Abstract
Members of Botryosphaeriales are commonly encountered as endophytes or pathogens of various plant hosts. The Botryosphaeriaceae represents the predominant family within this order, containing numerous species associated with canker and dieback disease on a wide range of woody hosts. During the course of routine surveys from various plant hosts in Thailand, numerous isolates of Botryosphaeriaceae, including Aplosporellaceae were collected. Isolates were subsequently identified based on a combination of morphological characteristics and phylogenetic analysis of a combined dataset of the ITS and EF1-α gene regions. The resulting phylogenetic tree revealed 11 well-supported clades, correlating with different members of Botryosphaeriales. Other than confirming the presence of taxa such as Lasiodiplodia theobromae, L. pseudotheobromae and Neofusicoccum parvum, new records for Thailand include Pseudofusicoccum adansoniae and P. ardesiacum. Furthermore, four novel species are described, namely Diplodia neojuniperi from Juniperus chinensis, Lasiodiplodia thailandica from Mangifera indica, Pseudofusicoccum artocarpi and Aplosporella artocarpi from Artocarpus heterophyllus, while a sexual morph is also newly reported for L. gonubiensis. Further research is presently underway to determine the pathogenicity and relative importance of these species on different woody hosts in Thailand.
Keywords: Aplosporella, Botryosphaeriaceae, Diplodia, Lasiodiplodia, multigene phylogeny, Pseudofusicoccum, sexual morph, systematic
Introduction
The Botryosphaeriaceae was introduced as family in the Botryosphaeriales by Schoch et al. (2006). Based on recent molecular phylogenetic studies, however, several species have been excluded from the Botryosphaeriaceae and allocated to different families within the order, namely Planistromellaceae (Kellermania) (Minnis et al. 2012), Phyllostictaceae (Phyllosticta) (Wikee et al. 2013), Aplosporellaceae (Aplosporella and Bagnisiella), Saccharataceae (Saccharata) and Melanopsaceae (Melanops) (Slippers et al. 2013). The Botryosphaeriaceae represents the predominant family of this order, and Phillips et al. (2013) provided phylogenetic support for 17 genera including Barriopsis, Botryobambusa, Botryosphaeria, Cophinforma, Diplodia, Dothiorella, Endomelanconiopsis, Lasiodiplodia, Macrophomina, Neodeightonia, Neofusicoccum, Neoscytalidium, Phaeobotryon, Pseudofusicoccum, Spencermartinsia, Sphaeropsis and Tiarosporella. Species of Botryosphaeriaceae have a cosmopolitan distribution on a wide range of plant hosts, encompassing endophytes, saprobes and plant pathogens (von Arx & Müller 1954, Slippers & Wingfield 2007). Recent studies have revealed that some of them are severe canker and dieback pathogens of a range of important crops such as Proteaceae cut-flowers (Denman et al. 2003, Marincowitz et al. 2008), Eucalyptus (Slippers et al. 2004b, 2007, Zhou et al. 2008), grapevines (van Niekerk et al. 2006, Urbez-Torres et al. 2012), oaks (Sánchez et al. 2003), pines (Mohali et al. 2007) and stone fruits (Damm et al. 2007a, Slippers et al. 2007, Quaglia et al. 2014). Furthermore, these fungi also cause fruit diseases, which are mainly associated with fruit and stem-end rot as reported in avocado (McDonald & Eskalen 2011), mango (Ismail et al. 2012, Marques et al. 2013) and olives (Lazzizera et al. 2008).
Members of the Botryosphaeriaceae are known to be widely distributed, occurring on a broad range of plant hosts in many countries, including Thailand (Trakunyingcharoen et al. 2014). Liu et al. (2012) accepted 29 genera in the Botryosphaeriales, reported six new species from Thailand, and introduced two new genera, namely Botryobambusa (B. fusicoccum) and Cophinforma (C. eucalypti). Furthermore, four new species were described, namely Auerswaldia dothiorella (= Dothiorella thailandica), A. lignicola (= Lasiodiplodia lignicola), Botryosphaeria fusispora and Phaeobotryosphaeria eucalypti (see Phillips et al. 2013). Other records for Thailand included Botryosphaeria agaves, Lasiodiplodia theobromae, Neodeightonia subglobosa and Neofusicoccum parvum (Liu et al. 2012). In addition, Lasiodiplodia pseudotheobromae was also newly associated with mango diseases in this country (Trakunyingcharoen et al. 2013).
Until relatively recently, species of Botryosphaeriaceae have been identified solely based on morphological characteristics (Denman et al. 2000, Xenopoulos & Tsopelas 2000). However, since conidial septation and pigmentation evolved more than once within different genera of the family (Slippers et al. 2013) and are strongly influenced by cultural conditions (Alves et al. 2006), misidentifications have proven to be rather common in the literature. In this regard, molecular phylogenetic studies have provided a powerful tool to accurately identify members of Botryosphaeriaceae based on a combination of different partial gene regions, including β-tubulin (TUB), translation elongation factor1-α (EF1-α), the internal transcribed spacers (ITS) of the nrDNA, and the small and large-subunit ribosomal rRNA genes (SSU and LSU) (Slippers et al. 2004a, 2005, 2013, Crous et al. 2006).
The aim of the present study was thus to identify species belonging to Aplosporellaceae and Botryosphaeriaceae collected from various plant hosts in Thailand by employing a polyphasic approach incorporating morphological, cultural and phylogenetic DNA data.
MATERIALS AND METHODS
Isolates and morphology
Both asymptomatic and symptomatic twigs and stems (associated with canker and dieback disease) were collected during February–June (2012) from various plant hosts located in Chiang Mai and Chiang Rai provinces of Thailand. The collected plant specimens included avocado (Persea americana), cananga (Cananga odorata), cassod (Senna siamea), Chinese juniper (Juniperus chinensis), coffee (Coffea arabica), Eucalyptus obliqua, fig (Ficus racemosa), golden shower (Cassia fistula), guava (Psidium sp.), Indian gooseberry (Phyllanthus emblica), longan (Dimocarpus longan), mango (Mangifera indica), Marian plum (Bouea burmanica), Para rubber (Hevea brasiliensis), palm (Veitchia merrillii), peacock flower (Caesalpinia pulcherrima), pine (Pinus kesiya), rose apple (Syzygium samarangense), sapodilla (Manilkara zapota), star gooseberry (Phyllanthus acidus), sweet osmanthus (Osmanthus fragrans) and wild Himalayan cherry (Prunus cerasoides). Samples were incubated in moist chambers at room temperature for 7–10 d to induce sporulation. Single propagule isolations were established on 2 % Potato Dextrose Agar (PDA) and incubated at room temperature for 7 d using the techniques explained by Crous et al. (1991). Isolates of Aplosporellaceae and Botryosphaeriaceae were primarily characterised based on colony morphology, together with morphology of their asexual and sexual morphs. To induce sporulation, isolates were inoculated onto sterile pine needles and placed on 2 % water agar (PNA; Smith et al. 1996) at 25 °C under near-ultraviolet light for 14–30 d. Fungal structures were mounted in clear lactic acid and studied under a Nikon Eclipse 80i compound microscope with differential inference contrast (DIC) illumination. Thirty measurements were made of each structure, and for spores the 95 % percentiles are presented, with extremes given between brackets. The isolates used in this present study are maintained in the culture collection of the CBS-KNAW Fungal Biodiversity Centre (CBS), Utrecht, The Netherlands (Table 1). Reference specimens were deposited in the CBS fungarium, and nomenclature and descriptions of taxonomic novelties in MycoBank (Crous et al. 2004).
Table 1.
Details and GenBank accession numbers of isolates of Botryosphaeriaceae included in this study. New isolates obtained in this study are indicated in bold, new GenBank sequence accession numbers in italics, and * represents ex-type isolates.
| Species | Accession no.1 | Substrate | Locality | Collector | GenBank2 | |
|---|---|---|---|---|---|---|
| ITS | EF1-α | |||||
| Aplosporella artocarpi | CPC 22791 | Artocarpus heterophyllus | Thailand | T. Trakunyingcharoen | KM006450 | KM006481 |
| A. prunicola | CBS 121167* | Prunus persica | South Africa | U. Damm | EF564376 | – |
| A. yalgorensis | MUCC 511* | Acacia cochlearis | Australia: Western Australia | K.M. Taylor | EF591926 | EF591977 |
| Barriopsis fusca | CBS 174.26* | Citrus sp. | Cuba | N.E. Stevens | EU673330 | EU673296 |
| Ba. iraniana | CBS 124698* | Mangifera indica | Iran | J. Abdollahzadeh & A. Javadi | FJ919663 | FJ919652 |
| Botryosphaeria dothidea | CBS 115476* | Prunus sp. | Switzerland | B. Slippers | AY236949 | AY236898 |
| Bo. fabicerciana | CBS 127193* | Eucalyptus sp. | China | M.J. Wingfield | HQ332197 | HQ332213 |
| Bo. ramosa | CBS 122069* | Eucalyptus camaldulensis | Australia: Western Australia | TI. Burgess | EU144055 | EU144070 |
| Botryosphaeria sp. | CPC 22789 | Bouea burmaxnica | Thailand | T. Trakunyingcharoen | KM006448 | KM006479 |
| Diplodia africana | CBS 120835* | Prunus persica | South Africa | U. Damm | EF445343 | EF445382 |
| Di. agrifolia | CBS 132777* | Quercus agrifolia | USA | S. Lynch & A. Eskalen | JN693507 | JQ517317 |
| Di. bulgarica | CBS 124254* | Malus sylvestris | Bulgaria | S.G. Bobev | GQ923853 | GQ923821 |
| Di. corticola | CBS 112549* | Quercus suber | Portugal | A. Alves | AY259100 | AY573227 |
| Di. cupressi | CBS 168.87* | Cupressus sempervirens | Israel | Z. Solel | DQ458893 | DQ458878 |
| Di. malorum | CBS 124130* | Malus sylvestris | Portugal | A.J.L. Phillips | GQ923865 | GQ923833 |
| Di. mutila | CBS 112553 | Vitis vinifera | Portugal | A.J.L. Phillips | AY259093 | AY573219 |
| Di. neojuniperi | CPC 22753* | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006431 | KM006462 |
| CPC 22754 | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006432 | KM006463 | |
| CPC 22802 | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006457 | KM006488 | |
| Di. olivarum | CBS 121887* | Olea europaea | Italy | S. Frisullo | EU392302 | EU392279 |
| Di. pseudoseriata | CBS 124906* | Blepharocalyx salicifolius | Uruguay | C. Perez | EU080927 | EU863181 |
| Di. quercivora | CBS 133852* | Quercus canariensis | Tunisia | B.T Linaldeddu | JX894205 | JX894229 |
| Di. rosulata | CBS 116470* | Prunus africana | Ethiopia | A. Gure | EU430265 | EU430267 |
| Di. sapinea | CBS 109726 | Pinus patula | Indonesia | M.J. Wingfield | DQ458896 | DQ458881 |
| CBS 393.84* | Pinus nigra | Netherlands | H.A. van der Aa | DQ458895 | DQ458880 | |
| Di. seriata | CBS 112555* | Vitis vinifera | Portugal | A.J.L. Phillips | AY259094 | AY573220 |
| Di. tsugae | CBS 418.64* | Tsuga heterophylla | Canada | A. Funk | DQ458888 | DQ458873 |
| Dothiorella iberica | CBS 115041* | Quercus ilex | Spain | J. Luque | AY573202 | AY573222 |
| Do. longicollis | CBS 122068* | Lysiphyllum cunninghamii | Australia: Western Australia | TI. Burgess | EU144054 | EU144069 |
| Do. thailandica | CBS 133991* | Dead bamboo culm | Thailand | D.Q. Dai, J.K. Liu & K.D. Hyde | JX646796 | JX646861 |
| Endomelanconiopsis endophytica | CBS 120397* | Theobroma cacao | Panama | E. Rojas, L. Mejia & Z. Maynard | EU683656 | EU683637 |
| E. microspora | CBS 353.97* | Soil | Papua New Guinea | H.A. van der Aa | EU683655 | EU683636 |
| Lasiodiplodia citricola | CBS 124707* | Citrus sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945354 | GU945340 |
| L. crassispora | CBS 118741* | Eucalyptus urophylla | Venezuela | S. Mohali | DQ103552 | DQ103559 |
| L. egyptiacae | CBS 130992* | Mangifera indica | Egypt | A.M. Ismail | JN814397 | JN814424 |
| L. euphorbicola | CMM 3609* | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234543 | KF226689 |
| L. gilanensis | CBS 124704* | – | Iran | J. Abdollahzadeh & A. Javadi | GU945351 | GU945342 |
| L. gonubiensis | CBS 115812* | Syzygium cordatum | South Africa | D. Pavlic | DQ458892 | DQ458877 |
| L. gonubiensis (sexual morph) | CPC 22781 | Phyllanthus emblica | Thailand | T. Trakunyingcharoen | KM006443 | KM006474 |
| L. hormozganensis | CBS 124709* | Olea sp. | Iran | J. Abdollahzadeh & A. Javadi | GU945355 | GU945343 |
| L. iraniensis | CBS 124710* | Salvadora persica | Iran | J. Abdollahzadeh & A. Javadi | GU945348 | GU945336 |
| L. jatrophicola | CMM 3610* | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234544 | KF226690 |
| L. macrospora | CMM 3833* | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234557 | KF226718 |
| L. mahajangana | CBS 124927* | Terminalia catappa | Madagascar | J. Roux | FJ900597 | FJ900643 |
| L. margaritacea | CBS 122519* | Adansonia gibbosa | Australia: Western Australia | TI. Burgess | EU144050 | EU144065 |
| L. missouriana | CBS 128311* | Vitis vinifera | USA | K. Striegler & G.M. Leavitt | HQ288225 | HQ288267 |
| L. parva | CBS 456.78* | Soil from Cassava-field | Columbia | O. Rangel | EF622083 | EF622063 |
| L. plurivora | CBS 120832* | Prunus salicina | South Africa | U. Damm | EF445362 | EF445395 |
| L. pseudotheobromae | CBS 116459* | Gmelina arborea | Costa Rica | J. Carranza-Velazquez | EF622077 | EF622057 |
| CPC 22756 | Osmanthus fragrans | Thailand | T. Trakunyingcharoen | KM006434 | KM006465 | |
| CPC 22758 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607141 | KJ607151 | |
| CPC 22759 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607142 | KJ607152 | |
| CPC 22760 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607143 | KJ607153 | |
| CPC 22761 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607144 | KJ607154 | |
| CPC 22762 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607145 | KJ607155 | |
| CPC 22770 | Persea americana | Thailand | T. Trakunyingcharoen | KJ607146 | KJ607156 | |
| CPC 22771 | Persea americana | Thailand | T. Trakunyingcharoen | KJ607147 | KJ607157 | |
| CPC 22776 | Psidium sp. | Thailand | T. Trakunyingcharoen | KM006438 | KM006469 | |
| CPC 22777 | Coffea arabica | Thailand | T. Trakunyingcharoen | KM006439 | KM006470 | |
| CPC 22778 | Psidium sp. | Thailand | T. Trakunyingcharoen | KM006440 | KM006471 | |
| CPC 22779 | Dimocarpus longan | Thailand | T. Trakunyingcharoen | KM006441 | KM006472 | |
| CPC 22783 | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193638 | KJ193682 | |
| CPC 22784 | Ficus racemosa | Thailand | T. Trakunyingcharoen | KM006444 | KM006475 | |
| CPC 22787 | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193639 | KJ193683 | |
| CPC 22788 | Bouea burmanica | Thailand | T. Trakunyingcharoen | KM006447 | KM006478 | |
| CPC 22790 | Syzygium samarangense | Thailand | T. Trakunyingcharoen | KM006449 | KM006480 | |
| CPC 22792 | Phyllanthus acidus | Thailand | T. Trakunyingcharoen | KM006451 | KM006482 | |
| CPC 22793 | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193640 | KJ193684 | |
| CPC 22794 | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193641 | KJ193685 | |
| CPC 22799 | Cananga odorata | Thailand | T. Trakunyingcharoen | KM006455 | KM006486 | |
| CPC 22801 | Dimocarpus longan | Thailand | T. Trakunyingcharoen | KM006456 | KM006487 | |
| CPC 22803 | Juniperus chinensis | Thailand | T. Trakunyingcharoen | KM006458 | KM006489 | |
| L. rubropurpurea | CBS 118740* | Eucalyptus grandis | Australia | TI. Burgess & G. Pegg | DQ103554 | DQ103572 |
| L. subglobosa | CMM 3872* | Jatropha curcas | Brazil | A.R. Machado & O.L. Pereira | KF234558 | KF226721 |
| L. thailandica | CPC 22755 | Phyllanthus acidus | Thailand | T. Trakunyingcharoen | KM006433 | KM006464 |
| CPC 22795* | Mangifera indica | Thailand | T. Trakunyingcharoen | KJ193637 | KJ193681 | |
| L. theobromae | CBS 111530 | Leucospermum sp. | USA: Hawaii | J.E. Taylor | EF622074 | EF622054 |
| CBS 164.96* | Fruit along coral reef coast | Papua New Guinea | A. Aptroot | AY640255 | AY640258 | |
| CPC 22766 | Pinus kesiya | Thailand | T. Trakunyingcharoen | KM006436 | KM006467 | |
| CPC 22780 | Manilkara zapota | Thailand | T. Trakunyingcharoen | KM006442 | KM006473 | |
| CPC 22798 | Syzygium samarangense | Thailand | T. Trakunyingcharoen | KM006454 | KM006485 | |
| L. venezuelensis | CMW 13513 | Acacia mangium | Venezuela | S. Mohali | DQ103549 | DQ103570 |
| L. viticola | CBS 128313* | Vitis vinifera | USA | R.D. Cartwright & W.D. Gubler | HQ288227 | HQ288269 |
| Lasiodiplodia sp. | CPC 22800 | Mangifera indica | Thailand | T Trakunyingcharoen | KJ193643 | KJ193687 |
| Neofusicoccum arbuti | CBS 116131* | Arbutus menziesii | USA | M. Elliot | GU251152 | GU252284 |
| Nf. australe | CMW 6837* | Acacia sp. | Australia | M.J. Wingfield | AY339262 | AY339270 |
| Nf. luteum | CBS 110299* | Vitis vinifera | Portugal | A.J.L. Phillips | AY259091 | AY573217 |
| Nf. parvum | CMW 9081* | Populus nigra | New Zealand | G.J. Samuels | AY236943 | AY236888 |
| CPC 22751 | Prunus cerasoides | Thailand | T. Trakunyingcharoen | KM006429 | KM006460 | |
| CPC 22752 | Prunus cerasoides | Thailand | T. Trakunyingcharoen | KM006430 | KM006461 | |
| CPC 22757 | Eucalyptus obliqua | Thailand | T. Trakunyingcharoen | KM006435 | KM006466 | |
| Nf. ribis | CBS 115475* | Ribes sp. | USA | B. Slippers & G. Hudler | AY236935 | AY236877 |
| Neoscytalidium hyalinum | CBS 312.90 | Homo sapiens | Netherlands | R. Benne | KJ193679 | KJ193723 |
| Ns. novaehollandiae | CBS 122071* | Crotalaria sp. | Australia: Western Australia | TI. Burgess & M.J. Wingfield | EF585540 | EF585580 |
| Phaeobotryon cupressi | CBS 124700* | Cupressus sempervirens | Iran | M.A. Aghajani | FJ919672 | FJ919661 |
| Ph.mamane | CBS 122980* | Sophora chrysophylla | USA: Hawaii | W. Gams | EU673332 | EU673298 |
| Phyllosticta citricarpa | CBS 111.20 | Citrus sp. | Australia | – | FJ538314 | FJ538372 |
| Pseudofusicoccum adansoniae | CBS 122055* | Adansonia gibbosa | Australia: Western Australia | TI. Burgess & M.J. Wingfield | EF585523 | EF585571 |
| CPC 22763 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607148 | KJ607158 | |
| CPC 22764 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607149 | KJ607159 | |
| CPC 22765 | Hevea brasiliensis | Thailand | T. Trakunyingcharoen | KJ607150 | KJ607160 | |
| CPC 22767 | Dimocarpus longan | Thailand | T. Trakunyingcharoen | KM006437 | KM006468 | |
| CPC 22786 | Cassia fistula | Thailand | T. Trakunyingcharoen | KM006446 | KM006477 | |
| CPC 22797 | Senna siamea | Thailand | T. Trakunyingcharoen | KM006453 | KM006484 | |
| Ps. ardesiacum | CBS 122062* | Adansonia gibbosa | Australia: Western Australia | TI. Burgess & M.J. Wingfield | EU144060 | EU144075 |
| CPC 22785 | Caesalpinia pulcherrima | Thailand | T. Trakunyingcharoen | KM006445 | KM006476 | |
| CPC 22804 | Veitchia merrillii | Thailand | T. Trakunyingcharoen | KM006459 | KM006490 | |
| Ps. artocarpi | CPC 22796 | Artocarpus heterophyllus | Thailand | T. Trakunyingcharoen | KM006452 | KM006483 |
| Ps. kimberleyense | CBS 122058* | Acacia synchronicia | Australia: Western Australia | TI. Burgess & M.J. Wingfield | EU144057 | EU144072 |
| Ps. olivaceum | CBS 124939* | Pterocarpus angolensis | South Africa | J. Roux | FJ888459 | FJ888437 |
| Ps. stromaticum | CBS 117448 | Eucalyptus-hybrid | Venezuela | S. Mohali | AY693974 | AY693975 |
| Ps. violaceum | CBS 124936* | Pterocarpus angolensis | South Africa | J. Mehl & J. Roux | FJ888474 | FJ888442 |
1 CBS = CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands; CMM = Phytopathogenic Fungi of the Universidade Federal Rural de Pernambuco; CMW = Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa; CPC = Culture Collection of P.W. Crous, housed at CBS; MUCC = Murdoch University Culture Collection, Perth, Australia.
2 ITS = internal transcribed spacers and intervening 5.8S nrDNA; EF1-α = partial translation elongation factor 1-alpha gene.
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from the 7–10-d-old mycelium growing on 2 % malt extract agar (MEA) using the UltraClean® Microbial DNA Isolation Kit (MOBIO Laboratories, Inc, Carlsbad, USA) following the manufacturer’s instructions. The internal transcribed spacer (ITS) and intervening 5.8S nrRNA gene region of the nuclear rDNA were amplified using primers ITS5 and ITS4 (White et al. 1990). The partial sequences of the EF1-α gene region was amplified using primers EF1-728F (Carbone & Kohn 1999) and EF-2 (O’Donnell et al. 1998). However, for some isolates in the genus Lasiodiplodia, this region was amplified using primers EF1-688F and EF1-1251R as described by Alves et al. (2008). Master mixes for amplification followed Ismail et al. (2012). The amplifications were conducted in a thermal cycler using the following amplification conditions: initial denaturation at 95 °C for 5 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 52 °C for 30 s, extension at 72 °C for 1 min, and a final extension step at 72 °C for 7 min.
The amplified fragments were sequenced in both directions with the same primers used for amplification, using the BigDye Terminator v. 3.1 Cycle Sequencing Kit following the manufacturer’s instructions. The sequencing reactions were run on an ABI PRISM™ 3730 DNA automated sequencer (Perkin-Elmer Applied BioSystems, Foster City, CA, USA).
Phylogenetic analyses
The generated nucleotide sequences were edited, and adjustments were made manually where necessary with MEGA v. 5.1 (Tamura et al. 2011). The consensus sequences were aligned using the online version of MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/). New sequences from this study were deposited in GenBank, and were analysed together with additional sequences of species in Botryosphaeriaceae obtained from GenBank (Table 1). The phylogenetic analysis was performed on the combined dataset of the ITS and EF1-α regions using PAUP v. 4.0b10 (Swofford 2003) for Maximum Parsimony (MP) and MrBayes v. 3.2.1 (Ronquist & Huelsenbeck 2003) for Bayesian Inference (BI), respectively. Trees were rooted to Phyllosticta citricarpa (CBS 111.20).
The MP analysis was performed using the heuristic search option with 1 000 random stepwise additions, and tree bisection and reconnection (TBR) as branch swapping algorithm (Swofford & Begle 1993). All characters were unordered and had equal weight and gaps were treated as missing data. Branches of zero length were collapsed and all multiple equally parsimonious trees were saved. The robustness of the equally most parsimonious trees was calculated using 1 000 bootstrap replications (Hillis & Bull 1993). Other calculated values for parsimony included tree length (TL), consistency index (CI), retention index (RI), rescaled consistency index (RC) and homoplasy index (HI).
The BI was performed by two independent runs of Markov Chain Monte Carlo (MCMC) algorithms (Larget & Simon 1999) to construct the phylogenetic tree. Four MCMC chains were run simultaneously, with heating parameter set at 0.3, under a general time-reversible (GTR) (Rodriguez et al. 1990) substitution model with rate variation of gamma-distribution (G), and proportion of invariable site (I) with a dirichlet state frequency parameters determined using MrModel Test v. 2.2 (Nylander 2004). The analyses were run for 100 000 000 generations until the average standard deviation of split frequencies came below 0.01, with trees saved every 1 000 generations. The first 25 % of saved trees were discarded as the ‘burn-in’ phase and the posterior probabilities (Rannala & Yang 1996) were calculated from the remaining trees.
Results
PCR amplification, sequencing and phylogenetic analyses
The generated amplicons of the ITS region were ± 570 bpusing the ITS5 and ITS4 primer pair. The generated amplicons of EF1-α region were ± 500 and 700 bp using the set of primers EF1-728F and EF-2, and EF1-688F and EF1-1251R, respectively. The sequences of the two amplified regions were aligned and analysed using MP and BI. New sequences in this study were deposited in GenBank as shown in Table 1 and the alignment and tree were deposited in TreeBASE.
The two datasets were congruent, and therefore combined. The alignment of the combined dataset of the ITS (628 characters; 283 unique site patterns) and EF1-α (392 characters; 344 unique site patterns) region consisted of 112 taxa with 1 020 characters including gaps, of which 361 characters were constant, 141 characters were variable and parsimony uninformative and 518 characters were parsimony informative. The heuristic search resulted in 1 000 equally most parsimonious trees with TL = 2 493, CI = 0.523, RI = 0.892, RC = 0.466 and HI = 0.477. The BI analysis lasted 2 820 000 generations and produced 5 642 trees of which 4 232 trees were sampled to produce a 50 % majority rule consensus Bayesian tree with nearly identical overall topology to the equally most parsimonious trees (Bayesian tree not shown, but posterior probability values are mapped to the parsimony tree presented in Fig. 1). The first of 1 000 equally most parsimonious trees, which showed the same overall topology, is shown in Fig. 1 with bootstrap support values and posterior probabilities indicated at the branch nodes. The parsimonious tree revealed 11 well-supported clades corresponding to established genera. Members of the Botryosphaeriaceae are indicated in Clades I–X. Clade I represents species of genus Phaeobotryon, Clade II species of Barriopsis and Clade III species of Diplodia. Clade IV is the dominant clade representing species of Lasiodiplodia. Clade V represents species of Neofusicoccum, Clade VI species of Botryosphaeria, Clade VII species of Neoscytalidium and Clade VIII species of Dothiorella. Clade IX represents species of Pseudofusicoccum, which are closely related to species in Endomelanconiopsis in Clade X. Clade XI represents species of Aplosporella (Aplosporellaceae), while Phyllosticta citricarpa (CBS 111.20) a member of Phyllostictaceae, was used as outgroup in this phylogenetic analysis.
Fig. 1.
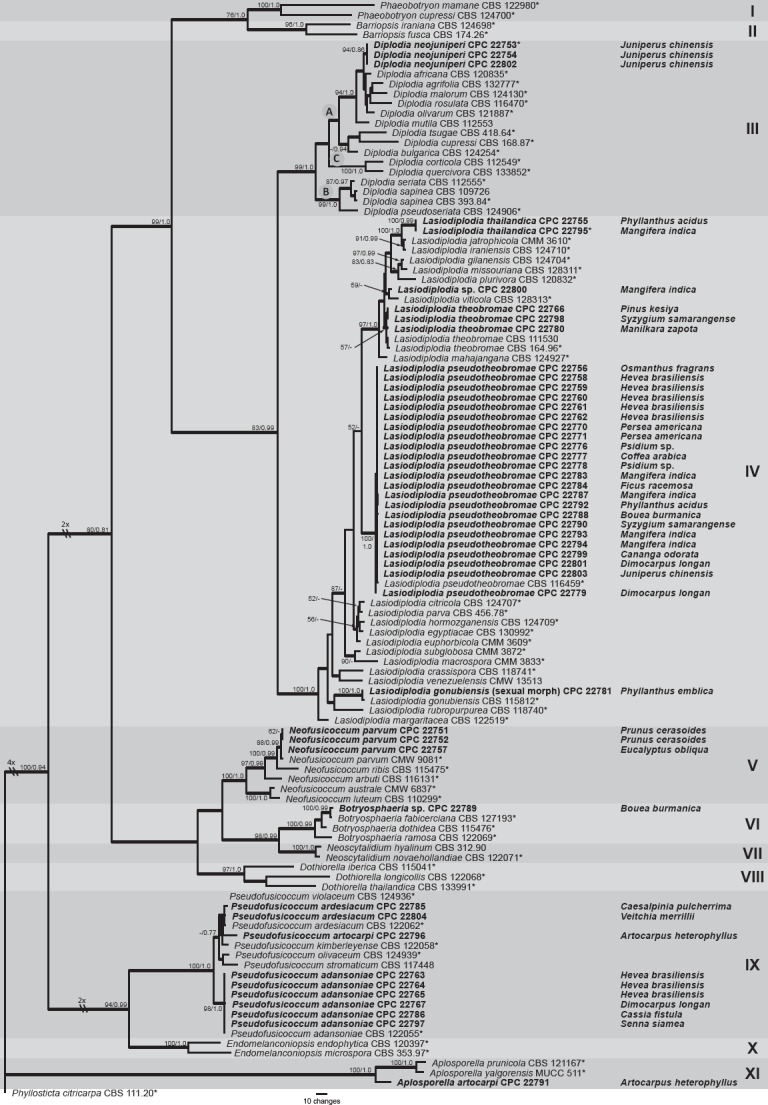
The first of 1 000 equally most parsimonious trees (TL = 2 493; CI = 0.523; RI = 0.892; RC = 0.466) resulting from a parsimony analysis of the combined ITS and EF1-α sequence alignment. The bootstrap support values (integers; to the left of the forward slash) and posterior probability values (≤ 1; to the right of the forward slash) are indicated at the nodes and the scale bar represents the number of changes. Thickened branches reflect those branches present in the strict consensus tree. Genera are indicated by different coloured blocks and provided with clade numbers in Roman numerals to the right of the tree. Species and strains from Thailand pertinent to this study are shown in bold and hosts from Thailand are printed in the middle of the tree, in line with the corresponding strain. The tree was rooted to Phyllosticta citricarpa (CBS 111.20).
The isolates obtained from Thailand clustered into six clades that included Aplosporella, Diplodia, Fusicoccum, Lasiodiplodia, Neofusicoccum and Pseudofusicoccum. The genus Lasiodiplodia contained several species and appeared to be the dominant group collected from Thailand in this study. New species identified in present study are described below and include Aplosporella artocarpi (Clade XI), Diplodia neojuniperi (Clade III), Lasiodiplodia thailandica (Clade IV) and Pseudofusicoccum artocarpi (Clade IX). In addition, this study is the first report of a sexual morph for L. gonubiensis.
Isolates and morphology
Members of Aplosporellaceae and Botryosphaeriaceae obtained from Thailand clustered in six phylogenetic clades, with each clade correlating with distinct morphological features of specific genera. The isolates formed asexual structures on sterile pine needles on WA within 2–4 wk of incubation. However, no sexual morph could be induced on any of the media tested.
Taxonomy
Aplosporella artocarpi T. Trakunyingcharoen, L. Lombard & Crous, sp. nov. — MycoBank MB810167; Fig. 2
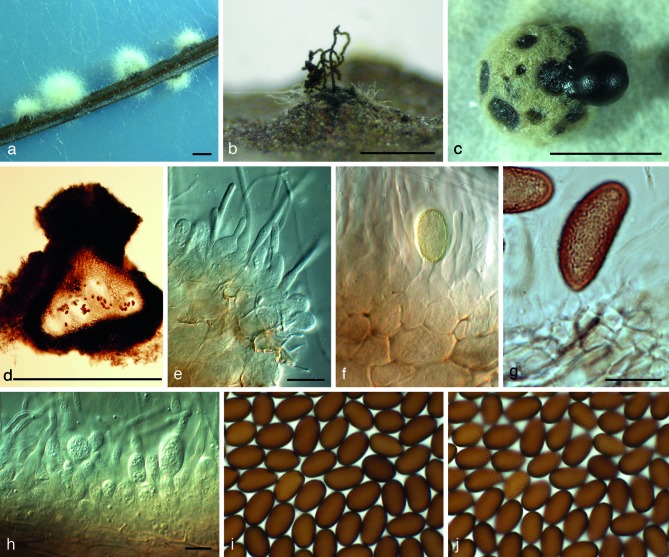
Fig. 2.
Aplosporella artocarpi (CBS 138651). a, b. Conidiomata sporulating on PNA; c. sporulation on PDA; d. vertical section through conidioma; e, f, h. conidiogenous cells and paraphyses; g. conidiogenous cells giving rise to conidium; i, j. brown conidia with surface ornamentation. — Scale bars: a–d = 550 μm, e–j = 10 μm.
Etymology. The name refers to the host genus from which it was collected, Artocarpus.
Conidiomata pycnidial, semi-immersed, mostly solitary, dark brown to black, with globose base, (350–)540–550(–650) × (490–)540–600(–700) μm, outer layers composed of dark brown textura angularis, becoming thin-walled and hyaline toward the inner region, multilocular, with (2–)4–5(–6) locules. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, subcylindrical, discrete, holoblastic, proliferating percurrently, forming 1–2 annellations near the apex, originating from the hyaline, inner conidiomatal wall, 3–5 × 2–4 μm. Paraphyses hyaline, cylindrical, with bluntly rounded apical cells, (13–)23–55(–60) × 2–3 μm. Conidia hyaline, ellipsoid to ovoid, smooth, moderately thick-walled, with granular content, aseptate, becoming pale brown before conidiomatal discharge, sometimes while still attached to the conidiogenous cells, becoming brown with rough outer surface, (17–)18–21(–22) × (9–)10–11 μm.
Culture characteristics — Colonies with white aerial mycelium on PDA, slightly fluffy, turning smokey grey with age, darker grey at the centre, sometimes mycelium turning yellowish to green at the colony margin, and forming conidiomata at the colony margin after 7–10 d; colonies turn dark grey to olivaceous green in reverse.
Habitat — Asymptomatic twig of Artocarpus heterophyllus. Known distribution — Chiang Mai province, Thailand.
Material examined. THAILAND, Chiang Mai province, on leaf of Juniperus chinensis, Feb. 2012, T. Trakunyingcharoen (holotype CBS H-21932, culture ex-type CPC 22753 = CBS 138652). Additional isolates are listed in Table 1.
Notes — Although the genus Aplosporella has previously been treated as a member of the Botryosphaeriaceae (Damm et al. 2007b), it was recently placed in its own family Aplosporellaceae (Slippers et al. 2013). Aplosporella artocarpi has been introduced as new species based on its distinct phylogenetic position and morphological features. Although conidial dimensions of A. artocarpi (17–)18–21(–22) × (9–)10–11 μm overlap with those of A. prunicola (17–)19–22(–25) × (9–)10–12(–18) μm and A. yalgorensis (16–)18–22(–26) × (7–)8–13(–14) μm (Damm et al. 2007b, Taylor et al. 2009), conidia of A. artocarpi are shorter and narrower than conidia of these two species. In addition, conidia of A. artocarpi are narrower than those of A. embeliae (18–22 × 12–16 μm), wider than A. subhyalina (18–22 × 4–6 μm) and longer than A. beaumontiana (13–20 × 10–11.5 μm) and A. clerodendri (12–16 × 8–10 μm) (Pande & Rao 1995). Nothing is presently known about the host specificity of species of Aplosporella, and thus far very few species are known from culture.
Diplodia neojuniperi T. Trakunyingcharoen, L. Lombard & Crous, sp. nov. — MycoBank MB810168; Fig. 3
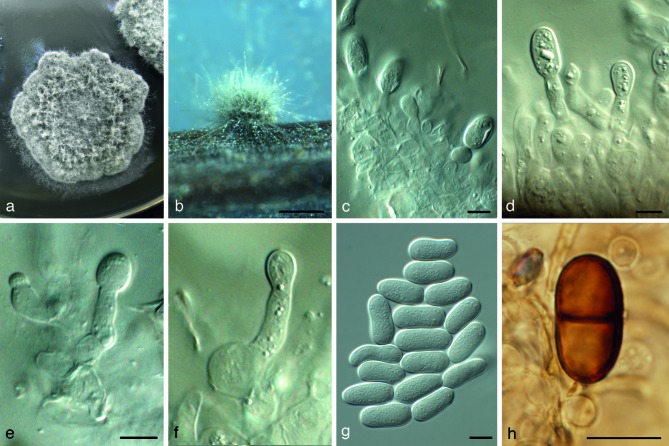
Fig. 3.
Diplodia neojuniperi (CBS 138652). a. Colony sporulating on MEA; b. colony sporulating on PNA; c–f. conidiogenous cells giving rise to conidia; g. hyaline conidia; h. mature, 1-septate, brown conidium. — Scale bars: b = 300 μm, all others = 10 μm.
Etymology. The name refers to its morphological similarity to D. juniperi.
Conidiomata pycnidial, immersed, dark brown to black, with globose base, solitary, unilocular, 230–330 × 200–320 μm, outer layers composed of dark brown textura angularis, becoming thin-walled and hyaline toward the inner region. Ostiole central, circular, up to 50 μm diam, papillate, with neck up to 20 μm tall. Conidiophores (when present) hyaline, cylindrical, 0–1-septate, 8–10 × 2.5–3 μm, mostly reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, thin-walled, discrete, cylindrical to ampulliform, holoblastic, proliferating to form 1–2 annellations or proliferating at the same level with visible periclinal thickening, generated from the hyaline inner wall of conidiomata, 9–12 × 2.5–3 μm. Paraphyses absent. Conidia hyaline, ellipsoid, unicellular, with granular content, 1 μm thick-walled, becoming pale brown with single median septum after discharge from the pycnidia, but rarely observed, (17–)18–21(–22) × (9–)10–11 μm.
Culture characteristics — Colonies forming two distinct zones on PDA. The first with grey mycelium, moderately dense and fluffy at the centre, while olivaceous grey mycelium flattening at the margin, erose or dentate, greenish olivaceous to black olivaceous in reverse after 7 d.
Habitat — Associated with leaf blight symptoms on Juniperus chinensis.
Known distribution — Chiang Mai province, Thailand.
Material examined. THAILAND, Chiang Mai province, on leaf of Juniperus chinensis, Feb. 2012, T. Trakunyingcharoen (holotype CBS H-21932, culture ex-type CPC 22753 = CBS 138652). Additional isolates are listed in Table 1.
Notes — Species of Diplodia are presently classified in three subclades (A–C) based on the molecular data and morphological characteristics (Phillips et al. 2013). Subclade A is a dominant group and consists of a number of species which include D. africana, D. agrifolia, D. bulgarica, D. cupressi, D. malorum, D. mutila, D. olivarum, D. rosulata, D. tsugae and now also D. neojuniperi. These species have hyaline conidia, which become pigmented and 1-septate after conidial discharge. Species of subclade B include D. sapinea and D. seriata. They have hyaline conidia, which turn pigmented at an early stage while still in their conidiomata, sometimes even while still attached to their conidiogenous cells. Although members of subclade C (D. corticola and D. quercivora) have similar morphological characteristics to those in subclade A, their conidia are much larger (Table 2).
Table 2.
A comparison of conidial morphology of Diplodia spp.
| Species | Group sensu Phillips et al. (2013) | Conidial dimensions (μm) | Reference |
|---|---|---|---|
| D. africana | A | (17–)25.5–33(–34) × (10–)12–14(–15) | Damm et al. (2007a) |
| D. agrifolia | A | (21.5–)27–36.5 × (12–)14.5–18 | Lynch et al. (2013) |
| D. bulgarica | A | (22.5–)24–27(–28) × (14.5–)15.5–18(–18.5) | Phillips et al. (2012) |
| D. corticola | C | (23.5–)26–34.5(–46) × (9–)12–16(–18.5) | Alves et al. (2004) |
| D. cupressi | A | (21.5–)23.5–28.5(–30.5) × (12–)13.5–15(–16) | Alves et al. (2006) |
| D. malorum | A | (24–)26–32(–36) × (12–)13–17.5(–18.5) | Phillips et al. (2012) |
| D. mutila | A | (23.5–)24.5–27(–27.5) × (12.5–)13–14(–14.5) | Montagne (1834) |
| D. neojuniperi | A | (17—)18–21(–22) × (9–)10—11 | Present study |
| D. olivarum | A | (21.5–)22–27.5(–28.5) × (10–)11–13.5(–14.5) | Lazzizera et al. (2008) |
| D. quercivora | C | (22.75–)28.14(–30.41) × (11.32–)13.08(–14.36) | Linaldeddu et al. (2013) |
| D. rosulata | A | (21–)25–32(–36) × (10–)11–17.5(–19.5) | Gure et al. (2005) |
| D. sapinea | B | (25.5–)30.5–52.5(–54) × (10–)12.5–20(–21) | Fuckel (1870) |
| D. seriata | B | (21.5–)22–27(–28) × (11–)11.5–14.5(–15.5) | de Notaris (1845) |
| D. tsugae | A | 36–41 × 18–22 | Phillips et al. (2012) |
To distinguish D. neojuniperi from other species within subclade A, it needs to be compared to its closest neighbours, D. Africana (based on MP analysis) and D. mutila (based on the BI analysis; data not shown). According to its conidial morphology, conidia of D. neojuniperi are much smaller than those of D. Africana and D. mutila (Table 2). Saccardo (1884) reported D. juniperi from Juniperus in Europe (conidia 18–20 × 8–10 μm). However, an examination of the type specimen (BR-Myc 148292,76) by Alves et al. (2006) failed to reveal any Diplodia material, and all fresh collections only rendered D. cupressi (conidia 21.5–30.5 × 12–16 μm). For this reason, as well as its geographic separation and wider conidial dimensions, the material from Thailand is described here as a novel species on juniper.
Lasiodiplodia gonubiensis Pavlic, Slippers & MJ. Wingf., Stud. Mycol. 50: 318. 2004. — Fig. 4
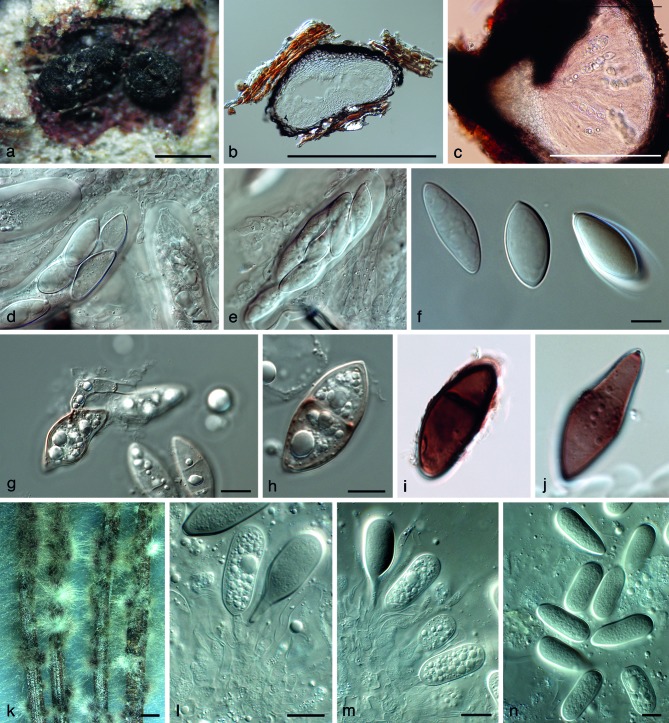
Fig. 4.
Lasiodiplodia gonubiensis (CBS 138654). a. Ascomata imbedded in host tissue; b, c. section through ascomata; d, e; asci. f–j. hyaline, young ascospores, that become brown and septate with age; k. conidiomata forming on PNA; l, m. conidiogenous cells giving rise to conidia; n. conidia. — Scale bars: a, b = 450 μm, c = 225 μm, all others = 10 μm.
Ascomata perithecial, immersed under bark, sometimes semiimmersed, solitary to aggregated, dark brown to black, unilocular, with globose base, (400–)530–600(–750) × (330–)400– 500(–570) μm, outer layers composed of dark brown textura angularis, becoming thin-walled and hyaline toward the inner region. Ostiole central, circular, up to 80 μm diam, neck papillate, (50–)100–120(–150) μm tall. Pseudoparaphyses hyaline, cylindrical, smooth, thin-walled, multi-septate, 73–125 × 3–4 μm. Asci hyaline, bitunicate with thick endotunica and well-developed apical chamber, clavate and stalked, sessile, originating from the inner hyaline wall of ascoma, (120–)150–180(–200) × (22.5–)27.5–30(–32.5) μm, containing (4–)6–8 ascospores. Ascospores hyaline, broadly fusiform to rhomboid to limoniform, moderately thick-walled with granular content, widest in the middle, ascospores with hyaline apiculus at both or either end, rarely becoming 1–2-septate with age, ascospores turn pale brown, 1–2-septate within ascoma or shortly after discharge, (32.5–)35–37.5(–40) × (16–)17.5–20(–25) μm. The asexual morph was described in full by Pavlic et al. (2004).
Culture characteristics — Colonies with white, fluffy aerial mycelium on PDA, becoming smoky-grey to olivaceous-grey, moderately dense at the centre of colony, a part of the colony forming wool-like aerial mycelium; reverse becoming greenish grey to olivaceous-grey after 7 d. Cultures remain sterile.
Habitat — Symptomless twigs of Phyllanthus emblica.
Known distribution — The reserved forest of Aomkoi district, Chiang Mai province, Thailand.
Material examined. THAILAND, Chiang Mai province, on twigs of Phyllanthus emblica, Apr. 2012, T. Trakunyingcharoen (CBS H-21934, culture CPC 22781 = CBS 138654). Additional isolates are listed in Table 1.
Notes — Ascospores of L. gonubiensis are normally aseptate, and rarely turn pale brown and 1–2-septate when mature like Barriopsis, Phaeobotryon and Sphaeropsis. In contrast, ascospores of L. gonubiensis are distinct from the sexual morphs of Dothiorella and Spencermartinsia, which are 1-septate. Ascospores of L. gonubiensis have a terminal apiculus, which is not found in Barriopsis. Thus ascospores of L. gonubiensis are morphologically distinct from other sexual morphs in the Botryosphaeriaceae based on its size, the presence of a terminal apiculus, and by becoming pigmented and septate upon ascospore release (Table 3).
Table 3.
Morphological comparison of Botryosphaeriaceae with dark ascospores.
| Species | Ascospores | Reference | ||
|---|---|---|---|---|
| Septation | Apiculus | Dimensions (pm) | ||
| Barriopsis fusca | aseptate | absent | (30–)31–36.5(–38.5) × (15.5–)16–18.5(–21) | Phillips et al. (2008) |
| Dothiorella iberica | 1 –septate | absent | (17.5–)22.5–23.5(–29) × (8.5–)10–10.5(–12.5) | Phillips et al. (2005) |
| Lasiodiplodia gonubiensis | rarely 1–2-septate | present | (32.5–)35–37.5(–40) × (16–)17.5–20(–25) | Present study |
| Phaeobotryon mamane | 2-septate | present | (30–)37–40(–45) × (11–)13–15(–16) | Phillips et al. (2008) |
| Spencermartinsia viticola | 1 -septate | present | (19–)22.5–23.5(–27) × (8.5–)10.5–11(–14.5) | Saccardo (1880) |
| Sphaeropsis visci | aseptate | present | (27.5–)31–37.5(–38.5) × (14.5–)15–19(–19.5) | Phillips et al. (2008) |
Lasiodiplodia thailandica T. Trakunyingcharoen, L. Lombard & Crous, sp. nov. — MycoBank MB810169; Fig. 5
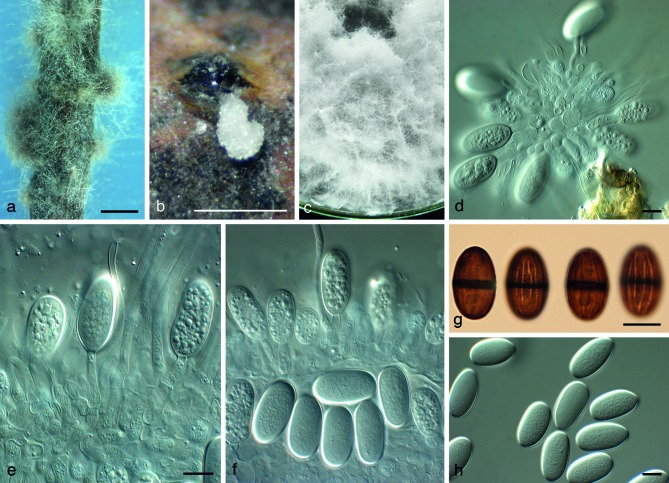
Fig. 5.
Lasiodiplodia thailandica (CBS 138653). a, b. Colony sporulating on PNA; b. fluffy aerial mycelium on PDA; d–f. conidiogenous cells giving rise to conidia; g. brown, 1-septate conidia; h. young, hyaline conidia. — Scale bars: a, b = 300 μm, all others = 10 μm.
Etymology. Name refers to the country where this fungus was collected, Thailand.
Conidiomata pycnidial, semi-immersed, solitary, rarely aggregated, dark brown to black, unilocular, with globose base, 310–330 × 300–370 μm, outer layers composed of dark brown textura angularis, becoming thin-walled and hyaline toward the inner region, hyphal hairs brown, septate, 60–150 × 5–6 μm, with rounded tips covering the outer wall of fruiting body. Ostiole central, circular, 40–60 μm diam; conidiomata papillate, neck 60–110 μm tall. Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline, smooth, thin-walled, discrete, cylindrical, holoblastic, proliferating percurrently from hyaline inner conidiomatal wall, 8–9 × 2–4 μm. Paraphyses hyaline, smooth, thin-walled, cylindrical, originating from the hyaline inner cells of pycnidial wall, the basal cells often slightly swollen, the apical cells with end-rounded tip, 1–3-septate, 25–51 × 1–1.5 μm. Conidia initially hyaline, wall 1.2–1.5 μm thick, ellipsoid, with granular content, unicellular, (20–)22–25(–26) × (12–)13–15(–16) μm, a few conidia turning pale brown with a single median septum and longitudinal striations after discharge from the pycnidia, but most of the discharged conidia remain hyaline.
Culture characteristics — Colonies with white fluffy mycelium on PDA, slightly mycelium dense and flattening at the centre, mycelium turning smoky-grey to olivaceous-grey with age, mycelium turning greenish olivaceous to black-olivaceous in reverse after 7 d.
Habitat — Symptomless twigs of Mangifera indica.
Known distribution — Chiang Mai province, Thailand.
Materials examined. THAILAND, Chiang Mai province, on twigs of Mangifera indica, May 2012, T. Trakunyingcharoen (holotype CBS-H 21933, culture ex-type CPC 22795 = CBS 138760); on petiole of Phyllanthus acidus, Feb. 2012, T. Trakunyingcharoen, CBS 138653 = CPC 22755. Additional isolates are listed in Table 1.
Notes — Conidia of L. thailandica are much smaller than conidia of L. crassispora, L. gilanensis, L. gonubiensis, L. macrospora, L. paraphysaria, L. plurivora, L. pseudotheobromae, L. rubropurpurea, L. theobromae, L. thomasiana and L. venezuelensis. However, conidia of L. thailandica are again larger than those of L. euphorbicola, L. hormozganensis, L. iraniensis, L. mahajangana, L. margaritacea, L. missouriana, L. parva, L. ricini, L. subglobosa and L. viticola. Although the conidial size of L. thailandica shows some overlap with L. citricola, L. egyptiacae and L. jatrophicola, these taxa are distinct based on the characteristics of their paraphyses. Paraphyses of L. thailandica are much shorter and narrower than those of the latter three species. Lasiodiplodia thailandica is phylogenetically closely related to L. iraniensis and L. jatrophicola. However, these species differ morphologically based on the dimensions of their conidia and paraphyses (Table 4). Furthermore, only some conidia of L. thailandica become pigmented after conidial discharge, which again separates it from most other Lasiodiplodia species.
Table 4.
A morphological comparison of Lasiodiplodia spp.
| Species | Conidial dimensions (μm) | Paraphyses | Reference | |
|---|---|---|---|---|
| Septation | Size (μm) | |||
| L. citricola | (20–)22–27(–31) × (10.9–)12–17(–19) | 1–5-septate | 125 × 3–4 | Abdollahzadeh et al. (2010) |
| L. crassispora | 27–30(–33) × 14–17 | septate | 45.7 × 2.7 | Burgess et al. (2006) |
| L. egyptiacae | 20–24 × 11–12 | aseptate | 57 × 2–3 | Ismail et al. (2012) |
| L. euphorbicola | 15–23 × 9–12 | septate | 76 × 2–4 | Machado et al. (2014) |
| L. gilanensis | (25.2–)28–35(–38.8) × (14.4–)15–18(–19) | 1–3-septate | 95 × 2–4 | Abdollahzadeh et al. (2010) |
| L. gonubiensis | (28–)32–36(–39) × (14–)16–18.5(–21) | aseptate | 38.1 × 2.3 | Pavlic et al. (2004) |
| L. hormozganensis | (15.3–)18–24(–25.2) × 11–14 | 1–7-septate | 83 × 2 –4 | Abdollahzadeh et al. (2010) |
| L. iraniensis | (15.3–)17–23(–29.7) × 11–14 | 1–6-septate | 127 × 2–4 | Abdollahzadeh et al. (2010) |
| L. jatrophicola | 22–26 × 14–17 | 0(–1)-septate | 70 × 3 | Machado et al. (2014) |
| L. macrospora | 28–35 × 15–17 | septate | 105 × 3–4 | Machado et al. (2014) |
| L. mahajangana | (13.5–)15.5–19(–21.5) × (10–)11.5–13(–14) | aseptate | 43 × 3 | Begoude et al. (2010) |
| L. margaritacea | (12–)14–17(–19) × (10–)11–12(–12.5) | 1–2-septate | 37.1 × 2.2 | Pavlic et al. (2008) |
| L. missouriana | (16.1—)17.4—19.6(—21) × (8.1–)8.9–10.6(–11.8) | aseptate | 55 × 2–3 | Urbez-Torres et al. (2012) |
| L. paraphysaria | 30–32 × 15–16 | 1-septate | 90–100 × 3 | Saccardo & Sydow (1899) |
| L. parva | (15.5–)16–23.5(–24.5) × (10–)10.5–13(–14.5) | septate | 105 × 3–4 | Alves et al. (2008) |
| L. plurivora | (22–)26.5–32.5(–35) × (13–)14.5–17(–18.5) | 1–6-septate | 130 × 2–5 | Damm et al. (2007a) |
| L. pseudotheobromae | (22.5–)23.5–32(–33) × (13.5–)14–18(–20) | mostly aseptate | 58 × 3–4 | Alves et al. (2008) |
| L. ricini | 16–19 × 10–11 | 1-septate | 25–35 × 2 | Saccardo (1915) |
| L. rubropurpurea | 24–33 × 13–17 | aseptate | 42.4 × 2.6 | Burgess et al. (2006) |
| L. subglobosa | 16–23 × 11–17 | aseptate | 41 × 2–3 | Machado et al. (2014) |
| L. thailandica | (20–)22–25(–26) × (12–)13–15(–16) | 1–3-septate | 51 × 1–1.5 | Present study |
| L. theobromae | (19–)21–31(–32.5) × (12–)13–15.5(–18.5) | septate | 55 × 3–4 | Alves et al. (2008) |
| L. thomasiana | 28–30 × 11–12 | – | 80–90 × 1.5 | Saccardo & Trotter (1913) |
| L. venezuelensis | 26–33 × 12–15 | septate | 28.3 × 3.5 | Burgess et al. (2006) |
| L. viticola | (16.8–)18.2–20.5(–22.9) × (7.9–)8.8–10.1(–10.7) | aseptate | 60 × 2–3 | Urbez-Torres et al. (2012) |
Pseudofusicoccum artocarpi T. Trakunyingcharoen, L. Lombard & Crous, sp. nov. — MycoBank MB810170; Fig. 6
Fig. 6.
Pseudofusicoccum artocarpi (CBS 138655). a. Sporulation on PNA; b, c. aggregated conidiomata on host tissue; d. colony on MEA with fluffy aerial mycelium; e. section through conidiomata; f. conidia becoming brown and septate with age; g. young conidia. — Scale bars: b, c, e = 250 μm, f, g = 10 μm.
Etymology. The name refers to the host genus from which it was collected, Artocarpus.
Conidiomata pycnidial, semi-immersed, solitary to aggregated, mostly aggregated, dark brown, unilocular, rarely multilocular, with globose base, (350–)400–520(–550) × (130–)170–280 (–360) μm, covered by pale brown, septate hyphal hairs, that turn brown with age, (60–)80–180(–250) × 2–3(–4) μm; outer layers composed of dark brown textura angularis, becoming thin-walled and hyaline toward the inner region, 5–6 cells thick. Ostiole central, circular, rarely with 2 ostioles, ostiole 40–50 μm diam; conidiomata papillate, neck length 80–100(–140) μm tall. Conidiophores hyaline, cylindrical, 9–11 × 2 μm, 0–2-septate. Conidiogenous cells hyaline, smooth, thin-walled, discrete, cylindrical, holoblastic, proliferating percurrently near apex, formed from the hyaline inner wall of the conidiomata, 8–11(–13) × ( 2–)3–4 μm. Paraphyses absent. Conidia hyaline, bacilliform to ellipsoid, straight to slightly curved, both apex and base blunt to broadly round, moderately thick-walled, with granular content, aseptate, (33–)34–43(–46) × 7–8(–9) μm, becoming pale brown and (1–)3–5(–7)-septate with age, sometimes conidia turn pale brown and septate while still attached to the conidiogenous cells.
Culture characteristics — Colonies with white, cottony aerial mycelium on PDA, moderately dense and flattening at the centre, becoming smoky-grey with age, and turning olivaceous-grey in reverse after 7 d.
Habitat — Asymptomatic twig of Artocarpus heterophyllus. Known distribution — Chiang Mai province, Thailand.
Material examined. THAILAND, Chiang Mai province, on twigs of Artocarpus heterophyllus, May 2012, T. Trakunyingcharoen (holotype CBS H-21935, culture ex-type CPC 22796 = CBS 138655). Additional isolates are listed in Table 1.
Notes — Conidia of P. artocarpi are clearly longer than those of other Pseudofusicoccum species, with conidia becoming pale brown and up to 7-septate with age (Table 5). These morphological characters are rather typical, and can easily be used to distinguish P. artocarpi from other Pseudofusicoccum species (Pavlic et al. 2008).
Table 5.
A comparison of conidial dimensions of Pseudofusicoccum spp.
| Species | Conidial dimensions (μm) | Reference |
|---|---|---|
| P. adansoniae | (19–)21–24(–26) × (3.5–)4.5–6(–6.5) | Pavlic et al. (2008) |
| P. ardesiacum | (17.5–)21–29(–32) × (6.3–)7–8(–9) | Pavlic et al. (2008) |
| P. artocarpi | (33–)34–43(–46) × 7–8(–9) | Present study |
| P. kimberleyense | (24–)28–33(–34) × (6.5–)7–8(–8.5) | Pavlic et al. (2008) |
| P. olivaceum | (17.9–)19.9–25.7(–30.4) × (5.9–)6.3–7.7(–8.9) | Mehl et al. (2011) |
| P. stromaticum | (19–)20–23(–24) × (4–)5–6 | Mohali et al. (2006) |
| P. violaceum | (26.5–)29.8–36.1(–39.6) × (8.0–)8.7–10.3(–11.6) | Mehl et al. (2011) |
Discussion
Schoch et al. (2006) introduced the order Botryosphaeriales with the Botryosphaeriaceae as a single family within the order. Based on subsequent research, however, six families are presently recognised within Botryosphaeriales, namely Aplosporellaceae, Botryosphaeriaceae, Melanopsaceae, Phyllostictaceae, Planistromellaceae and Saccharataceae (Liu et al. 2012, Minnis et al. 2012, Slippers et al. 2013, Wikee et al. 2013). In the present study, species of Aplosporellaceae and Botryosphaeriaceae obtained from various host plants in Thailand were identified as corresponding to the genera Aplosporella, Botryosphaeria, Diplodia, Lasiodiplodia, Neofusicoccum and Pseudofusicoccum based on morphology and DNA phylogeny.
Although members of these genera are commonly encountered, not much is known about the host specificity and relative importance of the majority of species that have been described to date. Species of Diplodia represent important pathogens that can cause blight, canker, dieback and rot diseases on numerous host plants (Burgess et al. 2004, Lazzizera et al. 2008, Linaldeddu et al. 2011). Several Diplodia species have been reported to be associated with Cupressus and Juniperus, including D. cupressi (Alves et al. 2006) and D. mutila (Tisserat et al. 1988, Flynn & Gleason 1993). In the present study an undescribed Diplodia species morphologically similar to D. juniper was isolated from Juniperus chinensis in Thailand, and described here as D. neojuniperi. The genus Lasiodiplodia represents one of the most well-known genera in the Botryosphaeriaceae, with species recorded on a broad host range in tropical and subtropical regions (Punithalingam 1976). Accordingly, isolates of Lasiodiplodia spp. appeared to represent the most dominant clade, and have the widest distribution and host range of all isolates collected in Thailand. Species of Lasiodiplodia obtained from Thailand were identified as L. pseudotheobromae, L. theobromae and L. viticola. Furthermore, L. thailandica was introduced as novel taxon, and the sexual morph of L. gonubiensis was also reported for the first time. Lasiodiplodia gonubiensis was first introduced by Pavlic et al. (2004) for endophytic isolates in leaves and branches of native Syzygium cordatum from South Africa. The collection of its sexual morph in Thailand is not totally unexpected, as it seems that generally species of Lasiodiplodia have wide geographical distributions (Phillips et al. 2013).
The genus Neofusicoccum was first introduced by Crous et al. (2006), and includes many species that are important pathogens causing several plant diseases globally (Slippers et al. 2005, de Oliveira Costa et al. 2010, Thomidis et al. 2011, Ni et al. 2012). Although some species such as N. arbuti and N. protearum appear to be largely host-specific (Phillips et al. 2013), most species of the genus have wide host ranges and geographical distributions. The genus Pseudofusicoccum was introduced by Crous et al. (2006) for species resembling Fusicoccum (= Botryosphaeria), but being distinct by having a persistent mucous sheath surrounding their conidia. Although Pavlic et al. (2008) suggested that all species of Pseudofusicoccum could be native to Australia, species such as P. olivaceum and P. violaceum were introduced on Pterocarpus angolensis native to South Africa, and appear to represent the first Pseudofusicoccum spp. not native to Australia (Mehl et al. 2011). In the present study P. adansoniae and P. ardesiacum were also collected from native and non-native plants of Thailand, and this is the first report of these species in this region. Pseudofusicoccum adansoniae was obtained from Senna siamea, Cassia fistula, Dimocarpus longan and Hevea brasiliensis, while P. ardesiacum was isolated from Veitchia merrillii and Caesalpinia pulcherrima. Furthermore, a seventh species of Pseudofusicoccum was newly described here as P. artocarpi from Artocarpus heterophyllus. Conidia of P. artocarpi are much longer than any of the presently known species, and also turn pale brown and multiseptate with age.
Aplosporella was previously included as a member of the Botryosphaeriaceae based on the molecular analyses conducted by Damm et al. (2007b). It was only recently excluded from this family and allocated to the Aplosporellaceae (Slippers et al. 2013). Historically, species of Aplosporella have mostly been described based on their host occurrence (Damm et al. 2007b). In the present study, A. artocarpi was introduced as a novel species from Artocarpus heterophyllus in Thailand. No species of Aplosporella have thus far been described from Artocarpus, and A. artocarpi is also distinct from all taxa presently known from their DNA sequence data. However, the genus is clearly under-represented, and many more collections would be required in an attempt to understand its host specificity and potential pathogenicity.
In general, species of Botryosphaeriaceae have cosmopolitan distributions and broad host ranges. They are commonly encountered as endophytes and opportunistic pathogens, causing a range of important plant diseases leading to economic losses in many regions of the world (Slippers & Wingfield 2007). Very little has been known about botryosphaeriaceous fungi in Thailand until the recent study of Liu et al. (2012). Likewise, the present study adds four new species and three new records from native and non-native plant hosts in Thailand as belonging to the Aplosporellaceae and Botryosphaeriaceae. Further studies on the ecology, epidemiology, distribution and pathogenicity of these taxa is now urgently required to provide a better understanding about the importance and potential impact that these fungi may have on woody hosts in Thailand.
Acknowledgments
This work was financially supported by the Laboratory of Evolutionary Phytopathology, CBS-KNAW Fungal Biodiversity Centre (CBS), Utrecht, The Netherlands, the Thailand Research Fund (DBG5380011 and MRG5580163) and the Royal Golden Jubilee PhD Programme (Grant No. PHD/0353/2552). The first author is grateful to Nattapol Srithongkum and Nattawut Pattamas for their help during collection trips. Special thanks are also extended to the staff at CBS for their kind suggestions and technical support.
REFERENCES
- Abdollahzadeh J, Javadi A, Goltapeh EM, et al. 2010. Phylogeny and morphology of four new species of Lasiodiplodia from Iran. Persoonia 25: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves A, Correia A, Luque J, et al. 2004. Botryosphaeria corticola, sp. nov. on Quercus species, with notes and description of Botryosphaeria stevensii and its anamorph, Diplodia mutila. Mycologia 96: 598–613. [PubMed] [Google Scholar]
- Alves A, Correia A, Phillips AJL. 2006. Multi-gene genealogies and morphological data support Diplodia cupressi sp. nov., previously recognized as D. pinea f. sp. cupressi, as a distinct species. Fungal Diversity 23: 1–15. [Google Scholar]
- Alves A, Crous PW, Correia A, et al. 2008. Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13. [Google Scholar]
- von Arx JA, Müller E. 1954. Die Gattungen der amerosporen Pyrenomyceten. Beiträge zur Cryptogamenflora der Schweiz II (I): 1–434. [Google Scholar]
- Begoude BAD, Slippers B, Wingfield MJ, et al. 2010. Botryosphaeriaceae associated with Terminalia catappa in Cameroon, South Africa and Madagascar. Mycological Progress 9: 101–123. [Google Scholar]
- Burgess TI, Sakalidis ML, Hardy GEStJ. 2006. Gene flow of the canker pathogen Botryosphaeria australis between Eucalyptus globulus plantations and native eucalypt forests in Western Australia. Australasian Ecology 31: 559–566. [Google Scholar]
- Burgess TI, Wingfield MJ, Wingfield BD. 2004. Global distribution of Diplodia pinea genotypes revealed using simple sequence repeat (SSR) markers. Australasian Plant Pathology 33: 513–519. [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Park RF. 1991. Mycosphaerella nubilosa a synonym of M. molleriana. Mycological Research 95: 628–632. [Google Scholar]
- Damm U, Crous PW, Fourie PH. 2007a. Botryosphaeriaceae as potential pathogens of Prunus species in South Africa, with descriptions of Diplodia africana and Lasiodiplodia plurivora sp. nov. Mycologia 99: 664–680. [DOI] [PubMed] [Google Scholar]
- Damm U, Fourie PH, Crous PW. 2007b. Aplosporella prunicola, a novel species of anamorphic Botryosphaeriaceae. Fungal Diversity 27: 35–43. [Google Scholar]
- Denman S, Crous PW, Groenewald JG, et al. 2003. Circumscription of Botryosphaeria species associated with Proteaceae based on morphology and DNA sequence data. Mycologia 95: 294–307. [PubMed] [Google Scholar]
- Denman S, Crous PW, Taylor JE, et al. 2000. An overview of the taxonomic history of Botryosphaeria, and a re-evaluation of its anamorphs based on morphology and ITS rDNA phylogeny. Studies in Mycology 45: 129–140. [Google Scholar]
- Flynn PH, Gleason ML. 1993. Isolation of Botryosphaeria stevensii, cause of Botryosphaeria canker, from rocky mountain juniper in Iowa. Plant Disease 77: 210. [Google Scholar]
- Fuckel L. 1870. Symbolae mycologicae: Beiträge zur Kenntniss der rheinischen Pilze. Jahrbücher des Nassauischen Vereins für Naturkunde 23–24: 1–459. [Google Scholar]
- Gure A, Slippers B, Stenlid J. 2005. Seed-borne Botryosphaeria spp. from native Prunus and Podocarpus trees in Ethiopia, with a description of the anamorph Diplodia rosulata sp. nov. Mycological Research 109: 1005–1014. [DOI] [PubMed] [Google Scholar]
- Hillis DM, Bull JJ. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology 42: 182–192. [Google Scholar]
- Ismail AM, Cirvilleri G, Polizzi G, et al. 2012. Lasiodiplodia species associated with dieback disease of mango (Mangifera indica) in Egypt. Australasian Plant Pathology 41: 649–660. [Google Scholar]
- Larget B, Simon D. 1999. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution 16: 750–759. [Google Scholar]
- Lazzizera C, Frisullo S, Alves A, et al. 2008. Morphology, phylogeny and pathogenicity of Botryosphaeria and Neofusicoccum species associated with drupe rot of olives in southern Italy. Plant Pathology 57: 948–956. [Google Scholar]
- Linaldeddu BT, Franceschini A, Alves A, et al. 2013. Diplodia quercivora sp. nov.: a new species of Diplodia found on declining Quercus canariensis trees in Tunisia. Mycologia 105: 1266–1274. [DOI] [PubMed] [Google Scholar]
- Linaldeddu BT, Scanu B, Maddau L, et al. 2011. Diplodia africana causing dieback on Juniperus phoenicea: a new host and first report in the northern hemisphere. Phytopathologia Mediterranea 50: 473–477. [Google Scholar]
- Liu JK, Phookamsak R, Mingkhuan M, et al. 2012. Towards a natural classification of Botryosphaeriales. Fungal Diversity 57: 149–210. [Google Scholar]
- Lynch SC, Eskalen A, Zambino PJ, et al. 2013. Identification and pathogenicity of Botryosphaeriaceae species associated with coast live oak (Quercus agrifolia) decline in southern California. Mycologia 105: 125–140. [DOI] [PubMed] [Google Scholar]
- Machado AR, Pinho DB, Pereira OL. 2014. Phylogeny, identification and pathogenicity of the Botryosphaeriaceae associated with collar and root rot of the biofuel plant Jatropha curcas in Brazil, with a description of new species of Lasiodiplodia. Fungal Diversity: doi 10.1007/s13225-013-0274-1. [Google Scholar]
- Marincowitz S, Groenewald JZ, Wingfield MJ, et al. 2008. Species of Botryosphaeriaceae occurring on Proteaceae. Persoonia 21: 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques MW, Lima NB, Morais Jr MA de, et al. 2013. Species of Lasiodiplodia associated with mango in Brazil. Fungal Diversity 61: 181–193. [Google Scholar]
- McDonald V, Eskalen A. 2011. Botryosphaeriaceae species associated with avocado branch cankers in California. Plant Disease 95: 1465–1473. [DOI] [PubMed] [Google Scholar]
- Mehl JWM, Slippers B, Roux J, et al. 2011. Botryosphaeriaceae associated with Pterocarpus angolensis (kiaat) in South Africa. Mycologia 103: 534–553. [DOI] [PubMed] [Google Scholar]
- Minnis AM, Kennedy AH, Grenier DB, et al. 2012. Phylogeny and taxonomic revision of the Planistromellaceae including its coelomycetous anamorphs: contributions towards a monograph of the genus Kellermania. Persoonia 29: 11–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohali S, Slippers B, Wingfield MJ. 2006. Two new Fusicoccum species from Acacia and Eucalyptus in Venezuela, based on morphology and DNA sequence data. Mycological Research 110: 405–413. [DOI] [PubMed] [Google Scholar]
- Mohali S, Slippers B, Wingfield MJ. 2007. Identification of Botryosphaeriaceae from Eucalyptus, Acacia and Pinus in Venezuela. Fungal Diversity 25: 103–125. [Google Scholar]
- Montagne JFC. 1834. Notice sur les plantes cryptogames récemment découvertes en France contenant aussi l’indication précis des localités de quelques espèces les plus rares de la flore française. Annales des Sciences Naturelles Botanique, sér. 2, 1: 295–307. [Google Scholar]
- Ni H-F, Yang H-R, Chen R-S, et al. 2012. New Botryosphaeriaceae fruit rot of mango in Taiwan: identification and pathogenicity. Botanical Studies 53: 467–478. [Google Scholar]
- van Niekerk JM, Fourie PH, Halleen F, et al. 2006. Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathology Mediterranea 45: 43–54. [Google Scholar]
- Notaris G de. 1845. Micromycetes italici novi vel minus cogniti. Decas 4. Memorie della Reale Accademia delle Scienze di Torino 7: 17–30. [Google Scholar]
- Nylander JAA. 2004. MrModel Test v. 2. Programme distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, et al. 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the Unites States of America 95: 2044–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira Costa VS de, Michereff SJ, Martins RB, et al. 2010. Species of Botryosphaeriaceae associated on mango in Brazil. European Journal Plant Pathology 127: 509–519. [Google Scholar]
- Pande A, Rao VG. 1995. The genus Aplosporella Speg. (= Haplosporella Speg.). Coelomycetes from India. Nova Hedwigia 60: 79–117. [Google Scholar]
- Pavlic D, Slippers B, Coutinho TA, et al. 2004. Lasiodiplodia gonubiensis sp. nov., a new Botryosphaeria anamorph from native Syzygium cordatum in South Africa. Studies in Mycology 50: 313–322. [Google Scholar]
- Pavlic D, Wingfield MJ, Barber P, et al. 2008. Seven new species of the Botryosphaeriaceae from baobab and other native trees in Western Australia. Mycologia 100: 851–866. [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Abdollahzadeh J, et al. 2013. The Botryosphaeriaceae: genera and species known from culture. Studies in Mycology 76: 51–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Correia A, et al. 2005. Two new species of Botryosphaeria with brown, 1-septate ascospores and Dothiorella anamorphs. Mycologia 97: 513–529. [DOI] [PubMed] [Google Scholar]
- Phillips AJL, Alves A, Pennycook SR, et al. 2008. Resolving the phylogenetic and taxonomic status of dark-spored teleomorph genera in Botryosphaeriaceae. Persoonia 21: 29–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AJL, Lopes J, Abdollahzadeh J, et al. 2012. Resolving the Diplodia complex on apple and other Rosaceae hosts. Persoonia 29: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punithalingam E. 1976. Botryodiplodia theobromae. CMI descriptions of pathogenic fungi and bacteria, No. 519. Commonwealth Mycological Institute, Kew, UK. [Google Scholar]
- Quaglia M, Moretti C, Buonaurio R. 2014. Molecular characterization of Diplodia seriata, a new pathogen of Prunus laurocerasus in Italy. Phytoparasitica 42: 189–197. [Google Scholar]
- Rannala B, Yang Z. 1996. Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Oliver JF, Marin A, et al. 1990. The general stochastic model of nucleotide substitutions. Journal of Theoretical Biology 142: 485–501. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Saccardo PA. 1884. Sylloge fungorum omnium hucusque cognitorum. Vol. III Padova. [Google Scholar]
- Saccardo PA. 1880. Fungi gallici, series II. Michelia 2: 38–135. [Google Scholar]
- Saccardo PA. 1915. Fungi ex insula Melita (Malta) lecti a Doct. Caruana-Gatto et Doct. G. Borg annis MCMXIII et MCMIV. Nuovo Giornale Botanico Italiano 22: 61. [Google Scholar]
- Saccardo PA, Sydow P. 1899. Supplementum Universale, Pars IV. Sylloge Fungorum 14: 938. [Google Scholar]
- Saccardo PA, Trotter A. 1913. Supplementum Universale, Pars IX. Sylloge Fungorum 22: 1012. [Google Scholar]
- Sánchez ME, Venegas J, Romero MA, et al. 2003. Botryosphaeria and related taxa causing oak canker in southwestern Spain. Plant Disease 87: 1515–1521. [DOI] [PubMed] [Google Scholar]
- Schoch CL, Shoemaker RA, Seifert KA, et al. 2006. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. [DOI] [PubMed] [Google Scholar]
- Slippers B, Boissin E, Phillips AJL, et al. 2013. Phylogenetic lineages in the Botryosphaeriales: a systematic and evolutionary framework. Studies in Mycology 76: 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slippers B, Crous PW, Denman S, et al. 2004a. Combined multiple gene genealogies and phenotypic characters differentiate several species previously identified as Botryosphaeria dothidea. Mycologia 96: 83–101. [PubMed] [Google Scholar]
- Slippers B, Fourie G, Crous PW, et al. 2004b. Speciation and distribution of Botryosphaeria spp. on native and introduced Eucalyptus trees in Australia and South Africa. Studies in Mycology 50: 343–358. [Google Scholar]
- Slippers B, Johnson GI, Crous PW, et al. 2005. Phylogenetic and morphological re-evaluation of the Botryosphaeria species causing diseases of Mangifera indica. Mycologia 97: 99–110. [DOI] [PubMed] [Google Scholar]
- Slippers B, Smit WA, Crous PW, et al. 2007. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit trees in South Africa and other regions of the world. Plant Pathology 56: 128–139. [Google Scholar]
- Slippers B, Wingfield MJ. 2007. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biology Reviews 21: 90–106. [Google Scholar]
- Smith H, Wingfield MJ, Crous PW, et al. 1996. Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. South African Journal of Botany 62: 86–88. [Google Scholar]
- Swofford DL. 2003. PAUP* Phylogenetic analysis using parsimony (*and other methods) version 4. Sinauer Associates, Sunderland, Massachusetts. [Google Scholar]
- Swofford DL, Begle DP. 1993. PAUP: Phylogenetic analysis using parsimony, ver. 3.1. User’s manual. Illinois Natural History Survey, Champaign, IL. [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. 2011. MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K, Barber PA, Hardy GEStJ, et al. 2009. Botryosphaeriaceae from tuart (Eucalyptus gomphocephala) woodland, including descriptions of four new species. Mycological Research 113: 337–353. [DOI] [PubMed] [Google Scholar]
- Thomidis T, Michailides TJ, Exadaktylou E. 2011. Neofusicoccum parvum associated with fruit rot and shoot blight of peaches in Greece. European Journal of Plant Pathology 131: 661–668. [Google Scholar]
- Tisserat NA, Rossman AY, Nus A. 1988. A canker disease of rocky mountain juniper caused by Botryosphaeria stevensii. Plant Disease 72: 699–701. [Google Scholar]
- Trakunyingcharoen T, Cheewangkoon R, To-anun C. 2013. Phylogeny and pathogenicity of fungal species in the family Botryosphaeriaceae associated with mango (Mangifera indica) in Thailand. International Journal of Agricultural Technology 9: 1535–1543. [Google Scholar]
- Trakunyingcharoen T, Cheewangkoon R, To-anun C, et al. 2014. Botryosphaeriaceae associated with diseases of mango (Mangifera indica). Australasian Plant Pathology 43: 425–438. [Google Scholar]
- Urbez-Torres JR, Peduto F, Striegler RK, et al. 2012. Characterization of fungal pathogens associated with grapevine trunk diseases in Arkansas and Missouri. Fungal Diversity 52: 169–189. [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA. [Google Scholar]
- Wikee S, Lombard L, Nakashima C, et al. 2013. A phylogenetic re-evaluation of Phyllosticta (Botryosphaeriales). Studies in Mycology 76: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenopoulos S, Tsopelas P. 2000. Sphaeropsis canker, a new disease of cypress in Greece. Forest Pathology 30: 121–126. [Google Scholar]
- Zhou XD, Xie YJ, Chen SF, et al. 2008. Diseases of eucalypt plantations in China: challenges and opportunities. Fungal Diversity 32: 1–7. [Google Scholar]