Highlights
-
•
We review the genetics of wheat resistance to Septoria tritici blotch, with a map of known genes.
-
•
Qualitative resistance is usually monogenic, genotype-specific and non-durable.
-
•
Quantitative resistance is generally polygenic with low specificity and greater durability.
-
•
Major requirements for resistance breeding are diverse germplasm and field sites with severe Septoria.
Keywords: Durable resistance, Gene-for-gene relationship, Genetic mapping, Plant breeding, Quantitative trait locus (QTL), Septoria tritici blotch
Abstract
This paper reviews current knowledge about genes for resistance to Septoria tritici blotch (STB) of wheat, caused by Zymoseptoria tritici (formerly Mycosphaerella graminicola). These genes can be placed into two classes, although a few may have characteristics of both classes. Qualitative resistance is controlled by genes which control large fractions of genetic variation, 21 of which have been discovered and mapped so far. Most of them have been shown to be genotype-specific, being effective against the minority of Z. tritici isolates which are avirulent, and Stb6 has been shown to control a gene-for-gene relationship. Most qualitative resistances are unlikely to be durable and some formerly effective genes have been overcome by the evolution of pathogen virulence. Quantitative resistance is generally controlled by genes with small-to-moderate effects on STB. They have generally weaker specificity than qualitative genes and have provided more durable resistance. 89 genome regions carrying quantitative trait loci (QTL) or meta-QTL have been identified to date. Some QTL have been mapped at or near loci of qualitative genes, especially Stb6, which is present in several sources of resistance. Another gene of particular interest is Stb16q, which has been effective against all Z. tritici isolates tested so far. In addition to resistance, the susceptibility of wheat cultivars to STB can also be reduced by disease escape traits, some of which may be undesirable in breeding. The fundamental requirements for breeding for STB-resistance are genetic diversity for resistance in wheat germplasm and a field trial site at which STB epidemics occur regularly and effective selection can be conducted for resistance combined with other desirable traits. If these are in place, knowledge of resistance genes can be applied to improving control of STB.
1. Types of resistance to Septoria tritici blotch
Resistance to S. tritici blotch (STB; caused by Zymoseptoria tritici, formerly Mycosphaerella graminicola) became a significant target trait in wheat breeding much more recently than diseases such as the rusts and powdery mildew. The potential threat posed by STB was drawn to international attention by a very damaging epidemic in North Africa in 1968–1969, which followed the introduction of semi-dwarf wheat cultivars and increased use of artificial fertiliser (Saari and Wilcoxson, 1974). Subsequently, STB increased in importance, particularly in semi-dwarf cultivars given high rates of nitrogen fertiliser (Wiese, 1987) and is now a potentially damaging disease throughout the temperate regions (HGCA, 2012; O’Driscoll et al., 2014; Fones and Gurr, 2015). Early work on genetics (reviewed by Goodwin, 2007) focussed on the discovery of sources of resistance for breeding and on cultivar-by-isolate interaction but substantial progress has been made in the last 20 years in the genetics of resistance. This is giving breeders a deeper understanding of effective approaches to improving resistance (Torriani et al., 2015) and will allow resistance genes to be isolated and their functions revealed.
As in many other plant diseases, wheat has essentially two types of resistance to STB, as shown in a large study of 236 wheat cultivars grown in the UK in the 1990s and their progenitors (Arraiano and Brown, 2006; Arraiano et al., 2009). Qualitative resistance is strong and is usually controlled by major genes with a large effect. These genes are generally effective against avirulent pathogens but not against other, virulent isolates. Their pattern of interaction with Z. tritici accords with the gene-for-gene relationship, as has been demonstrated for Stb6 (Brading et al., 2002). Quantitative resistance, by contrast, has a partial phenotype and is controlled by several-to-many genes with moderate-to-small effects. In many instances but not always, it is effective against all Z. tritici genotypes. Even though STB is almost always scored as a quantitative trait or at least on an ordinal scale, segregation of a qualitative gene can give rise to a large difference between two groups of progeny, resistant and susceptible (e.g. Stb6; Brading et al., 2002). This can be obscured, however, by segregation of minor genes which alter the level of STB symptoms and lead to intermediate phenotypes.
In certain cultivar-by-isolate interactions, adult-plant responses to Z. tritici do not necessarily reflect responses of seedlings to the pathogen (Kema and van Silfhout, 1997; Chartrain et al., 2004a). Many genotype-specific, qualitative resistances are independent of growth stage (Kema and van Silfhout, 1997; Arraiano et al., 2001a; Brown et al., 2001; Grieger et al., 2005) whereas the expression of partial resistance may depend on the plant’s growth stage (Chartrain et al., 2004a). Stb17 is an example of a gene with a quantitative effect on disease which is expressed in adult plants but not seedlings (Tabib Ghaffary et al., 2012), while genes on chromosome arm 5BS of Hobbit sib increased susceptibility only in adult plants (Arraiano et al., 2007a).
In this review, we survey the genes for STB-resistance reported to date (Fig. 1), including both qualitative (Table 1) and quantitative resistance (Table S1), beginning with the first genes to be named, Stb1, Stb2 and Stb3 (Wilson, 1985). Earlier reports of sources and genetics of resistance were summarised by Kema et al. (1996a, 1996b), Goodwin (2007) and Raman and Milgate (2012). We also describe how knowledge of the genetics of resistance can be applied to wheat breeding.
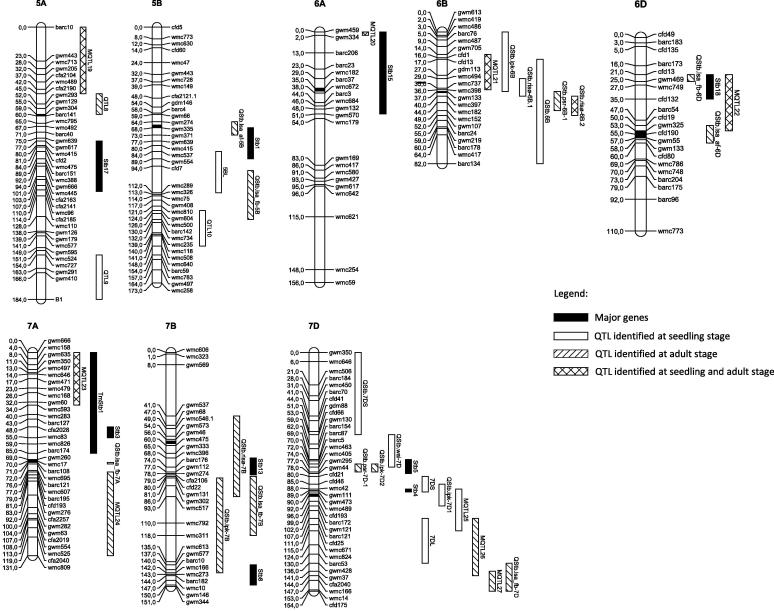
Fig. 1.
Location in the wheat genome of major genes, QTL and meta-QTL involved in resistance to Septoria tritici blotch. Loci have been projected on the simplified SSR consensus map of Somers et al. (2004). Five QTL from Table S1 were not included in the map due to a lack of shared markers between the original paper and the consensus map. Solid bars represent major genes (see Section 1 and Table 1) and other bars patterns indicate QTL identified at different plant growth stages.
Table 1.
Major genes for resistance of bread wheat (Triticum aestivum) to Septoria tritici blotch, with their chromosomal locations, nearest markers, Z. tritici isolates with which they were identified, growth stage at which plants were inoculated (S: seedling, A: adult) and resistant source line.
| Gene | Chromo-some | Associated markers (distance to gene) | Avirulent inoculum | Stage | Resistance source | References |
|---|---|---|---|---|---|---|
| Stb1 | 5BL | Xbarc74 (2.8cM), Xgwm335 (7.4cM) | IN95-Lafayette-1196-WW 1-4 & Purdue local (USA) | S, A | Bulgaria 88 | Adhikari et al. (2004a) |
| Stb2 | 1BS | Xwmc406 (6cM), Xwmc230 (5cM) | Paskeville local (Australia) (and IPO92034) | A | Veranopolis | Liu et al. (2013) |
| Stb3 | 7AS | Xwmc83 | Paskeville local isolate (Australia) | A | Israel 493 | Goodwin and Thompson (2011) |
| Stb4 | 7DS | Xgwm111 (0.7cM) | IN95-Lafayette-1196-WW-1-4, I-89, IPBr1 | S, A | Tadinia | Adhikari et al. (2004c) |
| Stb5 | 7DS | Xgwm44 (7.2cM) | IPO94269 | S, A | Synthetic 6x | Arraiano et al. (2001b) |
| Stb6 | 3AS | Xgwm369 (2cM) | IPO323 | S, A | Flame, Hereward | Brading et al. (2002) |
| Stb7 | 4AL | Xwmc313 (0.3 to 0.5cM), Xwmc219 (1cM) | MG2 (Canada) (and IPO87019) | S | ST6 | McCartney et al. (2003) |
| Stb8 | 7BL | Xgwm146 (3.5cM), Xgwm577 (5.3cM) | IN95-Lafayette-1196-WW 1-4 | A | Synthetic W7984 | Adhikari et al. (2003) |
| Stb9 | 2BL | Xfbb226 (3.6cM), Xwmc317, Xbarc0129 | IPO89011 | S | Courtot, Tonic | Chartrain et al. (2009) |
| Stb10 | 1Dc | Xgwm848 | IPO94269 and ISR8036 | S | Kavkaz-K4500 | Chartrain et al. (2005c) |
| Stb11 | 1BS | Xbarc008 (1cM) | IPO90012 | S | TE9111 | Chartrain et al. (2005a) |
| Stb12 | 4AL | Xwmc219 | ISR398 and ISR8036 | S | Kavkaz-K4500 | Chartrain et al. (2005c) |
| Stb13 | 7BL | Xwmc396 (7-9cM) | MG96-36, MG2 (Canada) | S | Salamouni | Cowling (2006) |
| Stb14 | 3BS | Xwmc500 (2cM), wmc632 (5cM) | MG2 (Canada) | S | Salamouni | Cowling (2006) |
| Stb15 | 6AS | Xpsr904 (14cM) | IPO88004 | S | Arina, Riband | Arraiano et al. (2007b) |
| StbSm3 | 3AS | barc321 (1.9cM) | MG96-36, MG2 (Canada) | S | Salamouni | Cuthbert (2011) |
| Stb16q | 3DL | Xgwm494 (4.3cM), Xbarc128 (9.9cM) | IPO88018 and IPO94218 | S, A | SH M3 | Tabib Ghaffary et al. (2012) |
| Stb17 | 5AL | Xhbg247 (3.1cM), Xgwm617 (38.3cM) | IPO88018 | A | SH M3 | Tabib Ghaffary et al. (2012) |
| Stb18 | 6DS | Xgpw5176, Xgpw3087 | IPO323, IPO98022, IPO89011, IPO98046 | S, A | Balance | Tabib Ghaffary et al. (2011) |
| StbWW | 1BS | Xbarc119b (0.9–4.1cM) | 79, 2, 1A | S | WW1842, WW2449, WW2451 | Raman et al. (2009) |
| TmStb1 | 7AmS | Xbarc174 (23.5cM) | IPO323 | S | MDR043 (T. monococcum) | Jing et al. (2008) |
2. Methods of studying resistance
As with genetic analysis of any trait, study of STB-resistance requires a method of scoring the phenotype which can be applied to large populations. Tests can be conducted at the seeding stage, with inoculation typically when seedlings are two weeks old. Both whole-seedling (Brading et al., 2002) and detached leaf assays (Arraiano et al., 2001a) are used, and require conditions with high relative humidity. They generally take around four weeks following inoculation. The advantage of using whole seedlings is that many plants can be tested but a disadvantage, especially in greenhouse trials, is that environmental conditions may not be strictly controlled, which can affect the development of disease (Arraiano et al., 2001a). Detached-leaf tests are particularly suitable when many isolates are to be tested, although this can also be done in a greenhouse (Kema et al., 1996a). Alternatively, plants can be tested at the adult stage in the glasshouse (Adhikari et al., 2003) or field (Kema and van Silfhout, 1997). Given the genotype-specificity of most qualitative resistances (see below), identification of single genes in field conditions requires plants to be inoculated with a Z. tritici isolate at a dose sufficient to make contamination by natural inoculum comparatively negligible (Kema and van Silfhout, 1997; Brown et al., 2001). The expression of symptoms following inoculation in adult plants proceeds at a broadly similar rate as in seedlings. When genetic analysis is conducted on naturally-infected trials, the genes identified are effective against the current local Z. tritici population. This is generally relevant to the practice of plant breeding but may have implications for repeatability if there is polymorphism at avirulence loci corresponding to segregating resistance genes.
In most genetic analyses of STB-resistance, the phenotype studied is formation of pycnidia, the asexual fruiting bodies of Z. tritici which form within necrotic tissue on the leaf. In this case, the data are fractions of leaf area covered by necrotic lesions bearing pycnidia. Some studies have reported other phenotypes in addition to pycnidium formation, including total necrotic leaf area with or without pycnidia, latent period and disease progress. These traits are scored visually, either by eye or, more recently, by computer-aided image analysis (Stewart and McDonald, 2014).
3. Qualitative resistance
Please refer to Table 1 for details of genes for STB-resistance in bread wheat (Triticum aestivum) and to Fig. 1 for their locations.
3.1. Stb1, Stb2 and Stb3
Stb1, Stb2 and Stb3 were the first genes for STB-resistance to be named (Wilson, 1985). Before then, it was generally thought that resistance to Z. tritici was a quantitative, polygenic trait. Although quantitative resistance is indeed considerably more important than qualitative resistance in wheat breeding, the discovery that significant amounts of resistance can be controlled by major genes opened the way to genetic analysis of STB and may offer an opportunity to improve resistance by ‘stacking’ or ‘pyramiding’ several Stb genes (Chartrain et al., 2004b; see Section 6.1).
Stb1, Stb2 and Stb3 have been mapped to chromosome arms 5BL (i.e. the long arm of chromosome 5B; Adhikari et al., 2004a), 1BS (the short arm of 1B; Liu et al., 2013) and 7AS (Goodwin and Thompson, 2011) respectively. Stb2 and Stb3 were originally mapped to chromosomes 3BS and 6DS respectively (Adhikari et al., 2004b) but those locations were corrected for the reasons given in the subsequent papers. Stb2 was found to map to the same region of 1BS as Stb11 (Chartrain et al., 2005a) but no test of allelism of the two genes has yet been done.
The sources of Stb1, Stb2 and Stb3 – cvv. Bulgaria 88, Veranopolis from Brazil, and Israel 493 respectively – all have Stb6 in addition (Chartrain et al., 2005b). The resistance of Bulgaria 88 was described as durable by Adhikari et al. (2004a) but it is not known if this refers to Stb1, Stb6 or both.
3.2. Stb4
Stb4 in cv. Tadinia was the first gene to be identified by controlled inoculation with a single isolate of Z. tritici, CA30 from California (Somasco et al., 1996). It was subsequently mapped to chromosome arm 7DS (7DS; Adhikari et al., 2004c), close to the locus of Stb5 (Arraiano et al., 2001b). Again, the allelism of Stb4 and Stb5 has not been tested.
3.3. Genes from synthetic hexaploid wheat: Stb5, Stb8, Stb16q and Stb17
The first gene for STB-resistance to be mapped was Stb5 (Arraiano et al., 2001b), which originated from a highly resistant synthetic hexaploid line, Synthetic 6x, derived from Triticum dicoccoides (AABB genomes) and Triticum tauschii (also known as Aegilops squarrosa; DD). Stb5 mapped to the pericentromeric region of chromosome arm 7DS, close to where Stb4 was mapped subsequently (Adhikari et al., 2004c). The mapping work used Z. tritici isolate IPO94269 from The Netherlands but Stb5 conferred resistance to all but one of the isolates tested. The location of Stb5 was greatly facilitated by the use of precise cytogenetic stocks (Arraiano et al., 2001b; Simon et al., 2001), which form a unique resource for wheat genetics to identify chromosomes carrying genes for traits of interest.
Synthetic hexaploids are a rich source of qualitative genes for resistance to STB and other diseases. Stb8 was identified in another synthetic line, W7984, bred by CIMMYT (the International Maize and Wheat Improvement Centre). It conferred resistance to an isolate from the USA and mapped to the long arm of chromosome 7B (Adhikari et al., 2003).
Two further genes, Stb16q and Stb17, were discovered in the synthetic hexaploid line M3 (Tabib Ghaffary et al., 2012). Stb16q on chromosome 3DL was designated as a quantitative (q) locus because it was not possible to determine if there was indeed a single gene at the locus. However, it controlled a high proportion of variation in necrotic leaf area, leaf area bearing pycnidia and latent period, and, alone among major Stb genes reported so far, conferred resistance at the seedling stage to all Z. tritici isolates tested, of which there were 20. It may be better regarded as a type of qualitative resistance. Stb17 on 5AL was detected only at the adult-plant stage and was less potent than Stb16q. The phenotype of Stb17 may fall between the qualitative and quantitative classes.
3.4. Stb6
The only qualitative gene for STB-resistance which has been shown to control a gene-for-gene relationship is Stb6, at the distal end of the short arm of chromosome 3A (Brading et al., 2002). This gene, which confers resistance to a Dutch Z. tritici isolate, IPO323, was first identified in the UK cvv. Flame and Hereward.
Stb6 is an especially notable gene as it was found to be present in most of the well-known sources of STB-resistance studied previously. Analysis of alleles of a simple-sequence repeat (SSR or microsatellite), Xgwm369, closely linked to Stb6, allied to analysis of wheat breeding pedigrees indicated that Stb6 had been introduced on at least six separate occasions into modern European germplasm, and was also present in Chinese Spring, a selection from a landrace which has been widely used in genetic studies of wheat (Chartrain et al., 2005b). It is the second most frequent Stb gene in European wheat, present in about 15% of cultivars tested (Arraiano and Brown, 2006).
At least five analyses of quantitative trait loci (QTL; Eriksen et al., 2003; Zwart et al., 2010; Tabib Ghaffary et al., 2011; Kelm et al., 2012; Goudemand et al., 2013) as well as an association genetic analysis (Kollers et al., 2013) have mapped field resistance to STB close to the Stb6 locus. A study of a large panel of UK and continental European cultivars found Stb6 to be associated with a reduction in STB symptoms in field conditions (Arraiano et al., 2009). This is consistent either with a minor gene for partial resistance to STB being closely linked to Stb6 or with Stb6 itself having a residual effect on field resistance even though virulence to Stb6 is almost fixed in the European population of Z. tritici (J.K.M.B., unpublished data).
Avirulence (AVR) to Stb6 was shown to be controlled by a single gene in Z. tritici IPO323 in a cross with IPO94269 in which virulence to several cultivars and breeding lines co-segregated (Kema et al., 2000; Brading et al., 2002). As resistance to IPO323 maps to the Stb6 locus in all these cultivars (Brading et al., 2002; Chartrain et al., 2005b), it is concluded that Stb6 is present in all of them and that it corresponds to a single AVR gene in IPO323.
3.5. Stb7 and Stb12
Stb7 on chromosome 4AL was first identified in cultivar ST6 as conferring resistance to the Canadian isolate MG2. It was mapped close to SSR locus Xwmc313 (McCartney et al., 2002, 2003). A gene for resistance to the Uruguayan isolate IPO87019 which mapped in the same location in the Portuguese line TE9111 (later released as cv. Nabão) was thought to be Stb7, an allele of Stb7 or a closely linked gene (Chartrain et al., 2005a). As the resistance gene in TE9111 was mapped by QTL analysis a precise location for the gene could not be achieved.
The CIMMYT breeding Kavkaz-K4500 L.6.A.4 (KK) carries Stb12, which is closely linked to Stb7 on chromosome 4AL. Stb12 provides resistance to isolate Isr398 from Israel but not to IPO87019 (Chartrain et al., 2005c). It was mapped by QTL analysis and was closer to Xgwm219 than to Xwmc313, which are ∼3cM apart. Of the 94 single-seed descent progeny of the cross of KK with the Isr398-susceptible cv. Shafir, four were resistant to ISR398 but susceptible to IPO97019, demonstrating the existence of two genes in that region. This is an example of Stb genes being clustered, a common feature of genes involved in plant defence.
3.6. Other qualitative genes in bread wheat
Stb9 on chromosome 2BL was mapped in the spring wheat cvv. Courtot and Tonic (Chartrain et al., 2009). It confers resistance to the Dutch isolate IPO89011.
Stb10 was also discovered in KK. Like Stb5, it conferred resistance to IPO94269 but it was clearly a different gene, located near the centromere of chromosome 1D (Chartrain et al., 2005c).
Stb11 on chromosome 1BS was identified and mapped in TE9111 and reported to confer resistance to isolate IPO90012 from Mexico (Chartrain et al., 2005a) but it may be widespread in global spring wheat breeding. When remapped, Stb2 was located close to or at the Stb11 locus (Liu et al., 2013). StbWW, identified in three populations in Australia, was also mapped on chromosome arm 1BS at or near Stb11 (Raman et al., 2009). These genes may all be Stb11, which may have spread in global wheat breeding by the movement of elite breeding lines from CIMMYT.
Stb13 on chromosome 7BL and Stb14 on 3BS were discovered in the Canadian cv. Salamouni (McCartney et al., 2002; Cowling, 2006). Both genes conferred resistance to MG2, like Stb7, while Stb13 also provided resistance to MG96-36 (Cowling et al., 2004; Cowling, 2006). Salamouni also has a third gene, designated StbSm3, which maps close to the Stb6 locus on chromosome 3AS but apparently distal to it (Cuthbert, 2011). No test of the allelism of StbSm3 and Stb6 has yet been conducted.
Stb15 on chromosome 6AS was identified as providing resistance to the Ethiopian isolate IPO88004 (Arraiano et al., 2007b). It is very common in European winter wheat, present in about 60% of cultivars tested (Arraiano and Brown, 2006) but, unlike the other widespread gene, Stb6, it is not associated with resistance in field conditions (Arraiano et al., 2009).
Stb18 on 6DS confers genotype-specific resistance in the French winter wheat cv. Balance (Tabib Ghaffary et al., 2011). It was expressed at the seedling stage but inconsistently in adult plants, being detected in one of two years of field trials of a population produced from Apache x Balance.
3.7. Qualitative resistance in durum wheat
Although STB is a severe disease of modern cultivars of durum wheat (Triticum durum), especially in North Africa, the genetics of STB-resistance in T. durum are poorly understood. In a search for sources of resistance in older, landrace cultivars, resistance to Z. tritici isolate Tun06 in a selection from the Agili landrace segregated as a single major gene (Ferjaoui et al., 2011). This gene was associated with AFLP markers but has not yet been assigned to a chromosome (Medini et al., 2014).
3.8. Resistance in Triticum monococcum
The diploid emmer wheat, Triticum monococcum, is highly resistant to Z. tritici. All accessions tested varied from very resistant to immune both to artificial resistance as seedlings and in five years of field trials. The genetics of resistance were studied in one accession, MDR043, and the gene TmStb1 was mapped to chromosome 7AmS (Table 1; Jing et al., 2008).
4. Quantitative resistance
Please refer to Table S1 for details of these genes and to Fig. 1 for their locations.
4.1. QTL in bi-parental crosses
In field trials, resistance to STB generally appears as a quantitative trait, largely additive in nature with some dominance, controlled by an oligogenic or polygenic system with moderate to high heritability in both durum wheat (van Ginkel and Scharen, 1987, 1988; Berraies et al., 2014) and bread wheat (Danon and Eyal, 1990; Jlibene et al., 1994; Simon et al., 1998; and papers cited in Section 4 and Table S1). QTL for resistance to STB at both seedling and adult stages are distributed throughout the genome of wheat (Table S1a). To date, 167 QTL of resistance against STB have been detected in a total of nineteen bi-parental mapping populations. From seven of these populations, 27 meta-QTL, i.e. refined QTL from multiple individual QTL, have been identified, integrating 105 individual QTL (Goudemand et al., 2013; Table S1b). Of 89 regions identified, 62 QTL and 27 meta-QTL, 27 were detected at the seedling stage, 48 at the adult stage and 14 at both stages. They included genome regions involved in the control of necrosis, pycnidium development and disease progress estimated as area under the disease progress curve (AUDPC). Two minor QTL controlling latent period have also been identified (Tabib Ghaffary et al., 2011).
All chromosomes except 5D carry at least one QTL or meta-QTL for STB-resistance. Nineteen QTL or meta-QTLs co-localised with genes involved in plant height (Rht: reduced height), heading date (Ppd: photoperiod-insensitivity) or both, among which six mapped closely to the Rht8 and Ppd-D1 (2DS), Rht-D1 (4DS), Ppd-A1 (2AS) and Rht-B1 (4BS). Three chromosome arms, 3BL, 6BS and 7DL, were especially involved in quantitative resistance to STB according to the number of QTLs identified. There are probably co-localisations with qualitative Stb genes for 22 QTL and 6 meta-QTL. QTL have frequently been mapped to the regions where Stb6 (3AS), Stb5/Stb4 (7DS) and Stb11/Stb2/StbWW (1BS) are located and less frequently to the regions of Stb1 (5BL), Stb9 (2BL), Stb7 and Stb12 (4AL), Stb13 (7BL), Stb14 (3BS) and Stb18 (6DS). Except for eight QTL identified in synthetic hexaploid wheat (Simon et al., 2004a; Zwart et al., 2010) the chromosome substitution line ‘Chinese-Spring’ (T. aestivum subsp. spelta 7D) (Simon et al., 2010) and a line from the USA (Mergoum et al., 2013), all QTL listed in Table S1 originated from European germplasm.
4.2. Association genetics
Association mapping studies have highlighted the presence of many regions of the genome in cultivated wheat and landraces associated with resistance to STB, including both some previously associated with STB-resistance and some not. From spray-inoculated field trials conducted over two years, Kollers et al. (2013) detected 68 SSR significantly associated with adult resistance in a panel of 372 European lines. Nine loci were significantly associated with all phenotyping parameters. Association QTL mapped to the loci of Stb1, 4, 6 and 8, implying that these genes or alleles of them may be present in European cultivars. In addition, several traits related to STB-resistance mapped at or near QTL identified previously.
In a study of 1055 elite hybrids and their corresponding 87 parental lines trialled in two locations which either had natural infection or were inoculated by spraying with a mixture of isolates, Miedaner et al. (2013) identified eight single-nucleotide polymorphisms (SNP) associated with STB resistance. Although half the SNP were not genetically mapped, the others were located on chromosomes 1B, 2B, 5B and 6A. The 5B locus may represent Stb1 or a QTL identified in the population Arina x Forno (Miedaner et al., 2012) and Steele-ND x ND 735 (Mergoum et al., 2013).
Finally, seven SNP at four loci were significantly associated with resistance in an association mapping study of a panel of 528 spring wheat landraces of worldwide origin phenotyped at the adult stage in growth chambers. These SNP mapped to chromosomes 3B, 6B and 7B and most likely relate to new resistance genes (Gurung et al., 2014).
4.3. Cytogenetics
In a cytogenetic analysis, the 5BS arm of Hobbit sib (Dwarf A) was found to promote susceptibility to STB in adult plants but not in seedlings. Formally, the data were also consistent with this chromosome arm carrying genes which suppress resistance but Hobbit sib has no known STB-resistance genes. The same chromosome arm has genes which increase resistance to yellow (stripe) rust and powdery mildew, which implies that there may be a trade-off between breeding for resistance to STB and to biotrophic fungi (Arraiano et al., 2007a).
5. Specificity of resistance
5.1. Specialisation of Z. tritici to host cultivars and species
Strong specificity in the interaction between cultivars of bread wheat (T. aestivum) and Z. tritici isolates was reported by Eyal et al. (1973) and confirmed in subsequent studies on seedlings (Ahmed et al., 1995; Ballantyne and Thomson, 1995; Kema et al., 1996a, 1996b; Arraiano et al., 2001a; Chartrain et al., 2004b; Grieger et al., 2005; Arraiano and Brown, 2006; Medini and Hamza, 2008; Czembor et al., 2011; Abrinbana et al., 2012) and adult plants (Kema and van Silfhout, 1997; Brown et al., 2001; Grieger et al., 2005). These interactions are akin to gene-for-gene relationships but this has only been demonstrated in Stb6 resistance to IPO323 (Kema et al., 2000; Brading et al., 2002).
Specialisation of Z. tritici isolates to either T. aestivum or T. durum has been reported by some workers (Eyal et al., 1973; Kema et al., 1996a, 1996b and papers cited therein; Zhan et al., 2004) but not others (Eyal, 1999; Medini and Hamza, 2008). Cultivar-by-isolate specificity within T. durum has also been reported (Kema et al., 1996a, 1996b; Medini and Hamza, 2008; Ghaneie et al., 2012). In a cross of aestivum-adapted and durum-adapted isolates of Z. tritici, the AvrStb6 locus for avirulence to Stb6 (Brading et al., 2002) was associated with part of the variation in ability of progeny isolates to infect T. durum. This suggests that resistance of these two wheat species to inappropriate specialised forms of Z. tritici may be controlled in part by qualitative resistance genes (Ware, 2006).
Most Z. tritici isolates used in research on STB are virulent to almost all Stb genes although some isolates with more than one functional avirulence phenotype are known. IN95-Lafayette-1196-WW-1-4 was avirulent to both Stb1 (Adhikari et al., 2004a) and Stb4 (Adhikari et al., 2004c) while the Paskeville isolate was avirulent to Stb2 and Stb3 (Adhikari et al., 2004b). Among the isolates avirulent to Stb5 in Synthetic 6x (Arraiano et al., 2001b) were IPO323, which is also avirulent to Stb6 (Brading et al., 2002), IPO89011 (avirulent to Stb9: Chartrain et al., 2009), IPO94269 (avirulent to Stb10: Chartrain et al., 2005c) and IPO001, which is avirulent to some UK cultivars (Arraiano and Brown, 2006). MG2 was avirulent to Stb7 (McCartney et al., 2002), Stb13 and Stb14 (Cowling, 2006). IPO323 may detect a second resistance gene in KK in addition to Stb6 (Chartrain et al., 2005b). Otherwise, the high frequency of virulence implies that there is little obstacle to Z. tritici mutating to virulence on Stb genes which control gene-for-gene interactions.
5.2. Evolution of virulence
As in other diseases with a gene-for-gene system, the specificity of qualitative resistance to avirulent Z. tritici genotypes leads to selection for virulence (loss of avirulence). In Oregon, USA, such a ‘breakdown’ of resistance due to pathogen adaptation happened rapidly, with a catastrophic effect on disease control, in cv. Gene in the 1990s (Cowger et al., 2000) and more gradually in cv. Foote in the 2000s (Krenz et al., 2008). Cv. Gene is resistant to IPO323 and IPO94269, which are avirulent to Stb6 and Stb10 respectively (Chartrain et al., 2004b) but it is not known if either of these genes was the one which was overcome by the fungus. Kema and van Silfhout (1997) reported that the resistance of cv. Obelisk became less effective in The Netherlands during the 1980s. Virulence to Stb4 evolved in Z. tritici in California at some time before 2000 (Jackson et al., 2000).
6. Breeding for resistance to S. tritici blotch
6.1. The use of qualitative STB-resistance in breeding
A superficially attractive option for breeding for resistance to STB, as in many other plant diseases, is to use qualitative genes with large effects on the pathogen. Synthetic hexaploid wheat may be a rich source of such genes (Arraiano et al., 2001b Adhikari et al., 2003; Dreisigacker et al., 2008; Tabib Ghaffary et al., 2012). A persistent difficulty in applying this strategy in plant breeding is that many (but not all) single genes which confer strong resistance conform to the gene-for-gene relationship, while most gene-for-gene resistances (but not all) are readily overcome by the target pathogens (Poland et al., 2008; Mundt, 2014).
Breeding for resistance to STB can benefit greatly from the long history of breeding crops to control other diseases. While reliance on qualitative genes may reduce STB in the short-term, this approach is unlikely to provide durable resistance. A relevant comparison is with the successive use of gene-for-gene resistances against powdery mildew of barley (Brown, 1994) or yellow (stripe) rust of wheat (Hovmøller and Justesen, 2007), a strategy which has been far from durable. The useful lifetime of qualitative genes can be extended by supporting them with high levels of ‘background’, usually durable, quantitative resistance (Palloix et al., 2009).
It is striking that some notable sources of STB-resistance, such as KK (Chartrain et al., 2004b, 2005c), Salamouni (Cowling, 2006; Cuthbert, 2011) and TE9111 (Chartrain et al., 2005a) have several qualitative resistances (Chartrain et al., 2004b). This suggests that ‘stacking’ or ‘pyramiding’ several Stb genes might improve the effectiveness of resistance, a strategy which has sometimes been effective in controlling crop diseases (Mundt, 2014). Against this, the fact that most known Z. tritici isolates are virulent to most Stb genes (Section 5.1) suggests that the resistance achieved by gene-stacking may not be durable.
6.2. Selection for durable resistance
Known individual Stb genes are not currently effective against Z. tritici populations in Europe (see Section 5.1; also Arraiano et al., 2009) and have not been durable (Section 5.2), although some are associated with minor quantitative resistance (Section 4.1). The majority of variation in field resistance to STB, therefore, must be controlled by quantitative resistance, as defined in Section 1, and the progress in breeding for STB-resistance over the last 30 years presumably happened by the gradual accumulation of minor genes (Torriani et al., 2015). This type of resistance appears to be more durable than qualitative resistance. Its effectiveness may be gradually eroded (Mundt et al., 2002; Krenz et al., 2008) but this happens much more slowly and to a lesser extent than the rapid evolution of virulence in a gene-for-gene interaction (Poland et al., 2008; Brown, 2015).
A significant problem in the genetics of STB is that, when a gene has a large effect on resistance to the current pathogen population and therefore seems desirable as a source of resistance in breeding, there is currently no way of determining from its phenotype or underlying mechanism whether or not it might be durable. Qualitative genes which control detection of a specific pathogen genotype in a gene-for-gene relationship are much less likely to be durable than those that enhance downstream defences. The latter class of gene includes Lr34 in wheat against biotrophic pathogens (Krattinger et al., 2009), STV11 in rice against Rice stripe virus (Wang et al., 2014) and several others. In rusts and powdery mildews, gene-for-gene interactions generate a hypersensitive response and reduce the infection type (IT) of pustules or colonies, whereas quantitative resistance tends to reduce the extent of symptoms rather than the IT (Boyd et al., 1995; Jagger et al., 2011). No such distinction can yet be made between the phenotypes of genotype-specific qualitative resistance and other, potentially more durable forms of resistance to STB. As Z. tritici is initially endophytic, becoming necrotrophic in its pathogenic phase (Orton et al., 2011; Sánchez-Vallet et al., 2015), the only distinction between compatible and incompatible interactions known at present is the amount of disease visible on the leaf (Chartrain et al., 2004b).
If a plant resistance gene introduced from a wild population or a genetically diverse landrace population is effective against the current pathogen population, it will not necessarily be durable. If it follows the gene-for-gene relationship, virulence may be rare because the resistance gene has a fitness cost or the disease is not severe at the place of origin of the resistance gene (Brown and Tellier, 2011). Stb16q is intriguing and potentially useful because it has a strong effect against all the large number of Z. tritici isolates with which it has been tested (Tabib Ghaffary et al., 2012). Until it is isolated, however, or unless an isolate virulent to Stb16q is discovered, it will not be possible to tell whether it controls a gene-for-gene resistance which is effective against the current pathogen population, or is part of the plant’s downstream defences and therefore may be durable.
Greater knowledge about mechanisms of STB-resistance would support wheat breeding, particularly by characterising the difference between gene-for-gene resistance and other kinds of resistance which may be more durable. This would help breeders to make informed decisions about the likely durability of resistance without the lengthy process of isolating the gene. A method of selecting both effective qualitative resistance, which may not be durable, and a good level of quantitative, possibly durable resistance in the same cultivar would be especially useful (Risser et al., 2011).
In addition to disease resistance, STB levels can also be reduced by traits that contribute to disease escape, which limits the spread of fungal inoculum within crops (van Beuningen and Kohli, 1990; Simon et al., 2004b; Arraiano et al., 2009). This typically happens in cultivars which are taller and later-heading, as both traits reduce the spread of spores to the upper leaves. Escape traits can be undesirable, however, because they can be maladaptive in terms of agronomic properties and yield.
A general approach to increasing quantitative resistance (to any disease) stems from viewing plant breeding as a greatly accelerated form of natural selection, in which variation in traits is selected by breeders and inherited by the next generation of cultivars. The three essential requirements for breeding for effective, durable STB-resistance are diverse germplasm, efficient breeding processes which generate new combinations of genes, and field trial sites with high levels of STB at which resistant cultivars with good agronomic properties can be selected reliably and consistently. Once these fundamentals are in place, targetted selection of cost-effective genes or combinations of genes (Grimmer et al., 2015; Torriani et al., 2015) can contribute to raising the level of STB-resistance in new wheat cultivars.
Acknowledgment
J.K.M.B. and L.C. are supported by the BBSRC Biotic Interactions Strategic Programme.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.fgb.2015.04.017.
Contributor Information
James K.M. Brown, Email: james.brown@jic.ac.uk.
Cyrille Saintenac, Email: cyrille.saintenac@clermont.inra.fr.
Supplementary material
References
- Abrinbana M., Mozafari J., Shams-Bakhsh M., Mehrabi R. Resistance spectra of wheat genotypes and virulence patterns of Mycosphaerella graminicola isolates in Iran. Euphytica. 2012;186:75–90. [Google Scholar]
- Adhikari T.B., Yang X., Cavaletto J.R., Hu X., Buechley G., Ohm H.W., Shaner G., Goodwin S.B. Molecular mapping of Stb1, a potentially durable gene for resistance to Septoria tritici blotch in wheat. Theor. Appl. Genet. 2004;109:944–953. doi: 10.1007/s00122-004-1709-6. [DOI] [PubMed] [Google Scholar]
- Adhikari T.B., Wallwork H., Goodwin S.B. Microsatellite markers linked to the Stb2 and Stb3 genes for resistance to Septoria tritici blotch in wheat. Crop Sci. 2004;44:1403–1411. [Google Scholar]
- Adhikari T.B., Cavaletto J.R., Dubcovsky J., Gieco J.O., Schlatter A.R., Goodwin S.B. Molecular mapping of the Stb4 gene for resistance to Septoria tritici blotch in wheat. Phytopathology. 2004;94:1198–1206. doi: 10.1094/PHYTO.2004.94.11.1198. [DOI] [PubMed] [Google Scholar]
- Ahmed H.U., Mundt C.C., Coakley S.M. Host-pathogen relationship of geographically diverse isolates of Septoria tritici and wheat cultivars. Plant. Pathol. 1995;44:838–847. [Google Scholar]
- Arraiano L.S., Brown J.K.M. Identification of isolate-specific and partial resistance to spetoria tritici blotch in 238 European wheat cultivars and breeding lines. Plant. Pathol. 2006;55:54–61. [Google Scholar]
- Arraiano L.S., Brading P.A., Brown J.K.M. A detached seedling leaf technique to study resistance to Mycosphaerella graminicola (anamorph Septoria tritici) in wheat. Plant. Pathol. 2001;50:339–346. [Google Scholar]
- Arraiano L.S., Worland A.J., Ellerbrook C., Brown J.K.M. Chromosomal location of a gene for resistance to Septoria trictici blotch (Mycosphaerella graminicola) in the hexaploid wheat ‘Synthetic 6x’. Theor. Appl. Genet. 2001;103:758–764. [Google Scholar]
- Arraiano L.S., Kirby J., Brown J.K.M. Cytogenetic analysis of the susceptibility of the wheat line Hobbit sib (Dwarf A) to Septoria tritici blotch. Theor. Appl. Genet. 2007;116:113–122. doi: 10.1007/s00122-007-0651-9. [DOI] [PubMed] [Google Scholar]
- Arraiano L.S., Chartrain L., Bossolini E., Slatter H.N., Keller B., Brown J.K.M. A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola. Plant. Pathol. 2007;56:73–78. [Google Scholar]
- Arraiano L.S., Balaam N., Fenwick P.M., Chapman C., Feuerhelm D., Howell P., Smith S.J., Widdowson J.P., Brown J.K.M. Contributions of disease resistance and escape to the control of Septoria tritici blotch of wheat. Plant. Pathol. 2009;58:910–922. [Google Scholar]
- Ballantyne B., Thomson F. Pathogenic variation in Australian isolates of Mycosphaerella graminicola. Aust. J. Agric. Res. 1995;46:921–934. [Google Scholar]
- Berraies S., Ammar K., Gharbi M.S., Yahyaoui A., Rezgui S. Quantitative inheritance of resistance to Septoria tritici blotch in durum wheat in Tunisia. Chil. J. Agric. Res. 2014;74:35–40. [Google Scholar]
- Boyd L.A., Smith P.H., Foster E.M., Brown J.K.M. The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f.sp. hordei and host responses. Plant J. 1995;7:959–968. [Google Scholar]
- Brading P.A., Verstappen E.C.P., Kema G.H.J., Brown J.K.M. A gene-for-gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology. 2002;92:439–445. doi: 10.1094/PHYTO.2002.92.4.439. [DOI] [PubMed] [Google Scholar]
- Brown J.K.M. Chance and selection in the evolution of barley mildew. Trends Microbiol. 1994;2:470–475. doi: 10.1016/0966-842x(94)90650-5. [DOI] [PubMed] [Google Scholar]
- Brown, J.K.M., 2015. Durable resistance of crops to disease: a Darwinian perspective. Annu. Rev. Phytopathol. http://dx.doi.org/10.1146/annurev-phyto-102313-045914. [DOI] [PubMed]
- Brown J.K.M., Tellier A. Plant-parasite coevolution: bridging the gap between genetics and ecology. Annu. Rev. Phytopathol. 2011;49:345–367. doi: 10.1146/annurev-phyto-072910-095301. [DOI] [PubMed] [Google Scholar]
- Brown J.K.M., Kema G.H.J., Forrer H.R., Verstappen E.C.P., Arraiano L.S., Brading P.A., Foster E.M., Fried P.M., Jenny E. Resistance of wheat cultivars and breeding lines to Septoria tritici blotch caused by isolates of Mycosphaerella graminicola in field trials. Plant. Pathol. 2001;50:325–338. [Google Scholar]
- Chartrain L., Brading P.A., Widdowson J.P., Brown J.K.M. Partial resistance to Septoria tritici blotch (Mycosphaerella graminicola) in the wheat cultivars Arina and Riband. Phytopathology. 2004;94:497–504. doi: 10.1094/PHYTO.2004.94.5.497. [DOI] [PubMed] [Google Scholar]
- Chartrain L., Brading P.A., Makepeace J.C., Brown J.K.M. Sources of resistance to Septoria tritici blotch and implications for wheat breeding. Plant. Pathol. 2004;53:454–460. [Google Scholar]
- Chartrain L., Joaquim P., Berry S.T., Arraiano L.S., Azanza F., Brown J.K.M. Genetics of resistance to Septoria tritici blotch in the Portuguese wheat breeding line TE9111. Theor. Appl. Genet. 2005;110:1138–1144. doi: 10.1007/s00122-005-1945-4. [DOI] [PubMed] [Google Scholar]
- Chartrain L., Brading P.A., Brown J.K.M. Presence of the Stb6 gene for resistance to Septoria tritici blotch (Mycosphaerella graminicola) in cultivars used in wheat-breeding programmes worldwide. Plant. Pathol. 2005;54:134–143. [Google Scholar]
- Chartrain L., Berry S.T., Brown J.K.M. Resistance of wheat line Kavkaz-K4500 L.6.A.4 to Septoria tritici blotch controlled by isolate-specific resistance genes. Phytopathology. 2005;95:664–671. doi: 10.1094/PHYTO-95-0664. [DOI] [PubMed] [Google Scholar]
- Chartrain L., Sourdille P., Bernard M., Brown J.K.M. Identification and location of Stb9, a gene for resistance to Septoria tritici blotch in wheat cultivars Courtot and Tonic. Plant. Pathol. 2009;58:547–555. [Google Scholar]
- Cowger C., Hoffer M.E., Mundt C.C. Specific adaptation by Mycosphaerella graminicola to a resistant wheat cultivar. Plant. Pathol. 2000;49:445–451. [Google Scholar]
- Cowling, S.G., 2006. Identification and Mapping of Host Resistance Genes to Septoria tritici Blotch of Wheat. Ph.D. thesis, University of Manitoba.
- Cowling S.G., Brûlé-Babel A.L., Somers D.J., Lamari L. Identification and mapping of host resistance genes to Septoria tritici blotch of wheat. Phytopathology. 2004;94:S22. [Google Scholar]
- Cuthbert, R., 2011. Molecular Mapping of Septoria tritici Blotch Resistance in Hexaploid Wheat (Triticum aestivum L.). Ph.D. Thesis, University of Manitoba.
- Czembor P.C., Radecka-Janusik M., Mańkowski D. Virulence spectrum of Mycosphaerella graminicola isolates on wheat genotypes carrying known resistance genes to Septoria tritici blotch. J. Phytopathol. 2011;159:146–154. [Google Scholar]
- Danon T., Eyal Z. Inheritance of resistance to two Septoria tritici isolates in spring and winter bread wheat cultivars. Euphytica. 1990;47:203–214. [Google Scholar]
- Dreisigacker S., Kishii M., Lage J., Warburton M. Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust. J. Agric. Res. 2008;59:413–420. [Google Scholar]
- Eriksen L., Borum F., Jahoor A. Inheritance and localisation of resistance to Mycosphaerella graminicola causing Septoria tritici blotch and plant height in the wheat (Triticum aestivum L.) genome with DNA markers. Theor. Appl. Genet. 2003;107:515–527. doi: 10.1007/s00122-003-1276-2. [DOI] [PubMed] [Google Scholar]
- Eyal Z. The Septoria tritici and Stagonospora nodorum blotch diseases of wheat. Eur. J. Plant Pathol. 1999;105:629–641. [Google Scholar]
- Eyal Z., Amiri Z., Wahl I. Physiologic specialization of Septoria tritici. Phytopathology. 1973;63:1087–1091. [Google Scholar]
- Ferjaoui S., Sbei A., Aouadi N., Hamza S. Monogenic inheritance of resistance to Septoria tritici blotch in durum wheat ‘Agili’. Int. J. Plant Breed. 2011;5:17–20. [Google Scholar]
- Fones H., Gurr S.J. The impact of Septoria tritici blotch disease on wheat: an EU perspective. Fungal. Genet. Biol. 2015;79:3–7. doi: 10.1016/j.fgb.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaneie A., Mehrabi R., Safaie N., Abrinbana M., Saidi A., Aghaee M. Genetic variation for resistance to Septoria tritici blotch in Iranian tetraploid wheat landraces. Eur. J. Plant Pathol. 2012;132:191–202. [Google Scholar]
- Goodwin S.B. Back to basics and beyond: increasing the level of resistance to Septoria tritici blotch in wheat. Australas. Plant Pathol. 2007;36:532–538. [Google Scholar]
- Goodwin S.B., Thompson I. Development of isogenic lines for resistance to Septoria tritici blotch in wheat. Czech J. Genet. Plant Breed. 2011;47:S98–S101. [Google Scholar]
- Goudemand E., Laurent V., Duchalais L., Tabib Ghaffary S.M., Kema G.H.J., Lonnet P., Margale E., Robert O. Association mapping and meta-analysis: two complementary approaches for the detection of reliable Septoria tritici blotch quantitative resistance in bread wheat (Triticum aestivum L.) Mol. Breed. 2013;32:563–584. [Google Scholar]
- Grieger A., Lamari L., Brûlé-Babel A. Physiologic variation in Mycosphaerella graminicola from western Canada. Can. J. Plant Pathol. 2005;72:71–77. [Google Scholar]
- Grimmer M.K., Boyd L.A., Clarke S.M., Paveley N.D. Pyramiding of partial disease resistance genes has a predictable, but diminishing, benefit to efficacy. Plant. Pathol. 2015 [Google Scholar]
- Gurung S., Mamidi S., Bonman J.M., Xiong M., Brown-Guedira G., Adhikari T.B. Genome-wide association study reveals novel quantitative trait loci associated with resistance to multiple leaf spot diseases of spring wheat. PLoS One. 2014;9:e108179. doi: 10.1371/journal.pone.0108179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HGCA, 2012. Septoria tritici in winter wheat. Topic Sheet 113. HGCA Publications, Kenilworth, UK.
- Hovmøller M.S., Justesen A.F. Rates of evolution of avirulence phenotypes and DNA markers in a northwest European population of Puccinia striiformis f. sp. tritici. Mol. Ecol. 2007;16:4637–4647. doi: 10.1111/j.1365-294X.2007.03513.x. [DOI] [PubMed] [Google Scholar]
- Jackson L.F., Dubcovsky J., Gallagher L.W., Wennig R.L., Heaton R.L., Vogt R.L., Gibbs L.K., Kirby D., Canevari M., Carlson H., Kearney T., Marsh B., Munier D., Mutters C., Orloff S., Schmierer J., Vargas R., Williams J., Wright S. Regional barley and common and durum wheat performance tests in California. Agron. Prog. Rep. 2000;272:1–56. [Google Scholar]
- Jagger L.J., Newell C., Berry S.T., MacCormack R., Boyd L.A. Histopathology provides a phenotype by which to characterize stripe rust resistance genes in wheat. Plant. Pathol. 2011;60:640–648. [Google Scholar]
- Jing H.C., Lovell D., Gutteridge R., Jenk D., Kornyukhin D., Mitrofanova O.P., Kema G.H.J., Hammond-Kosack K.E. Phenotypic and genetic analysis of the Triticum monococcum – Mycosphaerella graminicola interaction. New Phytol. 2008;179:1121–1132. doi: 10.1111/j.1469-8137.2008.02526.x. [DOI] [PubMed] [Google Scholar]
- Jlibene M., Gustafson J.P., Rajaram S. Inheritance of resistance to Mycosphaerella graminicola in hexaploid wheat. Plant Breed. 1994;112:301–310. [Google Scholar]
- Kelm C., Tabib Ghaffary S.M., Bruelheide H., Roder M.S., Miersch S., Weber W.E., Kema G.H.J., Saal B. The genetic architecture of seedling resistance to Septoria tritici blotch in the winter wheat doubled-haploid population Solitär x Mazurka. Mol. Breed. 2012;29:813–830. [Google Scholar]
- Kema G.H.J., van Silfhout C.H. Genetic variation for virulence and resistance in the wheat Mycosphaerella graminicola pathosystem. 3. Comparative seedling and adult plant experiments. Phytopathology. 1997;87:266–272. doi: 10.1094/PHYTO.1997.87.3.266. [DOI] [PubMed] [Google Scholar]
- Kema G.H.J., Annone J.G., Sayoud R., van Silfhout C.H., van Ginkel M., de Bree J. Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. 1. Interactions between pathogen isolates and host cultivars. Phytopathology. 1996;86:200–212. [Google Scholar]
- Kema G.H.J., Sayoud R., Annone J.G., van Silfhout C.H. Genetic variation for virulence and resistance in the wheat-Mycosphaerella graminicola pathosystem. 2. Analysis of interactions between pathogen isolates and host cultivars. Phytopathology. 1996;86:213–220. [Google Scholar]
- Kema G.H.J., Verstappen E.C.P., Waalwijk C. Avirulence in the wheat Septoria tritici leaf blotch fungus Mycosphaerella graminicola is controlled by a single locus. Mol. Plant-Microbe Interact. 2000;13:1375–1379. doi: 10.1094/MPMI.2000.13.12.1375. [DOI] [PubMed] [Google Scholar]
- Kollers S., Rodemann B., Ling J., Korzun V., Ebmeyer E., Argillier O., Hinze M., Plieske J., Kulosa D., Ganal M.W., Roder M.S. Genetic architecture of resistance to Septoria tritici blotch (Mycosphaerella graminicola) in European winter wheat. Mol. Breed. 2013;32:411–423. [Google Scholar]
- Krattinger S.G., Lagudah E.S., Spielmeyer W., Singh R.P., Huerta-Espino J., McFadden H., Bossolini E.S., Selter L.L., Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Krenz J.E., Sackett K.E., Mundt C.C. Specificity of incomplete resistance to Mycosphaerella graminicola in wheat. Phytopathology. 2008;98:555–561. doi: 10.1094/PHYTO-98-5-0555. [DOI] [PubMed] [Google Scholar]
- Liu Y.Y., Zhang L.L., Thompson I.A., Goodwin S.B., Ohm H.W. Molecular mapping re-locates the Stb2 gene for resistance to Septoria tritici blotch derived from cultivar Veranopolis on wheat chromosome 1BS. Euphytica. 2013;190:145–156. [Google Scholar]
- McCartney C.A., Brûlé-Babel A.L., Lamari L. Inheritance of race-specific resistance to Mycosphaerella graminicola in wheat. Phytopathology. 2002;92:138–144. doi: 10.1094/PHYTO.2002.92.2.138. [DOI] [PubMed] [Google Scholar]
- McCartney C.A., Brûlé-Babel A.L., Lamari L., Somers D.J. Chromosomal location of a race specific resistance gene to Mycosphaerella graminicola in the spring wheat ST6. Theor. Appl. Genet. 2003;107:1181–1186. doi: 10.1007/s00122-003-1359-0. [DOI] [PubMed] [Google Scholar]
- Medini M., Hamza S. Pathotype and molecular characterization of Mycosphaerella graminicola isolates collected from Tunisia, Algeria and Canada. J. Plant Pathol. 2008;90:65–73. [Google Scholar]
- Medini M., Ferjaoui S., Bahri B., Mhri W., Hattab S., Hamza S. Les méthodes d’analyse de ségrégation en mélange et d’association marqueur-trait révèlent des marqueurs AFLP communs de résistance à la septoriose chez un ancien blé dur de Tunisie. Biotech. Agron. Soc. Environ. 2014;18:3–10. [Google Scholar]
- Mergoum M., Harilal V.E., Singh P.K., Adhikari T.B., Kumar A., Ghavami F., Elias E., Alamri M.S., Kianian S.F. Genetic analysis and mapping of seedling resistance to Septoria tritici blotch in ‘Steele-ND’/’ND 735’ bread wheat population. Cereal Res. Comm. 2013;41:199–210. [Google Scholar]
- Miedaner T., Risser P., Paillard S., Schnurbusch T., Keller B., Hartl L., Holzapfel J., Korzun V., Ebmeyer E., Utz H.F. Broad-spectrum resistance loci for three quantitatively inherited diseases in two winter wheat populations. Mol. Breed. 2012;29:731–742. [Google Scholar]
- Miedaner T., Zha Y., Gowda M., Longin C.F.H., Korzun V., Ebmeyer E., Kazman E., Reif J.C. Genetic architecture of resistance to Septoria tritici blotch in European wheat. BMC Genomics. 2013;14:858. doi: 10.1186/1471-2164-14-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt C.C. Durable resistance: a key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014;27:446–455. doi: 10.1016/j.meegid.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundt C.C., Cowger C., Garrett K.A. Relevance of integrated disease management to resistance durability. Euphytica. 2002;124:245–252. [Google Scholar]
- Adhikari T.B., Anderson J.M., Goodwin S.B. Identification and molecular mapping of a gene in wheat conferring resistance to Mycosphaerella graminicola. Phytopathology. 2003;93:1158–1164. doi: 10.1094/PHYTO.2003.93.9.1158. [DOI] [PubMed] [Google Scholar]
- O’Driscoll A., Kildea S., Doohan F., Spink J., Mullins E. The wheat–Septoria conflict: a new front opening up? Trends Plant Sci. 2014;19:602–610. doi: 10.1016/j.tplants.2014.04.011. [DOI] [PubMed] [Google Scholar]
- Orton E.S., Deller S., Brown J.K.M. Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 2011;12:413–424. doi: 10.1111/j.1364-3703.2010.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palloix A., Ayme V., Moury B. Durability of plant major resistance genes to pathogens depends on the genetic background, experimental evidence and consequences for breeding strategies. New Phytol. 2009;183:190–199. doi: 10.1111/j.1469-8137.2009.02827.x. [DOI] [PubMed] [Google Scholar]
- Poland J.A., Balint-Kurti P.J., Wisser R.J., Pratt R.C., Nelson R.J. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 2008;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Raman H., Milgate A. Molecular breeding for Septoria tritici blotch resistance in wheat. Cereal Res. Commun. 2012;40:451–466. [Google Scholar]
- Raman R., Milgate A., Imtiaz M., Tan M.K., Raman H., Lisle C., Coombes N., Martin P. Molecular mapping and physical location of major gene conferring seedling resistance to Septoria tritici blotch in wheat. Mol. Breed. 2009;24:153–164. [Google Scholar]
- Risser P., Ebmeyer E., Korzun V., Hartl L., Miedaner T. Quantitative trait loci for adult-plant resistance to Mycosphaerella graminicola in two winter wheat populations. Phytopathology. 2011;101:1209–1216. doi: 10.1094/PHYTO-08-10-0203. [DOI] [PubMed] [Google Scholar]
- Saari E.E., Wilcoxson R.D. Plant disease situation of high-yielding dwarf wheats in Asia and Africa. Annu. Rev. Phytopathol. 1974;12:49–68. [Google Scholar]
- Sánchez-Vallet A., McDonald M.C., Solomon P.S., McDonald B.A. Is Zymoseptoria tritici a hemibiotroph? Fungal Genet. Biol. 2015 doi: 10.1016/j.fgb.2015.04.001. (THIS SPECIAL ISSUE) [DOI] [PubMed] [Google Scholar]
- Simon M.R., Perello A.E., Cordo C.A. Response to selection in F-2 populations of two wheat crosses for resistance to Septoria tritici. Cereal Res. Commun. 1998;26:275–280. [Google Scholar]
- Simon M.R., Ayala F.M., Corda C.A., Roder M.S., Börner A. Molecular mapping of quantitative trait loci determining resistance to Septoria tritici blotch caused by Mycosphaerella graminicola in wheat. Euphytica. 2004;138:41–48. [Google Scholar]
- Simon M.R., Worland A.J., Struik P.C. Influence of plant height and heading date on the expression of the resistance to Septoria tritici blotch in near-isogenic lines of wheat. Crop Sci. 2004;44:2078–2085. [Google Scholar]
- Simon M.R., Khlestkina E.K., Castillo N.S., Börner A. Mapping quantitative resistance to Septoria tritici blotch in spelt wheat. Eur. J. Plant Pathol. 2010;128:317–324. [Google Scholar]
- Simon M.R., Worland A.J., Cordo C.A., Struik P.C. Chromosomal location of resistance to Septoria tritici in seedlings of a synthetic hexaploid wheat, Triticum spelta and two cultivars of Triticum aestivum. Euphytica. 2001;119:149–153. [Google Scholar]
- Somasco O.A., Qualset C.O., Gilchrist D.G. Single-gene resistance to Septoria tritici blotch in the spring wheat cultivar ‘Tadinia’. Plant Breed. 1996;115:261–267. [Google Scholar]
- Somers D.J., Isaac P., Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.) Theor. Appl. Genet. 2004;109:1105–1114. doi: 10.1007/s00122-004-1740-7. [DOI] [PubMed] [Google Scholar]
- Stewart E.L., McDonald B.A. Measuring quantitative virulence in the wheat pathogen Zymoseptoria tritici using high-throughput automated image analysis. Phytopathology. 2014;104:985–992. doi: 10.1094/PHYTO-11-13-0328-R. [DOI] [PubMed] [Google Scholar]
- Tabib Ghaffary S.M., Robert O., Laurent V., Lonnet P., Margale E., van der Lee T.A.J., Visser R.G.F., Kema G.H.J. Genetic analysis of resistance to Septoria tritici blotch in the French winter wheat cultivars Balance and Apache. Theor. Appl. Genet. 2011;123:741–754. doi: 10.1007/s00122-011-1623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabib Ghaffary S.M., Faris J.D., Friesen T.L., Visser R.G.F., van der Lee T.A.J., Robert O., Kema G.H.J. New broad-spectrum resistance to Septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 2012;124:125–142. doi: 10.1007/s00122-011-1692-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torriani S.F.F., Melichar J.P.E., Mills C., Pain N., Sierotzki H., Courbot M. Zymoseptoria tritici: a major threat to wheat production, integrated approaches to control. Fungal Genet. Biol. 2015 doi: 10.1016/j.fgb.2015.04.010. (THIS SPECIAL ISSUE) [DOI] [PubMed] [Google Scholar]
- van Beuningen L.T., Kohli M.M. Deviation from the regression of infection on heading and height as a measure of resistance to Septoria tritici blotch in wheat. Plant Dis. 1990;74:488–493. [Google Scholar]
- van Ginkel M., Scharen A.L. Generation mean analysis and heritabilities of resistance to Septoria tritici in durum wheat. Phytopathology. 1987;77:1629–1633. [Google Scholar]
- van Ginkel M., Scharen A.L. Host-pathogen relationships of wheat and Septoria tritici. Phytopathology. 1988;78:762–766. [Google Scholar]
- Wang Q., Liu Y.Q., He J., Zheng X.M., Hu J.L. STV11 encodes a sulphotransferase and confers durable resistance to rice stripe virus. Nat. Commun. 2014;5:4768. doi: 10.1038/ncomms5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware, S.B., 2006. Aspects of Sexual Reproduction in Mycosphaerella Species on Wheat and Barley: Genetic Studies on Specificity, Mapping, and Fungicide Resistance. Ph.D. thesis, Wageningen University, The Netherlands.
- Wiese M.V., editor. Compendium of Wheat Diseases. American Phytopathological Society; St. Paul: 1987. 112 pp. [Google Scholar]
- Wilson R.E. Inheritance of resistance to Septoria tritici in wheat. In: Scharen A.L., editor. Septoria in Cereals: Proceedings of the Workshop. Montana State University; Bozeman: 1985. pp. 281–304. [Google Scholar]
- Zhan J., Kema G.H.J., McDonald B.A. Evidence for natural selection in the mitochondrial genome of Mycosphaerella graminicola. Phytopathology. 2004;94:261–267. doi: 10.1094/PHYTO.2004.94.3.261. [DOI] [PubMed] [Google Scholar]
- Zwart R.S., Thompson J., Milgate A., Bansal U., Williamson P., Raman H., Bariana H.S. QTL mapping of multiple foliar disease and root-lesion nematode resistances in wheat. Mol. Breed. 2010;26:107–124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.