope with rapid and frequent changes in their environment. These survival strategies may include spore formation, increased production of polysaccharides, biofilm formation or escape from biofilms (i.e., switching from a motile into a sessile form and vice versa), altered motility, change of metabolic capabilities, response to antibiotics, and many more. In general, these switches are initiated by environmental or bacterially produced signals that are perceived by a diverse array of regulators and delegated into the corresponding regulatory networks. This presumably results in altered transcription levels of different genes or operons, which eventually causes a change in the phenotype and a response to the environmental stimulus. Within this context, it is generally assumed that the majority of a population will undergo this switch within a short time period and that the population will show a mostly homogeneous expression profile. During the last decade, it became, however, evident that a certain fraction of cells in an isogenic population behaves differently from the others, even though the environment may not have changed significantly. The term “phenotypic heterogeneity” thereby describes usually “nongenetic” variations that are observed between individual cells in a genetically homogeneous (i.e., isogenic) population (1). Within this context, the term “bistability” describes the situation in which the population has bifurcated into coexisting cell types (2).
It has been postulated that the ecological advantage of producing nonconformists is linked to a spreading-the-risk strategy (3–6). Thereby, switching is a statistical event as a result of noise and can additionally be an evolved event on the basis of a genetically encoded regulatory mechanism or a result of small genetic variations as part of an evolutionary process (7–9). It is further assumed that stochastic switching can be favored over sensing-dependent switching if the environment changes infrequently (10).
In this minireview, we highlight recent findings with respect to phenotypic heterogeneity. We place special emphasis on quorum-sensing (QS)-dependent phenotypic heterogeneity. For a broader overview on the topic of phenotypic heterogeneity, we provide in Table 1 a wide range of published examples observed in Gram-positive and Gram-negative bacteria. Furthermore, Fig. 1 gives a first glance at heterogeneous gene expression observed under a microscope for a plant symbiont, a human pathogen, and an aquatic bacterium in isogenic laboratory cultures. The increasing number of published examples implies that phenotypic heterogeneity is a broadly observed phenomenon within the bacterial world. It should be considered an important component in many regulatory networks.
TABLE 1.
Recent examples of nongenetically determined phenotypic heterogeneity and bistability in bacteria
| Phenotypic heterogeneity and/or bistability trait(s) | Microorganism(s) | Reference(s) |
|---|---|---|
| Presence of persister cells, resistance to antibiotics and heavy metals | Staphylococcus sp., E. coli, S. Typhimurium, P. aeruginosa, and others | 37–39, 41 and references herein |
| SOS responsea | E. coli, C. glutamicum | 54, 89 |
| Response to peptide antibiotics | B. subtilis | 90 |
| Prophage induction | C. glutamicum, S. oneidensis, S. pneumoniae | 54–56 |
| Quorum sensing, presence of autoinducer synthesis genes | V. campbellii, V. fischeri, L. monocytogenes, D. shibae, P. syringae, P. putida, S. fredii, S. meliloti | 13, 19, 23–25, 29, 30, 32–34, 91 |
| Arabinose utilization | E. coli | 63, 64 |
| Polyhydroxybutyrate utilization | S. meliloti | 92 |
| Secretion of related genes | S. fredii | 30 |
| Quorum quenching genes | S. fredii | 30 |
| Motility, secondary-flagellum formation | S. putrefaciens, S. Typhimurium | 68, 93 |
| Biofilm escape, motility after putisolvin production | P. putida | 33 |
| Genomic island excision/transfer | M. loti, Pseudomonas knackmussii | 28, 94, 95 |
| Bacterial competence, DNA uptake | B. subtilis, S. mutans | 71–74 |
| Sporulation | B. subtilis | 9, 96, 97 |
| Colony heterogeneity | S. aureus | 7 |
| Increased lag phase | E. coli | 8 |
| Surface pilus formation | S. pneumoniae | 98 |
| Myo-inositol utilization | S. enterica | 99 |
| Biofilm formation | S. enterica, B. subtilis | 100, 101 |
| Antibiotic production | Streptomyces coelicolor | 102 |
The SOS response is in part linked to the formation of persister cells (see reference 39 and references therein).
FIG 1.
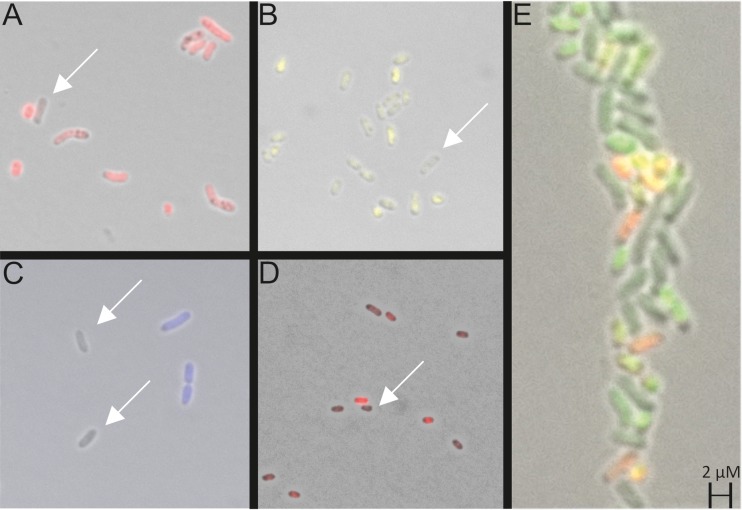
Phenotypic heterogeneous expression of various reporter gene fusions in plant-associated Sinorhizobium fredii NGR234, the human pathogen Stenotrophomonas maltophilia K279a, and the aquatic Janthinobacterium sp. strain HH102. Images are overlays of light and fluorescence microscopic pictures. Arrows indicate nonfluorescing cells. All strains carry low-copy-number plasmids with a promoter fused to a fluorescence protein. (A) S. fredii NGR234 carrying PngrI::dsRed2, a promoter fusion of the chromosomally encoded AI synthase gene in this organism (for further details, see reference 30); (B) S. fredii NGR234 carrying PtraI::mVenus, a promoter fusion of the second AI synthase gene of the organism in panel A; (C) S. maltophilia K279a carrying Pbla2::mCerulean, a promoter fusion of the beta lactamase 2 gene; (D) Janthinobacterium sp. HH102 carrying PvioA::mCherry, a promoter fusion of the vioA gene, the first gene of the violacein biosynthesis operon; (E) S. fredii NGR234 carrying PtraI::gfp and PngrI::dsRed2 promoter fusions on the same plasmid. All images were extracted from unpublished work from our laboratory.
BACTERIAL QS: A SYSTEM THOUGHT TO COORDINATE CELLS INSTEAD OF PRODUCING HETEROGENEOUS CELL BEHAVIOR
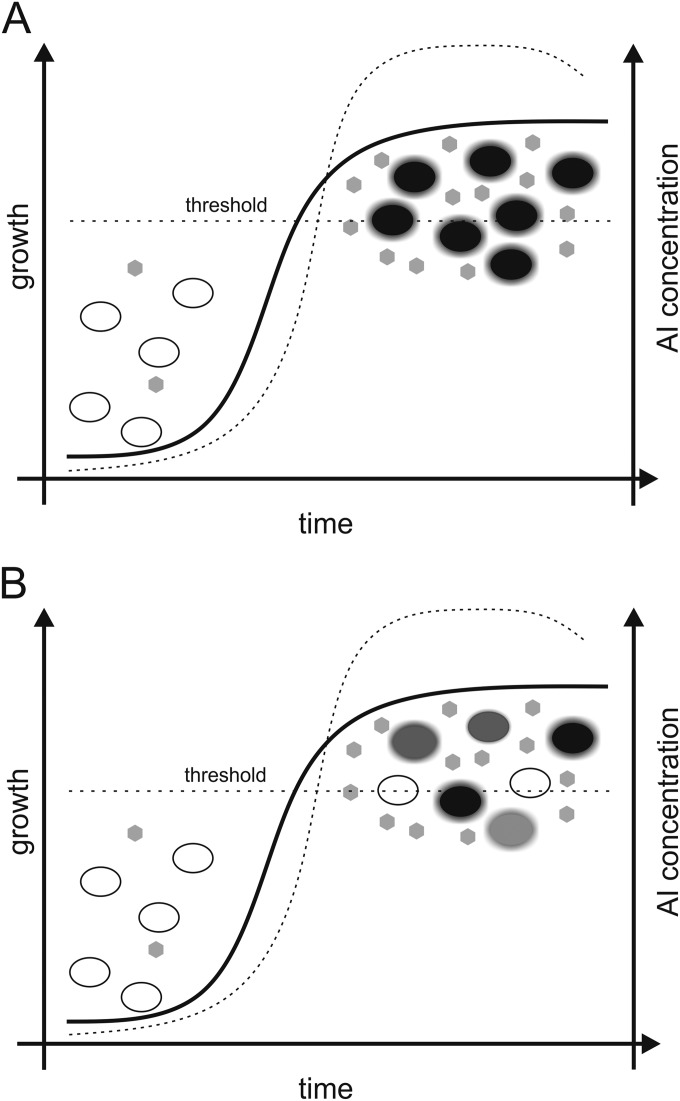
The term “quorum sensing” (QS) describes the intra- and interspecific cell-cell communication to sense population densities. For this, cells produce, release, and detect small diffusible molecules named autoinducers (AI). While Gram-negative bacteria release molecules like the homoserine lactones (HSLs), alpha-hydroxy-ketones, quinolone-like compounds, and others, Gram-positive bacteria produce and release autoinducing peptides. When a certain AI threshold concentration is reached, it is commonly accepted that the whole population collectively activates gene expression of QS-controlled genes in a well-coordinated manner (11, 12) (Fig. 2A). While this has been the classical view for many years, there is now growing evidence that QS-dependent responses of microorganism are not always homogenous (Fig. 2B).
FIG 2.
Two regulatory models that explain AI-dependent gene expression. (A) Classical textbook model of QS-dependent gene expression in Vibrio and other genera. Until recently, AI molecules were thought to be responsible for well-coordinated gene expression resulting in homogenous cell behavior. After reaching a certain threshold of AI concentration (dotted gray line) within a growing culture, all cells of the population turn on cell density-dependent genes like the lux genes in Vibrio and virtually all cells luminesce with the same signal intensity. (B) Only recently have independent research teams discovered a partly heterogeneous expression of QS-dependent genes in V. fischeri and V. campbellii. Not only did phenotypic heterogeneity in Vibrio result in the observation of on or off cells, cells also differed in the signal intensities throughout isogenic populations (13, 19, 24).
One of the first examples of a heterogeneous QS response has been reported for the model organism Vibrio fischeri and its QS-dependent bioluminescence (13). V. fischeri is a Gram-negative marine bacterium that frequently colonizes the light organs of fish and squid species (14, 15). Cell-cell communication in this microorganism has been studied very extensively over the last 4 decades, and it is perhaps one of the best-understood systems with respect to QS (16). V. fischeri encodes a single LuxI synthase that is involved in the synthesis of a 3-oxo-C6-HSL. Together with luxI, six genes are found in the same operon (luxICDABEG). Upstream of the lux operon, the corresponding regulator (i.e., luxR) is found (17). All together, these genes are involved in the biosynthesis of a luciferase protein, and their well-coordinated expression finally results in the observed bioluminescence through the reversible oxidation of a bacterially produced long-chain aldehyde and a reduced flavin mononucleotide. The expression of the V. fischeri lux operon is controlled through QS. At low cell densities, the lux genes are only weakly transcribed, whereas at high cell densities, the lux genes are transcribed at high levels. Light emission is observed only when a threshold concentration is reached, and until recently, it was assumed that virtually all cells would activate the lux operon's transcription, resulting in a cell density-dependent bioluminescence in isogenic populations (18) (Fig. 2A).
Interestingly, Perez and Hagan (13) provided evidence that individual cells differ widely in the onset of their bioluminescence and in their light intensities. Within their study, they convincingly demonstrated that on a time scale of 150 to 250 min, the bioluminescence of V. fischeri depends to a large extent on the exogenously added AI. However, the authors also described a large degree of cell-to-cell variability in their study. Obviously, many cells luminesced at very modest levels, while a small fraction of cells emitted much more brightly. The cells also responded on different time scales to the added AI. Based on these observations, the authors of that study postulate that the AI controls the overall behavior of the population but is less active at the single-cell level (13) (Fig. 2B).
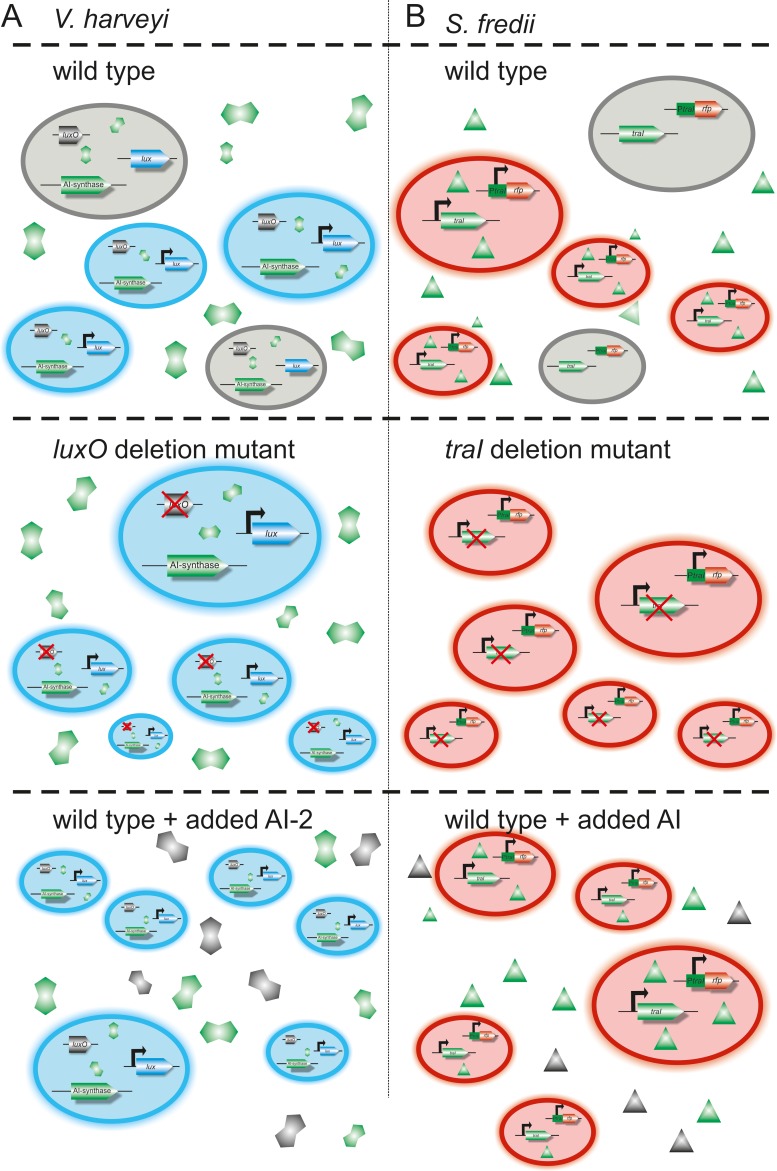
One of the first studies reporting phenotypic heterogeneity in the QS-dependent regulatory network within the genus Vibrio was published by Anetzberger and colleagues for Vibrio harveyi (recently reclassified as Vibrio campbellii) (19). In V. campbellii, expression of the lux operon is regulated through the concentration of the AI, which is an N-(3-hydroxy-butanol)-l-homoserine lactone (20). The bioluminescence in V. campbellii is heterogeneous, similar to the above-made observation for V. fischeri and in contrast to the general belief that a high AI concentration leads to synchronized gene expression in virtually all cells of genetically identical populations. Anetzberger and colleagues observed that despite high cell densities, only 69% of the cells produce bioluminescence. Additionally, the strength of the bioluminescence varies throughout the glowing cells. In the background of a luxO mutant, all cells luminesce (19) (Fig. 3A). LuxO is known to collect all information of the three AI-specific sensor kinases in V. campbellii and thereby to regulate the synthesis of four small RNAs that are involved in destabilizing the luxR transcript. LuxR acts as a master regulator in the QS-responsive network (21–23). Further studies implied that other AI-dependent genes are heterogeneously expressed in V. campbellii as well. Among these are the luxC, vscP, and vhp genes. AI-independent genes appear to be mostly homogeneously expressed in V. campbellii, in contrast to the heterogeneous expression of AI-dependent genes (24).
FIG 3.
QS-dependent phenotypic heterogeneity observed in the two model organisms Vibrio harveyi and Sinorhizobium fredii. (A, top) The wild type of V. harveyi shows phenotypic heterogeneity with respect to AI-controlled lux gene expression. (Middle) If the regulator luxO is deleted, all cells turn on. (Bottom) When externally added AI is present, all cells turn on. (B, top) The wild type of S. fredii shows phenotypic heterogeneity with respect to the AI synthase gene traI. (Middle) After deletion of the AI synthase, all cells turn on. (Bottom) Once externally added AI is present, all cells turn on. For further details, see references 23, 24, and 30.
Another interesting aspect of AI-linked heterogeneity was recently identified in the alphaproteobacterium Dinoroseobacter shibae (25). D. shibae is a member of the Roseobacter clade, which is highly abundant in marine habitats (26). Surprisingly, heterogeneous cell morphology in D. shibae is controlled in a QS-dependent manner. The D. shibae cell shape is highly variable, as it ranges from variously sized ovoid rods to long filaments, and this goes in parallel with a high copy number of chromosomes. Cells divide either by binary fission or by budding from the cell poles. D. shibae encodes in its genome three AI synthases and luxI homologues, which produce AI molecules ranging from C14- to C18-HSL. Unexpectedly, the deletion of only one of the three lux genes resulted in highly homogeneous cell morphologies. The deletion of the luxI1 gene abolished the synthesis of any AI molecule by this strain. Further, the authors of this study convincingly demonstrated that the phenotype of a luxI1 mutant could be restored by the addition of the corresponding C18-HSL at saturating concentrations. Additional transcriptome analyses in the background of this deletion mutant further supported the hypothesis of the AI-dependent morphological cell variability (25).
Within this framework, the QS-dependent DNA transfer and excision of a symbiotic island of Mesorhizobium loti has been observed. M. loti is a nitrogen-fixing alphaproteobacterium that is able to establish a symbiosis with legume plants. Its symbiotic genes (i.e., nod, nif, and fix) are found on a symbiotic island that has been designated ICEM/SymR7A (27). ICEM/SymR7A is excised and transferred in an AI concentration-dependent manner, during which the occurrence of excision events varies between single cells within an isogenic culture. In a laboratory culture of M. loti, the excision frequency of the symbiotic island was only 1% in the stationary growth phase compared to 40 to 70% in the exponential growth phase. The introduction of additional copies of traR, the regulator of the AI system, and the deletion of qseM, an antiactivator of the M. loti AI system, led to an upregulation of the symbiotic island excision to almost 100%. Additionally, the deletion of qseC, a modulator of qseM's transcription, resulted in a reduced excision frequency in individual cells and an increase in the level of the synthesized AI molecules (28).
The AI systems in Pseudomonas syringae and Xanthomonas campestris are heterogeneously expressed over a long time period of growth in laboratory cultures. The heterogeneity is not influenced by the addition of external AI, which leads to the conclusion that the heterogeneity of the AI systems of these two strains is not regulated in response to the AI concentration. The authors provided evidence that even the addition of 20 to 35 μM AI or diffusible signaling factor to the culture media did not significantly reduce the fraction of non-QS-responsive cells present in the population. In fact, they observed that in both organisms, the number of uninduced cells ranged from 18 to 25% (29). This is intriguing, since in the majority of all other studied organisms, the expression frequency at the single-cell level was affected by the concentration of the available AI.
In a recent report from our lab, we provided evidence that plant-released molecules can override AI signals that are involved in the regulation of phenotypic heterogeneity. For these studies, the alphaproteobacterium Sinorhizobium fredii NGR234 was chosen as a model organism. This organism is a plant symbiont that is able to fix nitrogen in the root nodules of many legumes. In batch cultures, the two AI systems, TraI/R and NgrI/R, are heterogeneously expressed (Fig. 1A and B) (30). Further work from our laboratory thereby implies that some cells turn on the traI gene and others the ngrI gene; however, only a few cells turn on both genes at the same time (Fig. 1E).
In contrast to the heterogeneous behavior of P. syringae and X. campestris, the heterogeneous behavior of the two S. fredii AI systems can be quenched by either the external addition of 50 μM AI or the deletion of the corresponding AI synthase gene (30). The upregulation of the AI systems in the background of the AI synthase deletion mutants was in part verified by population-wide transcriptome data (31). Not only the AI themselves but also the addition of crude plant root exudates or octopine as a plant-released molecule caused a more homogenous expression of the AI systems (30). Part of this study was confirmed by tests with the close relative Sinorhizobium meliloti using a sinI reporter gene fusion (32). Further investigations in S. fredii NGR234 indicated that quorum quenching genes and a type IV pilus gene cluster showed phenotypic heterogeneity that was independent of the influence of cell-cell communication signals (30).
Within this context, a recently published study on single-cell analyses of Pseudomonas putida strain IsoF implied that QS induces the production of the biosurfactant putisolvin, which triggers asocial motility of induced cells out of microcolonies. The biosurfactant synthesis is turned on by the stochastic switch of the AI system of P. putida. These observations lead to the assumption that in early growth stages, the QS signals are not released but rather directly bind to the AI receptors of the producing cell, resulting in an individually regulated gene expression. This in turn causes individual-based behavior in younger biofilms and not, as assumed for QS regulatory networks, to a group-based behavior. Removing cells with the highest AI concentration exerted a negative feedback on the left-behind cells (33).
Finally, it is noteworthy that heterogeneity with respect to QS is not restricted to Gram-negative bacteria. In the Gram-positive bacterium Listeria monocytogenes, a similar QS-dependent phenotypic heterogeneity could be observed (34). L. monocytogenes is a facultative intracellular food-borne pathogen that can cause severe infections in humans and animals. It is widely spread in the environment but often occurs in processed food samples (35). In this bacterium, the autoinducer is a peptide encoded by the agrD gene. agrD is found together with the agrB, -C, and -A genes in an operon. agrC encodes a two-component histidine kinase, AgrA a response regulator, and AgrB a protein that is involved in the processing of the precursor peptide agrD into a mature autoinducing peptide. Fusing the agr promoter region with a green fluorescent protein (GFP), Garmyn and colleagues provided evidence that the agr system is in general highly heterogeneously expressed in L. monocytogenes (34). They employed five different strains in their study, in which heterogeneity was observed with respect to the expression of a fused reporter for the agr genes. Therefore, the authors concluded that the agr system is in general subject to heterogeneous regulation. However, in different isolates, the percentages of “on” Agr cells significantly differed and ranged from 15 to 73%. It appeared that the overall ratio of off to on cells was strain dependent. The growth conditions, i.e., liquid culture or growth in biofilm flow cells, had a significant impact on the degree of heterogeneity as well. In fact, biofilm lifestyle appeared to increase the observed heterogeneity of L. monocytogenes cells.
All together, these many examples of QS-dependent phenotypic heterogeneity may demand a careful reviewing of the classical model of QS-dependent cell-cell communication in which virtually all cells are believed to be in either the on or the off mode (Fig. 2A). Thereby, the model might need only a better definition for the level of homogeneity, which is thought to occur in QS-dependent regulons (Fig. 2B).
PERSISTER CELLS AS A CLASSICAL MODEL FOR PHENOTYPIC HETEROGENEITY
A well-studied system with respect to phenotypic heterogeneity is the phenomenon of persister cells. This phenomenon was already observed in the mid-40s of the last century, when Bigger described the occurrence of Streptococcus pyogenes persister cells in an exponentially growing culture that had been treated with penicillin (36). In these experiments, a small fraction of a homogeneous population entered a distinct physiological state in which they were not killed by antibiotic treatment. Usually, these were less than 0.1% of the whole population. Much research has been focused on the exploration of the molecular keys linked to the persister phenomenon (37–41). Bertram and colleagues recently provided evidence that toxin-antitoxin (TA) systems play a key role in the regulatory network of persisters (42, 43). These systems consist of a toxin, which is normally a stable protein that interferes with vital cellular functions, and a cognate antitoxin, an unstable protein or RNA molecule, which regulates the toxin level. The most prominent example of a TA system controlling persistence is the Escherichia coli hipAB TA system. The newest results imply an inactivation of the Glu-tRNA synthase by the toxin HipA, which leads to an activation of RelA-mediated (p)pp(G)pp synthesis. The increased (p)pp(G)pp levels indirectly result in multidrug tolerance (44). The inactivation of HipA is mediated through the binding to HipB, which additionally leads to a conformational change of HipA (45). Essential to the regulatory effect of the hipAB system is a threshold concentration of HipA. When the threshold is exceeded, cells turn into persisters. This on and off switch is driven by the proteolysis of the antitoxin, by stochastic fluctuation, or by any change in the growth rate (39). Recently, the hipAB system of Shewanella oneidensis was characterized, and a ternary HipAB-DNA complex different from the one from E. coli was observed (46). Since persister cells are highly problematic with respect to antibiotic treatments of infectious diseases, much emphasis has been placed on the development of drugs that suppress the development and survival of persister cells (47–53).
PHENOTYPIC HETEROGENEOUS GENE EXPRESSION IS A WIDELY OBSERVED PHENOMENON
Phenotypic heterogeneity has been observed in a wide array of Gram-negative and Gram-positive bacteria. To give a better overview, we have assembled many published examples of phenotypic heterogeneity in Table 1. In the following section, we address a few selected phenomena that are subject to phenotypic heterogeneous regulation and have recently been uncovered.
SPONTANEOUS PROPHAGE INDUCTION AS A DRIVER OF PHENOTYPIC HETEROGENEITY
Recent studies with Corynebacterium glutamicum, Shewanella oneidensis, Salmonella sp., and Streptococcus pneumoniae indicated that lysis of a small number of cells by spontaneous prophage induction is beneficial to the remainder of the population and increases the overall fitness of the population with respect to survival and biofilm formation (54–57). In the Gram-positive, biotechnologically important microorganism C. glutamicum ATCC 13032, a spontaneously induced SOS response is partly responsible for the induction of prophages (54). The authors of that study observed a positive correlation between the spontaneous activation of the SOS response and the spontaneous induction of the prophage CGP. Interestingly, not every cell that turns on the SOS response induces prophage excision at the same time. An exceeded threshold of a certain, yet unknown, SOS response regulator is probably responsible for prophage induction in this bacterium (54).
S. pneumoniae is a potent biofilm producer that colonizes the upper respiratory tracts of humans and is the cause of many infections (58, 59). The majority of genomes of pneumococcal isolates showed a high prevalence of lysogenic bacteriophages (60, 61). Carrolo et al. convincingly demonstrated that biofilms formed by phage-infected bacteria are characterized by a higher-than-normal biomass and cell viability. Their data suggest that the DNA release of individual cells that are lysed after phage induction results in more-robust biofilms (55).
Likewise, the biofilm of S. oneidensis MR-1 is stabilized by external DNA (eDNA) that is released by individual cells after phage lysis. S. oneidensis MR-1 encodes three prophages in its genome, which are activated through UV radiation and other stresses, like ionizing radiation (62). Spontaneous phage lysis produces significant amounts of free eDNA, which plays a major role during surface attachment and development of three-dimensional biofilm structures, with which the different prophages contribute to different stages of biofilm formation (56).
METABOLIC ACTIVITIES ARE NOT ALWAYS UNIFORM IN BATCH CULTURES
Yet another recent example for heterogeneous expression of genes linked to metabolic traits concerns the arabinose utilization system in E. coli (63, 64). With quantitative time-lapse microscopy and by challenging E. coli cells with very low concentrations of arabinose, a time delay in the response of single cells to arabinose was discovered. This delay corresponded directly to the arabinose concentration. In this study, cells were trapped in microfluidic devices, and the authors showed that heterogeneity was linked to the uptake of externally supplied arabinose. The established stochastic model allowed the conclusion that the heterogeneous timing of gene induction was related to a broad distribution of uptake proteins expressed by the cell at the time of arabinose addition. Further, they showed that the off switching triggered by the sudden removal of arabinose is quite homogeneous and fast. The rather quick off switch was independent of internal arabinose degradation. Within this context, it is noteworthy that the usual noise generated is a result not only of regulation of the level of mRNA production but also of the level of enzyme activity per se. Therefore, fluctuations during the expression and folding of catabolically active enzymes can propagate and cause significant growth fluctuations in E. coli when it is grown on lactulose (65).
SOME CELLS MOVE DIFFERENTLY THAN OTHERS
Chemotaxis and motility have, as well, been reported to be subject to phenotypic heterogeneity, and it was suggested that the behavior of cells exhibited larger temporal variations at time scales ranging from seconds to minutes (66, 67). Within this context, Bubendorfer and colleagues recently reported on the secondary Shewanella putrefaciens CN-32 flagellar system, which is induced under planktonic conditions within a distinct subpopulation. This results in the formation of at least one lateral flagellum in addition to the primary polar flagellum and is advantageous to the cell with respect to bacterial spreading. Cells that express the secondary filaments were more fit with respect to spreading on soft-agar plates and through medium-filled channels despite having lower swimming speeds than their mono-flagellated counterparts. It is likely that the second flagellum is advantageous for directional movement and ultimately leads to a more efficient chemotaxis (68).
BACTERIAL COMPETENCE IS HETEROGENEOUS WITHIN ISOGENIC POPULATIONS
Bacterial competence is yet another process for which bistability and phenotypic heterogeneity can be observed. Natural competence is a complex process controlled by a regulatory network that ultimately allows the uptake of exogenous DNA from the environment. For detailed reviews, see references 69 and 70. Bacillus subtilis, Streptococcus pneumoniae, and Streptococcus mutans have been used as model organisms to characterize heterogeneity during this process. In all three bacterial strains, only a small fraction of the cells within an isogenic population will eventually be competent (71–74). For B. subtilis, a certain threshold concentration of ComK, the key competence-associated transcription factor, is responsible for an autostimulatory process that finally leads to genetic competence (74, 75).
Competence in S. pneumoniae is triggered by the secreted peptide pheromone competence-stimulating peptide (CSP), encoded by comC. CSP is bound by the receptor ComD, which forms a two-component signal transduction system with ComE and thereby responds to the external CSP concentration. Additionally, ComE activates the transcription of comAB and comCDE, inducing a positive-feedback loop that drives the cell toward the competent state. Cells that are not in the competent state are attacked and lysed by competent cells (reference 76 and references herein). This phenomenon is responsible for the observed heterogeneity.
Competence in S. mutans is regulated in a similar way. The competence-induced population of S. mutans segregates into autolysing cells and cells that become competent due to an imbalance in the CipB/CipI ratio. Besides these two cell fractions, a third fraction of cells that is not competent and carries low levels of ComE is observed (72, 73). The newest research results provide evidence that small peptides also play a key role in the competence regulation of S. mutans (77).
MEASURING PHENOTYPIC HETEROGENEITY AT A SINGLE-CELL LEVEL
Demonstrating the development of single cells within a population and thereby monitoring the fate of individual cells and their heterogeneous behavior over time are essential parameters for assessing the impact of phenotypic heterogeneity. Well-established methods for measuring phenotypic heterogeneity are time-lapse fluorescence microscopy and flow cytometric analyses or a combination of both. Additionally, the development of microfluidic devices and cell traps has been very helpful for data evaluation and single-cell tracking (72, 78–81).
With the exception of the studies on Vibrio in which naturally occurring bioluminescence was recorded, the majority of published studies have made use of fluorescence proteins (i.e., variants of the GFP) for the detection of gene expression at a single-cell level (Table 1). Mainly because of the high signal intensity needed for the detection of single cells for many of these studies, low-, medium-, or high-copy-number vectors were used. Therefore, the use of reporter genes in self-replicable plasmids might in part be problematic with respect to the regulation and expression of the target genes through the corresponding regulators, usually encoded on the bacterial chromosome in a single copy. This could be a problem especially when single-copy regulators are involved and their binding sites within the targeted promoter are outcompeted by the binding sites on the high-copy-number plasmids. Further, the maturation time of the fluorescing proteins employed will most likely have an impact on the outcome of the study (82, 83).
Besides the use of fluorescence proteins, the use of the fluorescing dyes Syto 9, propidium iodide, and 3,3′-diethyloxacarbocyanine iodide [DiOC2(3)] proved to be useful for the identification of subpopulations with reduced viability and membrane potential within C. glutamicum populations (84). Other studies employed in situ reverse transcription-PCRs at a single-cell level in combination with a single-cell beta-galactosidase assay to monitor heterogeneous expression of the lac operon in Salmonella enterica serovar Typhimurium (85). The detection of single mRNA molecules in individual cells might also be useful for analysis of phenotypic heterogeneity (86).
Within this context, culture conditions will also have a major impact on the outcome of any study, especially since it is well known that laboratory cultures cannot be considered truly adequate models for natural systems; besides, high variations of environmental parameters are known to occur with often-stable subpopulations in batch cultures. In particular, oxygen limitations and gradients of oxygen might affect the outcomes of the studies. This will be true for many of the GFP-based studies, as oxygenation of a tyrosine at position 67 is a rate-limiting step in its fluorescence (87). Further, is should be kept in mind that the bioluminescence of Vibrio also depends on oxygen availability due to the catalytic mechanism employed by the luciferase (88).
IS PHENOTYPIC HETEROGENEITY ALWAYS ONLY PHENOTYPIC?
With respect to the observation of phenotypic heterogeneity in a wide range of bacteria (Table 1), the question of how phenotypic the observed heterogeneity is may arise. It is likely that the observed phenomenon is in fact often related to genotypic changes caused by single nucleotide polymorphisms (SNPs) that potentially lead to a different phenotypic outcome for a certain subpopulation. One such example has been reported for the growth of Staphylococcus aureus on a laboratory medium with a high Mg2+ content (7). The parent strain divides into three subpopulations when growing on these plates: an orange center region (O) and a white (W) and a yellowish (Y) surrounding region. The transcriptional changes in the W strain match a point mutation in the RsbW anti-sigma factor. This leads to a loss of function of the kinase active residue, which in turn results in a nonfunctional σB complex. A σB deletion mutant shows the same QS hyperactivation phenotype as the W substrain, which leads to a growth advantage over the parent strain. Additionally, the Y strain revealed mutations in the GraRS and WalKR two-component systems causing a higher-than-normal resistance capacity against vancomycin and the bacteriocin Bsa (7). Yet another example of genomic mutations leading to phenotypic heterogeneity has recently been described for E. coli cultures that had been repeatedly treated with ampicillin. Sequencing of antibiotic-tolerant strains identified various mutations leading to stable subpopulations with altered growth behavior (8). These are only a very few and very recent examples of phenotypic heterogeneity that is caused by genetic changes instead of a stochastic or regulatory switch, and it is likely that others that uncover phenotypic as genotypic heterogeneity will follow.
Independently from the mechanisms through which phenotypic heterogeneity arises within a population, the many examples that have been published over the last 5 years indicate the wide occurrence and general importance of phenotypic heterogeneity for coordinated bacterial behavior. All together, these examples have significantly advanced our knowledge of the molecular keys involved in generating phenotypic heterogeneity within isogenic and homogeneous cultures. Because of the frequent occurrence of phenotypic heterogeneity, we may need to ask if truly homogenous and isogenic cultures exist at all. Within this framework, it is perhaps safe to speculate that phenotypic heterogeneity is a very common phenomenon that occurs in virtually any bacterial culture even though it is believed to be homogeneous, as this was the case for the QS-dependent processes.
ACKNOWLEDEGMENTS
This work was in part kindly funded by the Deutsche Forschungsgemeinschaft through grant STR451/7-1 within the SPP1617 priority program.
We are grateful to Thorsten Mascher for critical reading of the manuscript.
REFERENCES
- 1.Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annu Rev Genet 42:253–268. doi: 10.1146/annurev.genet.42.110807.091601. [DOI] [PubMed] [Google Scholar]
- 2.Dubnau D, Losick R. 2006. Bistability in bacteria. Mol Microbiol 61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- 3.Dhar N, McKinney JD. 2007. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol 10:30–38. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Smits WK, Kuipers OP, Veening JW. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat Rev Microbiol 4:259–271. doi: 10.1038/nrmicro1381. [DOI] [PubMed] [Google Scholar]
- 5.de Jong IG, Haccou P, Kuipers OP. 2011. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays 33:215–223. [DOI] [PubMed] [Google Scholar]
- 6.Veening JW, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 7.Koch G, Yepes A, Förstner KU, Wermser C, Stengel ST, Modamio J, Ohlsen K, Foster KR, Lopez D. 2014. Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158:1060–1071. doi: 10.1016/j.cell.2014.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 9.Siebring J, Elema MJH, Vega FD, Kovács ÁT, Haccou P, Kuipers OP. 2014. Repeated triggering of sporulation in Bacillus subtilis selects against a protein that affects the timing of cell division. ISME J 8:77–87. doi: 10.1038/ismej.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kussell E, Kishnoy R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waters CM, Bassler BL. 2006. The Vibrio harveyi quorum-sensing system uses shared regulatory components to discriminate between multiple autoinducers. Genes Dev 20:2754–2767. doi: 10.1101/gad.1466506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perez PD, Hagen SJ. 2010. Heterogeneous response to a quorum-sensing signal in the luminescence of individual Vibrio fischeri. PLoS One 5:e15473. doi: 10.1371/journal.pone.0015473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Visick KL, Foster J, Doino J, McFall-Ngai MJ, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–4586. doi: 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visick KL, McFall-Ngai MJ. 2000. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol 182:1779–1787. doi: 10.1128/JB.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruby EG. 1996. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol 50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 17.Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroph P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A 102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyashiro T, Ruby EG. 2012. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol 84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anetzberger C, Pirch T, Jung K. 2009. Heterogeneity in quorum sensing-regulated bioluminescence of Vibrio harveyi. Mol Microbiol 73:267–277. doi: 10.1111/j.1365-2958.2009.06768.x. [DOI] [PubMed] [Google Scholar]
- 20.Cao JG, Meighen EA. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem 264:21670–21676. [PubMed] [Google Scholar]
- 21.Freeman JA, Bassler BL. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol 31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- 22.Tu KC, Bassler BL. 2007. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Gene Dev 21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anetzberger C, Reiger M, Fekete A, Schell U, Stambrau N, Plener L, Kopka J, Schmitt-Kopplin P, Hilbi H, Jung K. 2012. Autoinducers act as biological timers in Vibrio harveyi. PLoS One 7:e48310. doi: 10.1371/journal.pone.0048310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anetzberger C, Schell U, Jung K. 2012. Single cell analysis of Vibrio harveyi uncovers functional heterogeneity in response to quorum sensing signals. BMC Microbiol 12:209. doi: 10.1186/1471-2180-12-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patzelt D, Wang H, Buchholz I, Rohde M, Gröbe L, Pradella S, Neumann A, Schulz S, Heyber S, Münch K, Münch R, Jahn D, Wagner-Döbler I, Tomasch J. 2013. You are what you talk: quorum sensing induces individual morphologies and cell division modes in Dinoroseobacter shibae. ISME J 7:2274–2286. doi: 10.1038/ismej.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner-Döbler I, Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60:255–280. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan JT, Ronson CW. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A 95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsay JP, Major AS, Komarovsky VM, Sullivan JT, Dy RL, Hynes MF, Salmond GPC, Ronson CW. 2013. A widely conserved molecular switch controls quorum sensing and symbiosis island transfer in Mesorhizobium loti through expression of a novel antiactivator. Mol Microbiol 87:1–13. doi: 10.1111/mmi.12079. [DOI] [PubMed] [Google Scholar]
- 29.Pradhan BB, Chatterjee S. 2014. Reversible non-genetic phenotypic heterogeneity in bacterial quorum sensing. Mol Microbiol 92:557–569. doi: 10.1111/mmi.12575. [DOI] [PubMed] [Google Scholar]
- 30.Grote J, Krysciak D, Schorn A, Dahlke RI, Soonvald L, Müller J, Hense BA, Schwarzfischer M, Sauter M, Schmeisser C, Streit WR. 2014. Evidence of autoinducer-dependent and -independent heterogeneous gene expression in Sinorhizobium fredii NGR234. Appl Environ Microbiol 80:5572–5582. doi: 10.1128/AEM.01689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krysciak D, Grote J, Rodriguez Orbegoso M, Utpatel C, Förstner K, Li L, Schmeisser C, Krishnan HB, Streit WR. 2014. RNA sequencing analysis of the broad-host-range strain Sinorhizobium fredii NGR234 identifies a large set of genes linked to quorum sensing-dependent regulation in the background of a traI and ngrI deletion mutant. Appl Environ Microbiol 80:5655–5671. doi: 10.1128/AEM.01835-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlüter JP, Czuppon P, Schauer O, Pfaffelhuber P, McIntosh M, Becker A. 2015. Classification of phenotypic subpopulations in isogenic bacterial cultures by triple promoter probing at single cell level. J Biotechnol 198:3–14. doi: 10.1016/j.jbiotec.2015.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Cárcamo-Oyarce G, Lumjiaktase P, Kümmerli R, Eberl L. 2015. Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat Commun 6:5945. doi: 10.1038/ncomms6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garmyn D, Gal L, Briandet R, Guilbaud M, Lemaître JP, Hartmann A, Piveteau P. 2011. Evidence of autoinduction heterogeneity via expression of the Agr system of Listeria monocytogenes at the single-cell level. Appl Environ Microbiol 77:6286–6289. doi: 10.1128/AEM.02891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuppler M, Loessner MJ. 2010. The opportunistic pathogen Listeria monocytogenes: pathogenicity and interaction with the mucosal immune system. Int J Inflamm 2010:704321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bigger JW. 1944. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497–500. doi: 10.1016/S0140-6736(00)74210-3. [DOI] [Google Scholar]
- 37.Conlon BP. 2014. Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells. Bioessays 36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- 38.Helaine S, Kugelberg E. 2014. Bacterial persisters: formation, eradication, and experimental systems. Trends Microbiol 22:417–424. doi: 10.1016/j.tim.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Maisonneuve E, Gerdes K. 2014. Molecular mechanisms underlying bacterial persisters. Cell 157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 40.Prax M, Bertram R. 2014. Metabolic aspects of bacterial persisters. Front Cell Infect Microbiol 4:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis K. 2010. Persister cells. Annu Rev Microbiol 64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 42.Bertram R, Schuster CF. 2014. Post-transcriptional regulation of gene expression in bacterial pathogens by toxin-antitoxin systems. Front Cell Infect Microbiol 4:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuster CF, Bertram R. 2013. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett 340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 44.Germain E, Castro-Roa D, Zenkin N, Gerdes K. 2013. Molecular mechanism of bacterial persistence by HipA. Mol Cell 52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen Y, Behiels E, Felix J, Elegheert J, Vergauwen B, Devreese B, Savvides SN. 2014. The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence. Nucleic Acids Res 42:10134–10147. doi: 10.1093/nar/gku665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay JP, Defraine V, Michiels J, Cenes Q, Aertsen A, Miller S, Lavigne R. 2014. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:3774–3784. doi: 10.1128/AAC.02668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murima P, McKinney JD, Pethe K. 2014. Targeting bacterial central metabolism for drug development. Chem Biol 21:1423–1432. doi: 10.1016/j.chembiol.2014.08.020. [DOI] [PubMed] [Google Scholar]
- 50.Lebeaux D, Chauhan A, Létoffé S, Fischer F, de Reuse H, Beloin C, Ghigo JM. 2014. pH-mediated potentiation of aminoglycosides kills bacterial persisters and eradicates in vivo biofilms. J Infect Dis 210:1357–1366. doi: 10.1093/infdis/jiu286. [DOI] [PubMed] [Google Scholar]
- 51.Marques CNH, Morozov A, Planzos P, Zelaya HM. 2014. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Appl Environ Microbiol 80:6976–6991. doi: 10.1128/AEM.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Starkey M, Lepine F, Maura D, Bandyopadhaya A, Lesic B, He J, Kitao T, Righi V, Milot S, Tzika A, Rahme L. 2014. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog 10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim JH, O'Brien K, Sharma R, Boshoff HIM, Rehren G, Chakraborty S, Wallach JB, Monteleone M, Wilson DJ, Aldrich CC, Barry CE III, Rhee KY, Ehrt S, Schnappinger D. 2013. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci U S A 110:19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nanda AM, Heyer A, Krämer C, Grünberger A, Kohlheyer D, Frunzke J. 2014. Analysis of SOS-induced spontaneous prophage induction in Corynebacterium glutamicum at the single-cell level. J Bacteriol 196:180–188. doi: 10.1128/JB.01018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carrolo M, Frias MJ, Rodrigues Pinto F, Melo-Cristino J, Ramirez M. 2010. Prophage spontaneous activation promotes DNA release enhancing biofilm formation in Streptococcus pneumoniae. PLoS One 5:e15678. doi: 10.1371/journal.pone.0015678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gödeke J, Paul K, Lassak J, Thormann KM. 2011. Phage-induced lysis enhances biofilm formation in Shewanella oneidensis MR-1. ISME J 5:613–626. doi: 10.1038/ismej.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bossi L, Fuentes JA, Mora G, Figuerosa-Bossi N. 2003. Prophage contribution to bacterial population dynamics. J Bacteriol 185:6467–6471. doi: 10.1128/JB.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sleeman KL, Griffiths D, Shackley F, Diggle L, Gupta S, Maiden MC, Moxon ER, Crook DW, Peto TEA. 2006. Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J Infect Dis 194:682–688. doi: 10.1086/505710. [DOI] [PubMed] [Google Scholar]
- 59.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O'Brien K, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. doi: 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramirez M, Severina E, Tomasz A. 1999. A high incidence of prophage carriage among natural isolates of Streptococcus pneumoniae. J Bacteriol 181:3618–3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Severina E, Ramirez M, Tomasz A. 1999. Prophage carriage as a molecular epidemiological marker in Streptococcus pneumoniae. J Clin Microbiol 37:3308–3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotechnol 20:1118–1123. doi: 10.1038/nbt749. [DOI] [PubMed] [Google Scholar]
- 63.Megerle JA, Fritz G, Gerland U, Jung K, Rädler JO. 2008. Timing and dynamics of single cell gene expression in the arabinose utilization system. Biophys J 95:2103–2115. doi: 10.1529/biophysj.107.127191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fritz G, Megerle J, Westermayer S, Brick D, Heermann R, Jung K, Rädler JO, Gerland U. 2014. Single cell kinetics of phenotypic switching in the arabinose utilization system of E. coli. PLoS One 9:e89532. doi: 10.1371/journal.pone.0089532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kiviet DJ, Nghe P, Walker N, Boulineau S, Sunderlikova V, Tans SJ. 2014. Stochasticity of metabolism and growth at the single-cell level. Nature 514:376–379. doi: 10.1038/nature13582. [DOI] [PubMed] [Google Scholar]
- 66.Korobkova E, Emonet T, Vilar JMG, Shimizu TS, Cluzel P. 2004. From molecular noise to behavioural variability in a single bacterium. Nature 428:574–578. doi: 10.1038/nature02404. [DOI] [PubMed] [Google Scholar]
- 67.Spudich JL, Koshland DE. 1976. Non-genetic individuality: chance in the single cell. Nature 262:467–471. doi: 10.1038/262467a0. [DOI] [PubMed] [Google Scholar]
- 68.Bubendorfer S, Koltai M, Rossmann F, Sourjik V, Thormann KM. 2014. Secondary bacterial flagellar system improves bacterial spreading by increasing the directional persistence of swimming. Proc Natl Acad Sci U S A 111:11485–11490. doi: 10.1073/pnas.1405820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mell JC, Redfield RJ. 2014. Natural competence and the evolution of DNA uptake specificity. J Bacteriol 196:1471–1483. doi: 10.1128/JB.01293-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seitz P, Blokesch M. 2013. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol Rev 37:336–363. doi: 10.1111/j.1574-6976.2012.00353.x. [DOI] [PubMed] [Google Scholar]
- 71.Hamoen LW, Venema G, Kuipers OP. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 72.Son MJ, Ahn SJ, Guo Q, Bume RA, Hagen SJ. 2012. Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol Microbiol 86:258–272. doi: 10.1111/j.1365-2958.2012.08187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lemme A, Gröbe L, Reck M, Tomasch J, Wagner-Döbler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J Bacteriol 193:1863–1877. doi: 10.1128/JB.01363-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smits WK, Eschervins CC, Susanna KA, Bron S, Kuipers OP, Hamoen LW. 2005. Stripping Bacillus: ComK auto-stimulation is responsible for the bistable response in competence development. Mol Microbiol 56:604–614. doi: 10.1111/j.1365-2958.2005.04488.x. [DOI] [PubMed] [Google Scholar]
- 75.van Sinderen D, Luttinger A, Kong L, Dubnau D, Venema G, Hamoen L. 1995. comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol Microbiol 15:455–462. doi: 10.1111/j.1365-2958.1995.tb02259.x. [DOI] [PubMed] [Google Scholar]
- 76.Johnsborg O, Havarstein LS. 2009. Regulation of natural genetic transformation and acquisition of transforming DNA in Streptococcus pneumoniae. FEMS Microbiol Rev 33:627–642. doi: 10.1111/j.1574-6976.2009.00167.x. [DOI] [PubMed] [Google Scholar]
- 77.Ahn SJ, Kaspar J, Kim JN, Seaton K, Burne RA. 2014. Discovery of novel peptides regulating competence development in Streptococcus mutans. J Bacteriol 196:3735–3745. doi: 10.1128/JB.01942-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Probst C, Grünberger A, Wiechert W, Kohlheyer D. 2013. Microfluidic growth chambers with optical tweezers for full spatial single-cell control and analysis of evolving microbes. J Microbiol Methods 95:470–476. doi: 10.1016/j.mimet.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 79.Probst C, Grünberger A, Wiechert W, Kohlheyer D. 2013. Polydimethylsiloxane (PDMS) sub-micron traps for single-cell analysis of bacteria. Micromachines 4:357–369. doi: 10.3390/mi4040357. [DOI] [Google Scholar]
- 80.Unthan S, Grünberger A, van Ooyen J, Gätgens J, Heinrich J, Paczia N, Wiechert W, Kohlheyer D, Noack S. 2014. Beyond growth rate 0.6: what drives Corynebacterium glutamicum to higher growth rates in defined medium? Biotechnol Bioeng 111:359–371. doi: 10.1002/bit.25103. [DOI] [PubMed] [Google Scholar]
- 81.Stratz S, Eyer K, Kurth F, Dittrich PS. 2014. On-chip enzyme quantification of single Escherichia coli bacteria by immunoassay-based analysis. Anal Chem 86:12375–12381. doi: 10.1021/ac503766d. [DOI] [PubMed] [Google Scholar]
- 82.Bevis BJ, Glick BS. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 83.Zhang J, Campbell RE, Ting AY, Tsien RY. 2002. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol 3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 84.Neumeyer A, Hübschmann T, Müller S, Frunzke J. 2013. Monitoring of population dynamics of Corynebacterium glutamicum by multiparameter flow cytometry. Microb Biotechnol 6:157–167. doi: 10.1111/1751-7915.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tolker-Nielsen T, Holmstrøm K, Boe L, Molin S. 1998. Non-genetic population heterogeneity studied by in situ polymerase chain reaction. Mol Microbiol 27:1099–1105. doi: 10.1046/j.1365-2958.1998.00760.x. [DOI] [PubMed] [Google Scholar]
- 86.Skinner SO, Sepúlveda LA, Xu H, Golding I. 2013. Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat Protoc 8:1100–1113. doi: 10.1038/nprot.2013.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. 1995. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci 20:448–455. doi: 10.1016/S0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- 88.Makemson JC. 1986. Luciferase-dependent oxygen consumption by bioluminescent vibrios. J Bacteriol 165:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamenšek S, Podlesek Z, Gillor O, Žgur-Bertok D. 2010. Genes regulated by the Escherichia coli SOS repressor LexA exhibit heterogenous expression. BMC Microbiol 10:283. doi: 10.1186/1471-2180-10-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kesel S, Mader A, Höfler C, Mascher T, Leisner M. 2013. Immediate and heterogeneous response of the LiaFSR two-component system of Bacillus subtilis to the peptide antibiotic bacitracin. PLoS One 8:e53457. doi: 10.1371/journal.pone.0053457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hense BA, Müller J, Kuttler C, Hartmann A. 2012. Spatial heterogeneity of autoinducer regulation systems. Sensors 12:4156–4171. doi: 10.3390/s120404156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ratcliff WC, Denison RF. 2011. Bacterial persistence and bet hedging in Sinorhizobium meliloti. Commun Integr Biol 4:98–100. doi: 10.4161/cib.14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. 2011. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc Natl Acad Sci U S A 108:20742–20747. doi: 10.1073/pnas.1108963108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. 2012. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet 8:e1002818. doi: 10.1371/journal.pgen.1002818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minoia M, Gaillard M, Reinhard F, Stojanov M, Sentchilo V, van der Meer JR. 2008. Stochasticity and bistability in horizontal transfer control of a genomic island in Pseudomonas. Proc Natl Acad Sci U S A 105:20792–20797. doi: 10.1073/pnas.0806164106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chung JD, Stephanopoulos G, Ireton K, Grossman AD. 1994. Gene expression in single cells of Bacillus subtilis: evidence that a threshold mechanism controls the initiation of sporulation. J Bacteriol 176:1977–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Veening JW, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A 105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Angelis G, Moschinoi M, Muzzi A, Pezzicoli A, Cesini S, Delany I, Lo Sapio M, Sinisi A, Donati C, Masignani V, Barocchi MA. 2011. The Streptococcus pneumoniae pilus-1 displays a biphasic expression pattern. PLoS One 6:e21269. doi: 10.1371/journal.pone.0021269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kröger C, Srikumar S, Ellwart J, Fuchs TM. 2011. Bistability in myo-inositol utilization by Salmonella enterica serovar Typhimurium. J Bacteriol 193:1427–1435. doi: 10.1128/JB.00043-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. 2010. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar Typhimurium. J Bacteriol 192:456–466. doi: 10.1128/JB.01826-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chai Y, Chu F, Kolter R, Losick R. 2008. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol 67:254–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mehra S, Charaniya A, Takano E, Hu WS. 2008. A bistable gene switch for antibiotic biosynthesis: the butyrolactone regulon in Streptomyces coelicolor. PLoS One 3:e2724. doi: 10.1371/journal.pone.0002724. [DOI] [PMC free article] [PubMed] [Google Scholar]