Abstract
Erianthus arundinaceus (E. arundinaceus) has many desirable agronomic traits for sugarcane improvement, such as high biomass, vigor, rationing ability, tolerance to drought, and water logging, as well as resistance to pests and disease. To investigate the introgression of the E. arundinaceus genome into sugarcane in the higher generations, intergeneric BC2 and BC3 progeny generated between Saccharum spp. and E. arundinaceus were studied using the genomic in situ hybridization (GISH) technique. The results showed that the BC2 and BC3 generations resulted from n + n chromosome transmission. Furthermore, chromosome translocation occurred at terminal fragments from the E. arundinaceus chromosome in some progeny of Saccharum spp. and E. arundinaceus. Notably, the translocated chromosomes could be stably transmitted to their progeny. This study illustrates the characterization of chromosome inheritance of the intergeneric BC2 and BC3 progeny between Saccharum spp. and E. arundinaceus. This work could provide more useful molecular cytogenetic information for the germplasm resources of E. arundinaceus, and may promote further understanding of the germplasm resources of E. arundinaceus for sugarcane breeders to accelerate its progress in sugarcane commercial breeding.
Introduction
Sugarcane (Saccharum spp.) is a large perennial grass that is indigenous to tropical and subtropical regions. As the most important sugar-producing crop worldwide, sugarcane has significant potential to contribute to the global sugar security and produces approximately 75% of the world’s raw sugar [1]. In addition, as a C4 plant, sugarcane is an efficient crop in converting solar energy into chemical energy. Therefore, it has been heralded as an alternative source of fuel and petrochemical feedstock for the production of first-generation bioethanol to alleviate the current energy crisis [2].
The genus Saccharum is an important member of the Poaceae family that consists of six species, including S. officinarum, S. spontaneum, S. robustum, S. barberi, S. sinense, and S. edule. Modern sugarcane cultivars are highly complex aneupolyploids, and most are primarily derived from interspecific hybridization between S. officinarum (2n = 80) and S. spontaneum (2n = 40–128) through nobilization [3]. This term was first coined by Dutch breeders Jesweit in Java during the early 1900s to denote the process of introgression of S. spontaneum into S. officinarum following hybridization and successive backcrossing. During the nobilization process, interspecific F1 hybrids were obtained from crosses between S. officinarum as the female parent and S. spontaneum as the male parent, and then were repeatedly backcrossed to S. officinarum as the female parent. Using this approach, progeny conserve the entire genome of S. officinarum in the first interspecific cross (F1) and the first backcross (BC1) [4]. Hence, the chromosome inheritance of progeny in F1 and BC1 exhibits 2n + n transmission. This not only allows for a quick recovery of the high sugar content from S. officinarum, but also integrates resistance genes to biotic and abiotic stresses from S. spontaneum [5]. Jesweit succeeded in the selective breeding of some new cultivars with high resistance to disease, and significantly contributed to the perseverance through the sugar crisis in Java at that time due to disease outbreaks [2]. POJ2878, hailed as the “wonder cane”, is one of the most successful examples of the utilization of nobilization.
However, due to the frequent utilization of a limited number of progenitors in sugarcane breeding programs, modern sugarcane cultivars have given rise to a sharp decline in genetic diversity [6,7]. Genetic erosion renders sugarcane increasingly vulnerable to resistance against biotic and abiotic stresses. As a result, the genetic potential for yield and quality improvement has hardly allowed for any advancement in the past several decades. Therefore, options for remedying the growing concern of a dearth of genetic variation has become an urgent and necessary task for sugarcane breeders. One efficient approach for combating this issue is by tapping into wild relatives to introduce favorable genes for increased productivity and better adaptability to a wide large range of growing conditions as well as providing more robust disease resistance. The genus Saccharum together with the four related genera, namely Erianthus, Miscanthus, Narenga, and Sclerostachya, comprise the “Saccharum complex” [8]. These four related genera serve as a rich gene pool for sugarcane improvement with tolerance to abiotic stresses and resistance to biotic stresses. As one of the most important wild relatives of sugarcane, Erianthus arundinaceus (E. arundinaceus) has many superior traits for sugarcane improvement [8–12]. It has already been considered to be one of the most popular germplasm sources for crossing utilization in sugarcane improvement. However, the taxonomy of Saccharum and Erianthus had been controversial for a long time [13]. Until there is a good deal of evidence that the genetic distance is large between Saccharum and Erianthus, according to morphological characteristics, chromosome number, and phylogenetic relationship, the evidence sheds light on the taxonomic relationship between Saccharum and Erianthus [8,14–17]. Despite the large genetic distance between Saccharum and Erianthus, genuine intergeneric F1 hybrids and their derivatives have been successfully generated in the past [9,11,18–21].
In sugarcane, there are various types of chromosome transmission such as n + n, 2n + n, n + 2n, and 2n + 2n [19,21–26]. Compared to the common type of chromosome transmission (n + n), the other three specific types of chromosome transmission (2n + n, n + 2n, and 2n + 2n) are derived from unilateral and bilateral sexual polyploidization. In the plant kingdom, sexual polyploidization, leading to unreduced gametes (2n gametes) with the somatic chromosome number rather than the gametophytic number (n gamete), is generally believed to be the predominant mechanism of polyploidization [27,28]. Based on the different gametogenesis, the unreduced gametes are divided into 2n egg gametes and 2n male gametes. 2n egg gametes typically originate from the unreduced ovule during megasporogenesis, whereas 2n male gametes are the result of the unreduced pollen during microsporogenesis [29]. The consequence of unilateral sexual polyploidization is 2n + n (result from the fertilization of unreduced ovule by normal haploid pollen) or n + 2n (result from the fertilization of normal ovule by unreduced pollen), while the result of bilateral sexual polyploidization is 2n + 2n (result from the fertilization of unreduced ovule by unreduced pollen). Several mechanisms have been described that the types of meiotic abnormalities responsible for the production of 2n gametes. Due to the different parental heterozygosity rate that each mechanism transmits to the progeny, the genetic consequences of different types of 2n gametes formation are highly divergent [28]. Hence, the use of 2n gametes, resulting in the different types of chromosome transmission during the establishment of sexual polyploids, is of prime importance to develop and conduct breeding strategies for crop improvement [30]. Indeed, it has already been proven effective for improvement of crops such as lily, potato, banana, and citrus [31–38]. In nobilization of S. spontaneum, the utilization of 2n gametes transmission from S. officinarum in interspecific crosses with S. spontaneum is such a typical example of the cytological peculiarity of 2n gametes.
Genomic in situ hybridization (GISH) is a powerful molecular cytogenetic tool to unravel the chromosome composition for the detection of different chromosomes sets derived from two or more distinct species and even recombinant chromosome segments in allopolyploids. During allopolyploid speciation and evolutionary process, the occurrence of chromosomal rearrangement is common, such as translocation and inversion. So far, this molecular cytogenetic technology has been widely used in investigating the chromosome composition and chromosomal translocation in a wide range of natural allopolyploids or artificial polyploid progeny [39–46]. Thus, knowledge of inferring the chromosome transmission from the chromosome composition in allopolyploid will make it possible to implement a strategy for developing useful varieties through breeding. Using GISH, much insight has been gained into sugarcane chromosomal inheritance and genomic recombination over the past several decades [11,19–21,24]. A previous study indicated that modern sugarcane cultivars possess approximately 120 chromosomes, with 70–80% derived from S. officinarum, 10–20% from S. spontaneum, and a few chromosomes derived from interspecific recombination [24].
In this study, two generations, including nine BC2 progeny and eight BC3 progeny, were characterized by GISH. The objectives were as follows: (1) to determine the chromosome transmission in these two generations, which can provide a reference for breeding strategies for further deployment of genes and traits from E. arundinaceus; and (2) to determine the presence of various types of intergeneric chromosomal translocation and obtain information on whether they can be inherited, which can provide a basic understanding for efficient utilization in sugarcane breeding.
Materials and Methods
Plant materials
The plant materials used in this study consisted of 17 progeny derived from two generations (BC2 and BC3) of intergeneric hybrids between Saccharum spp. and E. arundinaceus (Table 1). In the F1 generation, F1 hybrids between S. officinarum and E. arundinaceus were derived from crosses between Badila (S. officinarum, 2n = 80) as the female parent and HN 92–77 (E. arundinaceus, 2n = 60) or HN 92–105 (E. arundinaceus, 2n = 60) as the male parent. In the BC1 generation, F1 hybrids between S. officinarum and E. arundinaceus were used as the female parent. CP 84–1198 (2n = 120), a commercial cultivar containing germplasms from S. officinarum, S. spontaneum, S. barberi and S. robustum without contribution from E. arundinaceus, was used as the male parent [21]. In the BC2 and BC3 generation, all the female parents and the male parents are listed in detail in Table 1. Among them, ROC10, ROC20, ROC23, YT73-204, YT91-976, YT93-159, NJ57-416, and YC95-46 are the commercial cultivars containing germplasm from Saccharum spp. without contribution from E. arundinaceus, while “YCE” series are the progeny of E. arundinaceus. The progeny analyzed in this study were generated at the Hainan Sugarcane Breeding Station of Guangzhou Sugarcane Industry Research Institute. All plant materials were grown in the greenhouse at Fujian Agriculture and Forestry University.
Table 1. The intergeneric BC2 and BC3 progeny between Saccharum spp. and E. arundinaceus.
| Generation | Progeny | Female parent | Male parent |
|---|---|---|---|
| BC2 | YCE03-01 | NJ57-416 | YCE01-116 (BC1) |
| BC2 | YCE03-06 | YCE01-116 (BC1) | NJ57-416 |
| BC2 | YCE03-16 | YCE01-91 (BC1) | ROC23 |
| BC2 | YCE03-168 | YCE01-123 (BC1) | ROC10 |
| BC2 | YCE03-218 | YT73-204 | YCE01-105 (BC1) |
| BC2 | YCE03-249 | YCE01-69 (BC1) | YT73-204 |
| BC2 | YCE03-378 | ROC20 | YCE01-92 (BC1) |
| BC2 | YCE04-55 | YC95-46 | YCE01-102 (BC1) |
| BC2 | YCE05-179 | ROC20 | YCE01-134 (BC1) |
| BC3 | YCE05-64 | YT73-204 | YCE03-133 (BC2) |
| BC3 | YCE05-150 | YCE03-218 (BC2) | ROC10 |
| BC3 | YCE06-61 | ROC10 | YCE03-01 (BC2) |
| BC3 | YCE06-63 | ROC10 | YCE03-01 (BC2) |
| BC3 | YCE06-92 | YCE04-51 (BC2) | YT93-159 |
| BC3 | YCE06-111 | YCE03-168 (BC2) | YT93-159 |
| BC3 | YCE06-140 | YCE03-218 (BC2) | ROC10 |
| BC3 | YCE06-166 | YCE03-168 (BC2) | YT91-976 |
Note: “YCE” series are the progeny of E. arundinaceus, the other plant materials are the commercial cultivars containing germplasm from Saccharum spp. without contribution from E. arundinaceus.
Genomic in situ hybridization (GISH) procedure
Chromosome preparation and the GISH experiment were carried out according to the method described by D’hont et al. [9]. Genomic DNA from Badila (S. officinarum) and YN82-114 (S. spontaneum) was labelled with Biotin, the Biotin-labeled probe was detected with Avidin D, Rhodamine 600 (XRITC) and a Biotinylated anti-avidin antibody (Vector Laboratories, Burlingame, CA), respectively. Genomic DNA from HN92-77 or HN92-105 (E. arundinaceus) was labelled with Digoxigenin, and the Digoxigenin-labeled probe was detected with sheep-anti-Digoxin-FITC (Roche, Lewes, UK) and rabbit-anti-sheep-FITC secondary antibody (Roche, Lewes, UK). Chromosomes were then counterstained with 4′, 6-diamidino-2-phenylindole (DAPI) in a Vectashield anti-fade solution (Vector Laboratories, Burlingame, CA). FISH signals were captured using an AxioScope A1 Imager fluorescent microscope (Carl Zeiss, Gottingen, Germany). In this study, results are presented as the modal number and the range of chromosomes counting four to 22 metaphases for each progeny (Table 2). The images were processed using an AxioCam MRc5 and AxioVision v.4.7 imaging software (Carl Zeiss, Gottingen, Germany).
Table 2. Chromosome composition of the intergeneric BC2 and BC3 progeny between Saccharum spp. and E. arundinaceus.
| Generation | Progeny | Chromosome composition | No. of chromosomes | No. of metaphases analyzed | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Modal number | Range | Modal number | Range | Modal number | Range | Recombinants | Type of recombinants | ||||
| Total (2n cell) | Saccharum spp. | E. arundinaceus | |||||||||
| BC2 | YCE03-01 | 2n = 119 = 105S + 14E | 119 | 115–122 | 105 | 103–106 | 14 | 12–14 | 0 | — | 10 |
| BC2 | YCE03-06 | 2n = 119 = 105S + 14E | 119 | 118–123 | 105 | 104–107 | 14 | 13–15 | 0 | — | 16 |
| BC2 | YCE03-16 | 2n = 113 = 100S + 13E | 113 | 109–113 | 100 | 98–101 | 13 | 12–14 | 0 | — | 15 |
| BC2 | YCE03-168 | 2n = 110 = 100S + 9E + E/S | 111 | 108–113 | 100 | 99–103 | 10 | 8–10 | 1 | E/S | 10 |
| BC2 | YCE03-218 | 2n = 107 = 97S + 10E | 107 | 105–108 | 97 | 97–99 | 10 | 9–12 | 0 | — | 14 |
| BC2 | YCE03-249 | 2n = 110 = 97S + 13E | 110 | 107–113 | 97 | 96–100 | 13 | 12–15 | 0 | — | 22 |
| BC2 | YCE03-378 | 2n = 121 = 104S + 16E + S/E | 121 | 115–121 | 104 | 101–105 | 16 | 15–17 | 1 | S/E | 8 |
| BC2 | YCE04-55 | 2n = 111 = 98S + 13E | 111 | 109–114 | 98 | 96–100 | 13 | 11–13 | 0 | — | 18 |
| BC2 | YCE05-179 | 2n = 112 = 99S + 13E | 112 | 108–116 | 99 | 96–101 | 13 | 13–14 | 0 | — | 12 |
| BC3 | YCE05-64 | 2n = 118 = 107S + 6E + 2(E/S)+3(S/E) | 118 | 113–118 | 107 | 103–113 | 6 | 6 | 5 | 2(E/S)+3(S/E) | 4 |
| BC3 | YCE05-150 | 2n = 116 = 108S + 8E | 116 | 114–119 | 108 | 102–111 | 8 | 8 | 0 | — | 14 |
| BC3 | YCE06-61 | 2n = 114 = 107S + 7E | 114 | 109–115 | 107 | 102–108 | 7 | 7 | 0 | — | 20 |
| BC3 | YCE06-63 | 2n = 105 = 98S + 7E | 105 | 101–106 | 98 | 94–99 | 7 | 5–8 | 0 | — | 9 |
| BC3 | YCE06-92 | 2n = 118 = 109S + 7E + 2(E/S) | 118 | 115–119 | 109 | 102–112 | 7 | 7 | 2 | 2(E/S) | 10 |
| BC3 | YCE06-111 | 2n = 108 = 103S + 4E + E/S | 108 | 104–110 | 103 | 100–106 | 4 | 4 | 1 | E/S | 15 |
| BC3 | YCE06-140 | 2n = 112 = 106S + 5E + S/E | 112 | 108–112 | 106 | 100–109 | 5 | 5–6 | 1 | S/E | 11 |
| BC3 | YCE06-166 | 2n = 110 = 105S + 5E | 110 | 106–111 | 105 | 101–106 | 5 | 5 | 0 | — | 4 |
Note: Since small variation of chromosome counts can occur due to the loss or the overlapping of a few chromosomes from the preparation, the modal number of chromosomes and the range of total numbers of chromosomes in 2n cell are presented for the sugarcane clones analyzed. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively.
Results and Discussion
n + n chromosome transmission in BC2 and BC3 progeny
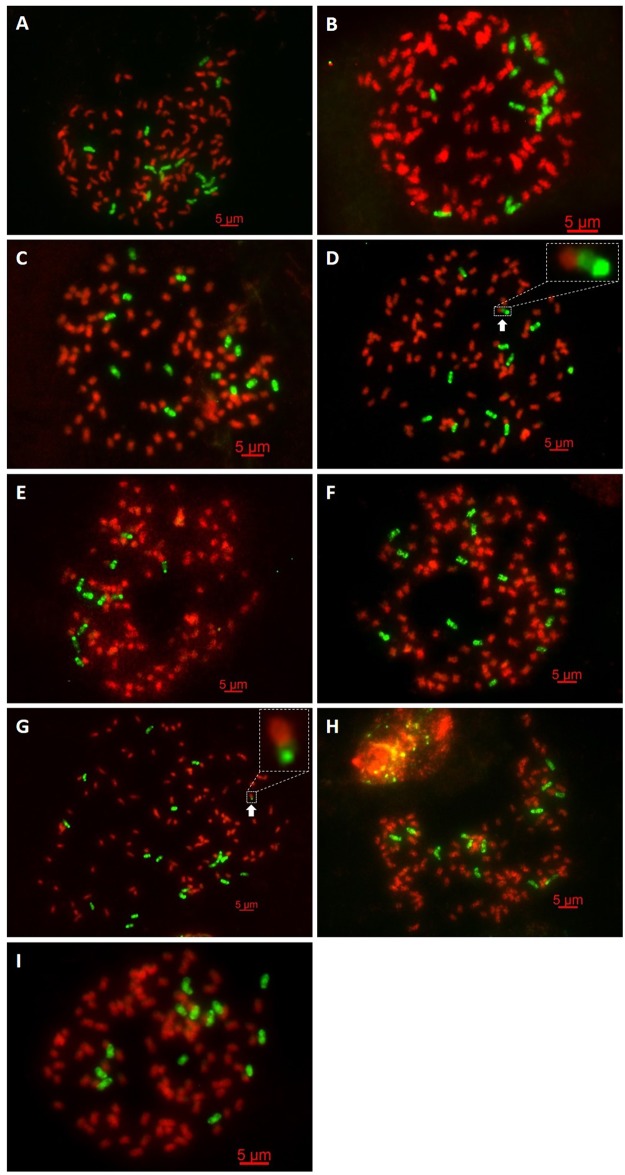
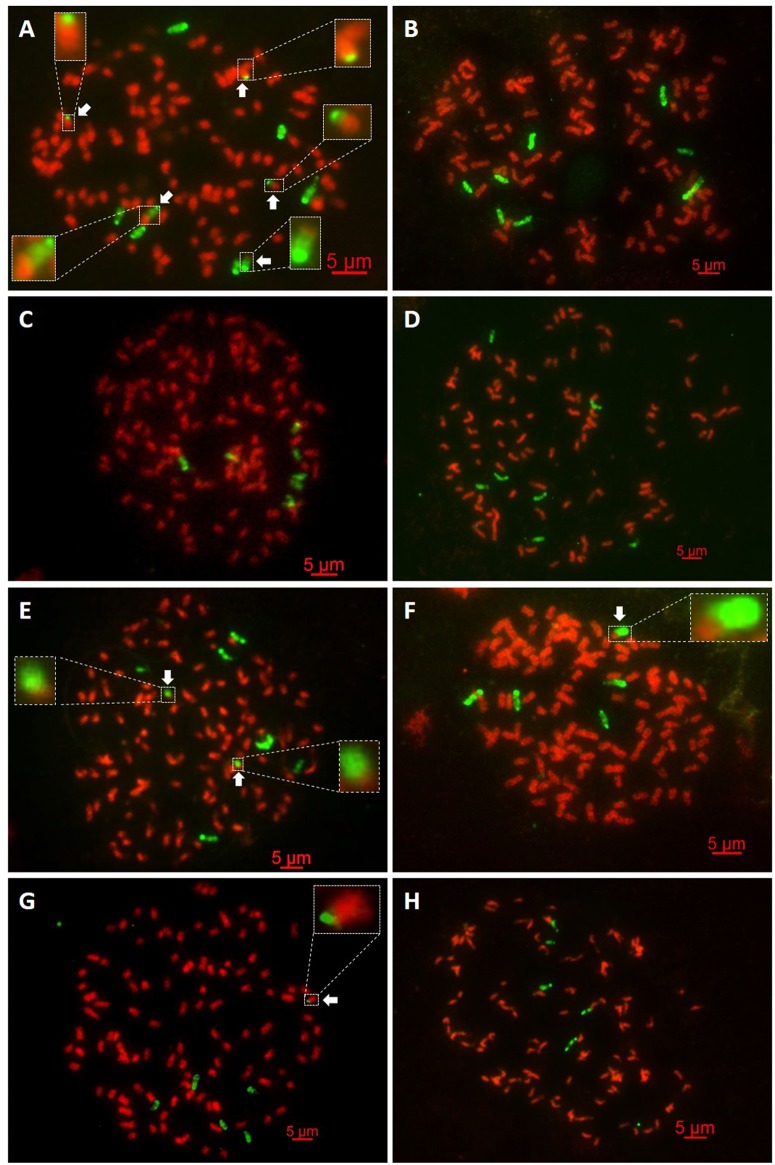
GISH experiments of nine BC2 progeny revealed plants with a total chromosome complement ranging from 107 to 121 chromosomes, of which 97–105 were derived from Saccharum spp. and 9–16 from E. arundinaceus, respectively (Table 2, Fig 1A–1I; Figs A-I in S1 File). Since 22–35 chromosomes were derived from E. arundinaceus in BC1 as parents (YCE01-69, YCE01-102, YCE01-92, YCE01-134, YCE01-105 and YCE01-116) [21], these results indicate that the number of E. arundinaceus chromosomes in the BC2 progeny was reduced by approximately half of the E. arundinaceus chromosomes of the BC1 parents. For instance, YCE01-116 (BC1) was the male parent of YCE03-01; YCE01-116 and YCE03-01 bore 28 and 14 chromosomes derived from E. arundinaceus, respectively [21]. This indicates that the BC2 progeny were products of n + n transmission. GISH experiments of eight BC3 progeny revealed plants with a total chromosome complement ranging from 105 to 118 chromosomes, of which 98–109 were derived from Saccharum spp. and 4–8 from E. arundinaceus, respectively (Table 2 and Fig 2A–2H; Figs A-H in S2 File). Since 9–14 chromosomes were derived from E. arundinaceus in BC2 as parents (YCE03-168, YCE03-218 and YCE03-01), these results indicate that approximately half of the E. arundinaceus chromosomes of the BC2 parents was transmitted to the BC3 progeny. For example, YCE03-01 (BC2) was the male parent of YCE06-63; YCE03-01 and YCE06-63 bore 14 and 7 chromosomes derived from E. arundinaceus, respectively. Our results indicate that the eight BC3 progeny were products of n + n transmission. Piperidis et al. [19,20] reported that the similar transmission was in some different intergeneric BC2 and BC3 progeny between Saccharum spp. and E. arundinaceus.
Fig 1. GISH analysis of the intergeneric BC2 progeny between Saccharum spp. and E. arundinaceus. Saccharum spp. chromosomes are visualized in red and E. arundinaceus chromosomes in green.
(A) YCE03-01: 2n = 119 = 105S + 14E; (B) YCE03-06: 2n = 119 = 105S + 14E; (C) YCE03-16: 2n = 113 = 100S + 13E; (D) YCE03-168: 2n = 111 = 100S + 10E + E/S; (E) YCE03-218: 2n = 107 = 97S + 10E; (F) YCE03-249: 2n = 110 = 97S + 13E; (G) YCE03-378: 2n = 121 = 104S + 16E + S/E; (H) YCE04-55: 2n = 111 = 98S + 13E; (I) YCE05-179: 2n = 112 = 99S + 13E. The arrowheads in Fig 1D and Fig 1G show the translocated chromosome. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
Fig 2. GISH analysis of the intergeneric BC3 progeny between Saccharum spp. and E. arundinaceus. Saccharum spp. chromosomes are visualized in red and E. arundinaceus chromosomes in green.
(A) YCE05-64: 2n = 118 = 107S + 6E + 2(E/S)+3(S/E); (B) YCE05-150: 2n = 116 = 108S + 8E; (C) YCE06-61: 2n = 114 = 107S + 7E; (D) YCE06-63: 2n = 105 = 98S + 7E; (E) YCE06-92: 2n = 118 = 109S + 7E + 2(E/S); (F) YCE06-111: 2n = 108 = 103S + 4E + E/S; (G) YCE06-140: 2n = 112 = 106S + 5E + S/E; (H) YCE06-166: 2n = 110 = 105S + 5E. The arrowheads in Fig 2A, Fig 2E, Fig 2F and Fig 2G show the translocated chromosome. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
In this study, YCE05-150 is a sibling line of YCE06-140 with five E. arundinaceus chromosomes. However, the number of E. arundinaceus chromosomes in YCE05-150 is eight. That is, more than half of the E. arundinaceus chromosomes in YCE03-218 was transmitted to YCE05-150. Given the results of chromosome count, the number of total chromosomes in YCE05-150, YCE03-218 (as the female parent) and ROC10 (as the male parent) were 116, 107 and 112, respectively. We can exclude the possibility that YCE05-150 was the product of 2n + n or n + 2n transmission. In fact, YCE05-150 was also the product of n + n transmission.
In nobilization of S. spontaneum, the interspecific F1 hybrids and BC1 progeny result from 2n + n transmission. The speedy process of nobilization is conducted to recover high biomass yield and sugar content from S. officinarum while retaining the stress tolerance characteristics from S. spontaneum. As a general rule, the improved varieties are obtained in the BC2 or BC3. However, the increasing times of nobilization may negatively impact the recovery of vigor and resistance to biotic or abiotic stresses. Molecular cytogenetic studies of E. arundinaceus indicated that chromosome transmission was n + n in F1, BC2, and BC3 generations, but was 2n + n in the BC1 generation [19–21]. Compared to the progress of nobilization for utilization of S. spontaneum, this slows down the progress of nobilization in utilization of E. arundinaceus and may require the improved varieties from the BC3, BC4, or even higher generations. Interestingly, Wu et al. [21] recently reported that an unexpected inheritance pattern of E. arundinaceus chromosomes resulted from more than a 2n + n transmission in the BC1 generation. This may lead to the presence of a larger number of new, multilocus allelic combinations and potentially creates a massive opportunity for selection of desirable traits in newly synthesized germplasm.
Smut caused by Sporisorium scitamineum is a destructive and worldwide disease of sugarcane, resulting in severe yield reductions and considerable loss in sugar content. Wild relatives represent potentially important sources of desirable genes for sugarcane improvement. As one of the most important wild relatives of sugarcane, E. arundinaceus has the potential to improve disease resistance in sugarcane. Introgressing from wild relatives into sugarcane is an effective approach to broadening the genetic basis of sugarcane germplasm. More strikingly, recent reports have proven that YCE05-179, a BC2 progeny, is resistant to sugarcane smut [47,48]. The rest of the other yet-to-be-identified progeny between Saccharum spp. and E. arundinaceus might provide superior lines resistant to abiotic and biotic stresses. It is necessary to screen the progeny between Saccharum spp. and E. arundinaceus for those most likely to improve commercial and agricultural traits.
Chromosomal translocation in the intergeneric BC2 and BC3 progeny between Saccharum spp. and E. arundinaceus
The current results obtained in this study together with those present in a previous study indicate that eight progeny harbored an intergeneric chromosomal translocation between Saccharum spp. and E. arundinaceus (Table 2, Fig 1D, 1G; Figs D, G in S1 File; and Fig 2A, 2E, 2F, 2G; Figs A, E, F, G in S2 File). In our previous study, out of 13 BC1 progeny analyzed, two BC1 progeny (YCE01-36 and YCE01-92) both carried an intergeneric translocated chromosome, and the chromosomal translocation occurred at a terminal fragment from the E. arundinaceus chromosome [21]. In this present study, out of nine BC2 progeny analyzed, two BC2 progeny (YCE03-168 and YCE03-378) both contained one translocated chromosome, and the chromosomal translocation occurred at a terminal fragment from the E. arundinaceus chromosome (Table 2, Fig 1D and 1G; Figs D and G in S1 File). Moreover, out of eight BC3 progeny analyzed, there were four BC3 progeny (YCE05-64, YCE06-92, YCE06-111, and YCE06-140) with chromosome translocations. Piperidis et al. [20] also reported the presence of intergeneric chromosomal translocations between Saccharum spp. and E. arundinaceus in BC3. In our study, YCE06-111 and YCE06-140 possessed an intergeneric translocated chromosome, and YCE06-92 and YCE05-64 carried two and five translocated chromosomes, respectively. The chromosomal translocation occurred at the terminal fragment from E. arundinaceus chromosomes in all these cases (Table 2, Fig 2A, 2E, 2F and 2G; Figs A, E, F, G in S2 File). Notably, multiple chromosome translocations tend to occur in YCE06-92 and YCE05-64, indicating that these two progeny were more prone to translocation. These results revealed that chromosome breakpoint tended to occur at the terminal fragment from Saccharum spp. chromosome and/or E. arundinaceus chromosome. But in essence, there are basically two distinct types of recombinant chromosome in these chromosomal translocation events: (1) the recombinant chromosome of Saccharum spp. with terminally located fragment of E. arundinaceus (S/E); and (2) the recombinant chromosome of E. arundinaceus with terminally located fragment of Saccharum spp. (E/S). In both types, S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. In the light of the GISH results in our previous and present study, the type of recombinant chromosome in YCE01-36, YCE01-92, YCE03-378 and YCE06-140 belongs to the former type, whereas that in YCE03-168, YCE06-92 and YCE06-111 belongs to the latter one. In addition, the type of recombinant chromosome in YCE05-64 is the admixture of S/E and E/S, including 2(E/S) and 3(S/E).
Translocated chromosomes are stably transmitted to the progeny
Based on the pedigree, YCE01-92 (BC1) is the female parent for YCE03-378 (BC2), and YCE03-168 (BC2) is the male parent for YCE06-111 (BC3), respectively. In YCE01-92 there is one recombinant chromosome which is transmitted to YCE03-378. Similarly, in YCE03-168 there is also one recombinant chromosome which is transmitted to YCE06-111. That is, according to the transgenerational inheritance of the translocated chromosome, we can conclude that these translocated chromosomes could be stably transmitted to the progeny in subsequent generations. We recently reported that chromosome translocations only occur in the terminal regions and not the centromeric regions. This finding demonstrated that the terminal regions of the E. arundinaceus and/or Saccharum spp. chromosomes are more actively involved in translocations than the centromeric regions. It is possible that this is because intercalary translocations require more chromosome breakage events than terminal translocations, and therefore rarely occur. In addition, recombination events occur in different generations and in different progeny, suggesting that translocation events are not restricted to an individual progeny.
Significance of intergeneric chromosome translocation for sugarcane improvement
Previous molecular cytogenetic studies have suggested that a few chromosomes were derived from interspecific recombination between S. officinarum and S. spontaneum in modern sugarcane cultivars [24]. In our study, despite a large genetic distance between Saccharum spp. and E. arundinaceus [15,16,49], the occurrence of intergeneric chromosomal translocations occurred within the BC1, BC2, and BC3 generations. These results suggest that intergeneric chromosome translocations can occur during an early generation. A considerable number of the recombinant chromosomes confirmed that homologous recombination occurs in the resulting progeny from Saccharum spp. and E. arundinaceus. Undoubtedly, the intergeneric chromosome translocations between Saccharum spp. chromosomes and E. arundinaceus chromosomes in this study represent a new genetic variation between these two genomes. From the point-of-view of sugarcane, intergeneric chromosome translocations can import E. arundinaceus chromosome segments or useful genes of E. arundinaceus into sugarcane. A translocation event can lead to a break in the genetic linkage, which increases the opportunity to segregate genetic variation and opens up the possibility for generating genetic and phenotypic novelty. Importantly, this kind of genetic variation may have a positive impact on sugarcane improvement.
Supporting Information
(A) Saccharum spp. chromosomes are visualized in red; (B) E. arundinaceus chromosomes are visualized in green; (C) All chromosomes are counterstained in blue; (D) A merged image is generated from the red and green channels. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
(DOC)
(A) Saccharum spp. chromosomes are visualized in red; (B) E. arundinaceus chromosomes are visualized in green; (C) All chromosomes are counterstained in blue; (D) A merged image is generated from the red and green channels. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
(DOC)
Acknowledgments
We thank Angélique D’hont, Nathalie Piperidis and George Piperidis for technical assistance. We thank Guangzhou Sugarcane Industry Research Institute for providing the plant materials used in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by National Natural Science Foundation of China (31401440, http://www.nsfc.gov.cn/), the earmarked fund for the Modern Agriculture Technology of China (CARS-20-1-5). This project is also supported by Guangxi Natural Science Foundation (2014GXNSFFA118002, http://gxnsf.gxsti.net/stms/login.jsp). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.FAOSTAT (2013). Available: http://faostat.fao.org.
- 2. Lam E, Shine J, Da Silva J, Lawton M, Bonos S, Calvino M, et al. Improving sugarcane for biofuel: engineering for an even better feedstock. GCB Bioenergy. 2009; 1(3): 251–255. [Google Scholar]
- 3. Jannoo N, Grivet L, Chantret N, Garsmeur O, Glaszmann JC, Arruda P, et al. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 2007; 50(4): 574–585. [DOI] [PubMed] [Google Scholar]
- 4. Bremer G. Problems in breeding and cytology of sugar cane. Euphytica. 1961; 10(1): 59–78. [Google Scholar]
- 5. Roach B. Nobilisation of sugarcane. Proc Int Soc Sugar Cane Technol. 1972; 14: 206–216. [Google Scholar]
- 6. Lu Y, D'Hont A, Walker D, Rao P, Feldmann P, Glaszmann J. Relationships among ancestral species of sugarcane revealed with RFLP using single copy maize nuclear probes. Euphytica. 1994; 78(1–2): 7–18. [Google Scholar]
- 7. Irvine J. Saccharum species as horticultural classes. Theor Appl Genet. 1999; 98(2): 186–194. [Google Scholar]
- 8. Amalraj VA, Balasundaram N. On the taxonomy of the members of "Saccharum complex". Genet Resour Crop Ev. 2006; 53(1): 35–41. [Google Scholar]
- 9. D'Hont A, Rao PS, Feldmann P, Grivet L, Islam-Faridi N, Taylor P, et al. Identification and characterisation of sugarcane intergeneric hybrids, Saccharum officinarum × Erianthus arundinaceus, with molecular markers and DNA in situ hybridisation. Theor Appl Genet. 1995; 91(2): 320–326. 10.1007/BF00220894 [DOI] [PubMed] [Google Scholar]
- 10. Rott P, Mohamed I, Klett P, Soupa D, de Saint-Albin A, Feldmann P, et al. Resistance to leaf scald disease is associated with limited colonization of sugarcane and wild relatives by Xanthomonas albilineans . Phytopathology. 1997; 87(12): 1202–1213. 10.1094/PHYTO.1997.87.12.1202 [DOI] [PubMed] [Google Scholar]
- 11. Piperidis G, Christopher MJ, Carroll BJ, Berding N, D'Hont A. Molecular contribution to selection of intergeneric hybrids between sugarcane and the wild species Erianthus arundinaceus . Genome. 2000; 43(6): 1033–1037. [PubMed] [Google Scholar]
- 12. Cai Q, Aitken KS, Fan YH, Piperidis G, Jackson P, McIntyre CL. A preliminary assessment of the genetic relationship between Erianthus rockii and the "Saccharum complex" using microsatellite (SSR) and AFLP markers. Plant Sci. 2005; 169(5): 976–984. [Google Scholar]
- 13. Jackson P, Henry RJ. Erianthus In: Kole C, editors. Wild Crop Relatives: Genomic and Breeding Resources. Berlin Heidelberg: Springer; 2011. pp. 97–107. [Google Scholar]
- 14. Burner DM. Cytogenetic analyses of sugarcane relatives (Andropogoneae: Saccharinae). Euphytica. 1991; 54(1): 125–133. [Google Scholar]
- 15. Nair NV, Nair S, Sreenivasan TV, Mohan M. Analysis of genetic diversity and phylogeny in Saccharum and related genera using RAPD markers. Genet Resour Crop Ev. 1999; 46(1): 73–79. [Google Scholar]
- 16. Alix K, Paulet F, Glaszmann J, D’Hont A. Inter-Alu-like species-specific sequences in the Saccharum complex. Theor Appl Genet. 1999; 99(6): 962–968. [Google Scholar]
- 17. Sobral BWS, Braga DPV, LaHood ES, Keim P. Phylogenetic analysis of chloroplast restriction enzyme site mutations in the Saccharinae Griseb. subtribe of the Andropogoneae Dumort. tribe. Theor Appl Genet. 1994; 87(7): 843–853. 10.1007/BF00221137 [DOI] [PubMed] [Google Scholar]
- 18. Ram B, Sreenivasan T, Sahi B, Singh N. Introgression of low temperature tolerance and red rot resistance from Erianthus in sugarcane. Euphytica. 2001; 122(1): 145–153. [Google Scholar]
- 19. Piperidis N, Chen J, Deng H, Wang L, Jackson P, Piperidis G. GISH characterization of Erianthus arundinaceus chromosomes in three generations of sugarcane intergeneric hybrids. Genome. 2010; 53(5): 331–336. 10.1139/g10-010 [DOI] [PubMed] [Google Scholar]
- 20. Piperidis N, Aitken K, Hermann S. Towards a reliable method to select potentially high value Erianthus hybrids. Int Sugar J. 2013: 1–9. [Google Scholar]
- 21. Wu J, Huang Y, Lin Y, Fu C, Liu S, Deng Z, et al. Unexpected inheritance pattern of Erianthus arundinaceus chromosomes in the intergeneric progeny between Saccharum spp. and Erianthus arundinaceus . PLoS One. 2014; 9(10): e110390 10.1371/journal.pone.0110390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burner DM, Legendre BL. Chromosome Transmission and Meiotic Stability of Sugarcane (Saccharum spp.) Hybrid Derivatives. Crop Sci. 1993; 33(3): 600–606. [Google Scholar]
- 23. Piperidis G, Piperidis N, D'Hont A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics. 2010; 284(1): 65–73. 10.1007/s00438-010-0546-3 [DOI] [PubMed] [Google Scholar]
- 24. D'Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet. 1996; 250(4): 405–413. [DOI] [PubMed] [Google Scholar]
- 25. Hermann SR, Aitken KS, Jackson PA, George AW, Piperidis N, Wei X, et al. Evidence for second division restitution as the basis for 2n+n maternal chromosome transmission in a sugarcane cross. Euphytica. 2012; 187(3): 359–368. [Google Scholar]
- 26. Deng ZH, Zhang MQ, Lin WL, Cheng F, Zhang CM, Li YC, et al. Analysis of Disequilibrium Hybridization in Hybrid and Backcross Progenies of Saccharum officinarum x Erianthus arundinaceus . Agr Sci China. 2010; 9(9): 1271–1277. [Google Scholar]
- 27. Lavia GI, Ortiz AM, Robledo G, Fernandez A, Seijo G. Origin of triploid Arachis pintoi (Leguminosae) by autopolyploidy evidenced by FISH and meiotic behaviour. Ann Bot. 2011; 108(1): 103–111. 10.1093/aob/mcr108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. De Storme N, Geelen D. Sexual polyploidization in plants cytological mechanisms and molecular regulation. New Phytol. 2013; 198(3): 670–684. 10.1111/nph.12184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mason AS, Pires JC. Unreduced gametes: meiotic mishap or evolutionary mechanism? Trends Genet. 2015; 31(1): 5–10. 10.1016/j.tig.2014.09.011 [DOI] [PubMed] [Google Scholar]
- 30. Ramanna MS, Jacobsen E. Relevance of sexual polyploidization for crop improvement—A review. Euphytica. 2003; 133(1): 3–18. [Google Scholar]
- 31. Barba-Gonzalez R, Lokker AC, Lim KB, Ramanna MS, Van Tuyl JM. Use of 2n gametes for the production of sexual polyploids from sterile Oriental x Asiatic hybrids of lilies (Lilium). Theor Appl Genet. 2004; 109(6): 1125–1132. [DOI] [PubMed] [Google Scholar]
- 32. Lim KB, Shen TM, Barba-Gonzalez R, Ramanna MS, Van Tuyl JM. Occurrence of SDR 2n gametes in Lilium hybrids. Breeding Sci. 2004; 54(1): 13–18. [Google Scholar]
- 33. Park TH, Kim JB, Hutten RCB, van Eck HJ, Jacobsen E, Visser RGF. Genetic positioning of centromeres using half-tetrad analysis in a 4x-2x cross population of potato. Genetics. 2007; 176(1): 85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jansky SH, Peloquin SJ. Advantages of wild diploid Solanum species over cultivated diploid relatives in potato breeding programs. Genet Resour Crop Ev. 2006; 53(4): 669–674. [Google Scholar]
- 35. Cuenca J, Aleza P, Navarro L, Ollitrault P. Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: application to citrus triploid progeny. Ann Bot. 2013; 111(4): 731–742. 10.1093/aob/mct032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cuenca J, Froelicher Y, Aleza P, Juarez J, Navarro L, Ollitrault P. Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv 'Fortune'. Heredity. 2011; 107(5): 462–470. 10.1038/hdy.2011.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aleza P, Juarez J, Cuenca J, Ollitrault P, Navarro L. Recovery of citrus triploid hybrids by embryo rescue and flow cytometry from 2x x 2x sexual hybridisation and its application to extensive breeding programs. Plant Cell Rep. 2010; 29(9): 1023–1034. 10.1007/s00299-010-0888-7 [DOI] [PubMed] [Google Scholar]
- 38. Ssebuliba RN, Tenkouano A, Pillay M. Male fertility and occurrence of 2n gametes in East African Highland bananas (Musa spp.). Euphytica. 2008; 164(1): 53–62. [Google Scholar]
- 39. Zhang HK, Bian Y, Gou XW, Dong YZ, Rustgi S, Zhang BJ, et al. Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. P Natl Acad Sci USA 2013; 110(48): 19466–19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harper J, Armstead I, Thomas A, James C, Gasior D, Bisaga M, et al. Alien introgression in the grasses Lolium perenne (perennial ryegrass) and Festuca pratensis (meadow fescue): the development of seven monosomic substitution lines and their molecular and cytological characterization. Ann Bot. 2011; 107(8): 1313–1321. 10.1093/aob/mcr083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manzanero S, Vega JM, Houben A, Puertas MJ. Characterization of the constriction with neocentric activity of 5RL chromosome in wheat. Chromosoma. 2002; 111(4): 228–235. [DOI] [PubMed] [Google Scholar]
- 42. An DG, Zheng Q, Zhou YL, Ma PT, Lv ZL, Li LH, et al. Molecular cytogenetic characterization of a new wheat-rye 4R chromosome translocation line resistant to powdery mildew. Chromosome Res. 2013; 21(4): 419–432. 10.1007/s10577-013-9366-8 [DOI] [PubMed] [Google Scholar]
- 43. Markova M, Michu E, Vyskot B, Janousek B, Zluvova J. An interspecific hybrid as a tool to study phylogenetic relationships in plants using the GISH technique. Chromosome Res. 2007; 15(8): 1051–1059. [DOI] [PubMed] [Google Scholar]
- 44. Zhang P, Li WL, Friebe B, Gill BS. The origin of a "zebra" chromosome in wheat suggests nonhomologous recombination as a novel mechanism for new chromosome evolution and step changes in chromosome number. Genetics. 2008; 179(3): 1169–1177. 10.1534/genetics.108.089599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mestiri I., Chague V, Tanguy AM, Huneau C, Huteau V, Belcram H, et al. Newly synthesized wheat allohexaploids display progenitor-dependent meiotic stability and aneuploidy but structural genomic additivity. New Phytol. 2010; 186(1): 86–101. 10.1111/j.1469-8137.2010.03186.x [DOI] [PubMed] [Google Scholar]
- 46. Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira SB, Chen G, et al. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 2001; 125(3): 1216–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Su Y, Guo J, Ling H, Chen S, Wang S, Xu L, et al. Isolation of a Novel Peroxisomal Catalase Gene from Sugarcane, which Is Responsive to Biotic and Abiotic Stresses. PloS One. 2014; 9(1): e84426 10.1371/journal.pone.0084426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Su YC, Xu LP, Xue BT, Wu QB, Guo JL, Wu LG, et al. Molecular cloning and characterization of two pathogenesis-related beta-1,3-glucanase genes ScGluA1 and ScGluD1 from sugarcane infected by Sporisorium scitamineum . Plant Cell Rep. 2013; 32(10): 1503–1519. 10.1007/s00299-013-1463-9 [DOI] [PubMed] [Google Scholar]
- 49. Alix K, Baurens FC, Paulet F, Glaszmann JC, D'Hont A. Isolation and characterization of a satellite DNA family in the Saccharum complex. Genome. 1998; 41(6): 854–864. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Saccharum spp. chromosomes are visualized in red; (B) E. arundinaceus chromosomes are visualized in green; (C) All chromosomes are counterstained in blue; (D) A merged image is generated from the red and green channels. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
(DOC)
(A) Saccharum spp. chromosomes are visualized in red; (B) E. arundinaceus chromosomes are visualized in green; (C) All chromosomes are counterstained in blue; (D) A merged image is generated from the red and green channels. S and E indicate Saccharum spp. chromosome and E. arundinaceus chromosome, respectively. S/E and E/S indicate Saccharum spp. centromere with E. arundinaceus chromosome segment and E. arundinaceus centromere with Saccharum spp. chromosome segment, respectively. Scale bars: 5 μm.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.