Abstract
Formation of foam cell macrophages, which sequester extracellular modified lipids, is a key event in atherosclerosis. How lipid loading affects macrophage phenotype is controversial, with evidence suggesting either pro- or anti-inflammatory consequences. To investigate this further, we compared the transcriptomes of foamy and non-foamy macrophages that accumulate in the subcutaneous granulomas of fed-fat ApoE null mice and normal chow fed wild-type mice in vivo. Consistent with previous studies, LXR/RXR pathway genes were significantly over-represented among the genes up-regulated in foam cell macrophages. Unexpectedly, the hepatic fibrosis pathway, associated with platelet derived growth factor and transforming growth factor-β action, was also over-represented. Several collagen polypeptides and proteoglycan core proteins as well as connective tissue growth factor and fibrosis-related FOS and JUN transcription factors were up-regulated in foam cell macrophages. Increased expression of several of these genes was confirmed at the protein level in foam cell macrophages from subcutaneous granulomas and in atherosclerotic plaques. Moreover, phosphorylation and nuclear translocation of SMAD2, which is downstream of several transforming growth factor-β family members, was also detected in foam cell macrophages. We conclude that foam cell formation in vivo leads to a pro-fibrotic macrophage phenotype, which could contribute to plaque stability, especially in early lesions that have few vascular smooth muscle cells.
Introduction
Atherosclerosis, the underlying cause of many vascular diseases, can be considered to be a healing response to multiple injurious stimuli that include endothelial dysfunction, lipid retention and inflammation (with activation of both innate and adaptive immune systems) [1,2]. Retention and oxidation of extracellular lipid in the vessel wall provokes the production of inflammatory mediators that recruit monocytes, which differentiate to macrophages [3,4]: it also creates neo-epitopes that can activate the immune system. Modified extracellular lipid is then taken up into foam cell macrophages (FCMs) [3] and smooth muscle cells (SMCs) [5,6]. Plaque macrophages and FCMs may also be differentially activated, adopting pro-inflammatory phenotypes similar to those designated M1 or ‘classically activated’ from in vitro experiments [7–9]. M1-like macrophages have been observed especially at the shoulder region of the fibrous cap of plaques. They express higher levels of many proteases, including matrix metalloproteinases (MMPs) [8], which can degrade the extracellular matrix leading to loss of collagen. This promotes subsequent cap rupture, thrombus formation and hence life-threatening myocardial infarctions or strokes [10,11]. Death of FCMs, because it releases lipid and enlarges the necrotic core of the plaque, increases mechanical stress on the plaque cap [12] and further promotes plaque rupture. Conversely, plaque macrophages with similarities to M2 or alternatively activated phenotypes have also been detected, especially in areas of plaques remote from the core, and in the adventitia [3,9,13]. These include so-called M2a, M2c/Mreg, Mhem and Mox macrophages that are anti-inflammatory, promote tissue repair and hence favour plaque stability [3,4,14–16].
The role of lipid uptake and foam cell formation per se in the activation of macrophage behaviour that provokes plaque rupture is controversial. Different in vitro studies suggested pro- or anti-inflammatory effects of treatment with oxidised lipids, whereas others showed no major influence on markers of M1 and M2 polarization [17–20]. Results from ex vivo studies of FCMs generated in vivo are also conflicting. Rabbit FCMs purified from subcutaneous granulomas showed increased activation of the nuclear factor-κB (NF-κB) pathway and production of several NF-κB dependent MMPs compared with non-foamy macrophages (NFMs) [21,22]. On the other hand, a recent paper that compared FCMs with NFMs isolated from the peritoneum of atherosclerosis-prone mice suggested that increased desmosterol levels in FCMs initiate a transcriptional programme mediated by the liver X receptor (LXR) that suppresses the expression of many pro-inflammatory genes [23]. To re-examine this controversy, we adapted the subcutaneous sponge model we used in rabbits [21,22] for use in mice, and compared the transcriptomes of FCMs and NFMs produced in fat-fed ApoE null or wild-type mice. We then validated the differential expression of selected genes at the protein level both in subcutaneous granulomas and in brachiocephalic artery plaques.
We found, unexpectedly, that foam cell formation under these conditions favoured neither M1 nor M2 polarization but instead induced a pro-fibrotic transcriptional programme that should favour plaque stability. Understanding how FCMs are different from NFMs could enhance our ability to treat or prevent atherosclerotic plaque development and subsequent acute events.
Materials and Methods
Animals
The housing and care of all animals and procedures used in these studies was in accordance with and under license of the Animals (Scientific Procedures) Act 1986 (London, United Kingdom), and conform to the Guide for the Care and Use of Laboratory Animals published by the U.S. National Institutes of Health (Publication No. 85–23, revised 1996). The study received local institutional review board (University of Bristol, Bristol, United Kingdom) approval. Homozygous ApoE−/− mice and wild-type (control) mice on a C57BL background were bred in the University of Bristol Animal Services Unit.
Surgical sponge implantation and harvest of foamy and non-foamy macrophages
Adult male ApoE−/− mice (10–25 weeks old) were changed from normal rodent diet to a high fat diet containing 23% fat from lard, supplemented with 0.15% (w/w) cholesterol (Special Diets Services, UK). Three weeks after commencing the high-cholesterol diet, the animals were anaesthetised with halothane and had six 0.5 cm3 pieces of sterile sponges (containing Matrigel (BD Biosciences, UK)) placed dorsally, subcutaneously under aseptic conditions and buprenorphine analgesia. The high-fat diet continued for 4 weeks post-surgery, when the sponges were recovered. For controls, age-matched male C57/129 mice were fed a normal chow diet before and after sponge insertion as above. Water and food were given ad libitum. Mice were euthanized by an overdose of halothane and the sponges removed using aseptic technique.
FCMs were purified from sponges removed from fat-fed ApoE null mice based on their lower buoyant density than other cells followed by differential adherence to plastic, whereas NFM were purified by differential adherence alone [21,22]. Purified cells were used for RNA extraction (Illumina array, RT-qPCR), viability (Trypan blue), lipid content (histochemistry) or identification of cell type (RT-qPCR, immunocytochemistry). Some whole sponges from ApoE-/- mice were fixed in 10% phosphate-buffered formalin and paraffin-embedded; 3 μm sections were then cut and used for immunohistochemistry.
Development of atherosclerosis
ApoE-/- male mice commenced a high-fat diet at 5 weeks of age. Twelve weeks later the animals were killed and perfusion fixed at normal pressure with phosphate-buffered saline (PBS) followed by 10% phosphate-buffered formalin. Brachiocephalic arteries were dissected out as described previously [24] and embedded in paraffin. Sections (3 μm thick) were prepared and used for immunohistochemistry.
Transcriptomic comparison of FCMs and NFMs
Total cellular RNA was extracted using the Qiagen RNease MiniKit (UK) according to the manufacturer’s instructions. RNA concentration was determined using NanoDrop spectrometry (NanoDrop Technologies, USA). RNA samples of high quality (A260/280>2) (n = 4) were compared using Illumina bead chips (MouseRef8 v2.0 Expression BeadChips, Illumina, USA) at 1 μg/sample.
Functional Annotation
All genes from the dataset that met the unadjusted P-value cut-off of 0.01 were uploaded to the Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com) system and included in the analysis. Each identifier was mapped to its corresponding object in the Ingenuity Knowledge Base. Functional analysis identified the biological functions and canonical pathway analysis identified the pathways that were most significant to the data set. Network maps were also generated within Ingenuity Pathway Analysis. Differentially expressed genes were overlaid onto a global molecular network developed from information contained in the Ingenuity Knowledge Base and networks of these molecules algorithmically generated based on their connectivity. Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). The intensity of the node colour indicates the degree of up- (red) or down- (green) regulation. Nodes are displayed using various shapes that represent the functional class of the gene product. Edges are displayed with various labels that describe the nature of the relationship between the nodes (e.g., P for phosphorylation, T for transcription).
Differentially expressed genes were also submitted to GO annotation and clustering using DAVID Bioinformatics Resources (National Institute of Allergy and Infectious Diseases (NIAID) 2008, NIH, http://david.abcc.ncifcrf.gov).
RNA isolation and quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) assays
The array results were validated and expanded on RNA samples prepared in the same way using RT-qPCR (n = 5–7) with selected primer pairs (Table 1). For reverse transcription, 100 ng of total RNA was used to make cDNA, using a Qiagen Quantitect Reverse Transcripase Kit according to the manufacturer’s instructions. Sequences of the PCR primer pairs used to amplify the respective cDNAs were designed using Ensembl and Primer3, and the specificity of the sequence confirmed using Nucleotide blast (NCBI). Samples were compared, either after normalising to a housekeeping gene (36B4) or as copies per ng RNA.
Table 1. Primer sequences used in this study.
| GENE | ALTERNATIVE NAME | SPECIES | PRIMER SEQUENCE |
|---|---|---|---|
| 36B4 | ribosomal protein, large, P0, RPLP0 | human | GCCCAGGGAAGACAGGGCGA GCGCATCATGGTGTTCTTGCCCA |
| NR2B2 | retinoid X receptor β, RXRβ | mouse | GGTGCTGACAGAGCTAGTGTCCAA TGCTTGCAATAGGTCTCCAGTGAG |
| NR2B1 | RXRα | mouse | AGGACAGTACGCAAAGACCTGACC ATGTTTGCCTCCACGTATGTCTCA |
| NR1H3 | liver X receptor α, LXRα | mouse | GCAGGACCAGCTCCAAGTAGAGAG CACAAAGGACACGGTGAAACAGTC |
| NR1H2 | LXRβ | mouse | GGCGGCCCAACTGCAGTGCAACAA GCAAAGCGTTGCTGGCGGGCATCT |
| SREBF1 | sterol regulatory element-binding transcription factor-1, SREBP1 | mouse | AGGCCATCGACTACATCCG TCCATAGACACATCTGTGCCTC |
| FASN | fatty acid synthase | mouse | GGCTCTATGGATTACCCAAGC CCAGTGTTCGTTCCTCGGA |
| MSR1 | macrophage scavenger receptor-1, SR-A1, SCARA1 | mouse | GCTGCCCTCATTGCTCTCT CTGGAAGCGTTCCGTGTCT |
| CD36 | thrombospondin receptor | mouse | GTACAGCCCAATGGAGCCA AACCCCACAAGAGTTCTTTCAAA |
| SCARB1 | scavenger receptor B1, SR-B1 | mouse | GGTGCGCCTCTGTTTCTCTC AGAACTACTGGCTCGATCTTCCCT |
| ABCA1 | ATP-binding cassette, sub-family A1 | mouse | CTCAGTTAAGGCTGCTGCTG TCAGGCGTACAGAGATCAGG |
| PPARγ | peroxisome proliferator-activated receptor-γ | mouse | TTGACAGGAAGGACAACGGACAAA TGTGATCTCTTGCACGGCTTCTAC |
| COL1α1 | collagen 1α1 | mouse | GATGATGCTAACGTGGTTCGTGAC CCATGTTGCAGTAGACCTTGATGG |
| COL4α1 | collagen 4α1 | mouse | CTGGCACAAAAGGGACGAG CGTGGCCGAGAATTTCACC |
| COL6α1 | collagen 6α1 | mouse | ATGTGACCCAACTGGTCAACTCAA AGCATGGAAGACAGAACACAGACG |
| COL8α1 | collagen 8α1 | mouse | TCATCATTTCCCTGAACTCTGTC CAAAGGCATGTGAGGGACTTG |
| BGN | biglycan | mouse | TAGCCAGTCCCTCCACAAACAAAT AGGAAGCTCCTTGATCCTCGTCTT |
| DCN | decorin | mouse | CACAACCTTGCTAGACCTGC GAAGTTCCTGGAGAGTTCTGG |
| CTGF | connective tissue growth factor | human | GGTGTACCGCAGCGGAGAGT GGGCCAAACGTGTCTTCCAG |
| BMP1 | bone morphogenic protein v1,2 | mouse | AGACCTTTATTCCCATGCCCAGTT TTCTTGGAGATGGTGTCGTCAGAG |
| THBS1 | thrombospondin-1 | mouse | GGGCAAAGAACTTGTCCAGACTGT ACTGGGTGACTTGTTTCCACATCA |
| TGFβ1 | transforming growth factor-β1 | mouse | CTCCCGTGGCTTCTAGTGC GCCTTAGTTTGGACAGGATCTG |
| FOS | FBJ murine osteosarcoma viral oncogene homolog | mouse | TTCGACCATGATGTTCTCG TTGGCACTAGAGACGGACAGA |
| FosB | FosB | mouse | CCCTCCTGCATATCTTTGTCCTGT CTGTCATTTCCTCGTTGGGTCTCT |
| JUN | jun proto-oncogene | mouse | TGACTGCAAAGATGGAAACGA CAGGTTCAAGGTCATGCTCTGT |
| JunB | JunB | mouse | TCACGACGACTCTTACGCAG CCTTGAGACCCCGATAGGGA |
| SMAD2 | Smad2 v1,2 | mouse | ATGTCGTCCATCTTGCCATTC AACCGTCCTGTTTTCTTTAGCTT |
| SMAD3 | Smad3 | mouse | TGCACAGCCACCATGAATTAC TCCATCTTCACTCAGGTAGCC |
| ACVR1 | activin A receptor-1/ALK2 v1,2,3 | mouse | AATGGTGAGCAATGGTATAGTG GGGTCTGAGAACCATCTGTTAGG |
| TBFBR1 | TGFβ receptor-1/ALK5 | mouse | CAGCTCCTCATCGTGTTGGTG GCACATACAAATGGCCTGTCTC |
| CTSC | cathepsin C | mouse | GCAGGTCATCTACAATGCAACCAG TGGAGCATAAATGCTTCTAAGGGA |
| CTSE | cathepsin E | mouse | GCAGGTCATCTACAATGCAACCAG TGGAGCATAAATGCTTCTAAGGGA |
| CTSB | cystatin b | mouse | AGGTGAAGTCCCAGCTTGAAT TCTGATAGGAAGACAGGGTCA |
| CSTF | cystatin f | mouse | TGTTCCAAAGATTTGATCTCCAG GTACCAGGGCTTTGCTGACAT |
| LYZ | lysozyme v1&2 | mouse | GAATGCCTGTGGGATCAATTGC GCTGCAGTAGAAGCACACCG |
| MMP2 | matrix metalloproteinase-2 | mouse | GGCTGACATCATGATCAACTTTGG GCCATCAGCCGTTCCCATACTTTAC |
| MMP9 | MMP9 | mouse | AGAGAGGAGTCTGGGGTCTGGTTT GAGAACACCACCGAGCTATCCACT |
| MMP12 | MMP12 | mouse | AATTACACTCCGGACATGAAGCGT GGCTAGTGTACCACCTTTGCCATC |
| MMP13 | MMP13 | mouse | ATGATGATGAAACCTGGACAAGCA ATAGGGCTGGGTCACACTTCTCTG |
| MMP14 | MMP14 | mouse | ACCACAAGGACTTTGCCTCTGAAG CACCGAGCTGTGAGATTCCCTTGA |
| MMP23 | MMP23 | mouse | CAAGGTTGGTGAGAGAGGGTAGGA AGGAGTAGGTGCTGAGAACACGCT |
| TIMP1 | tissue inhibitor of metalloproteinase-1 | mouse | AGGAACGAAATTTGCACATCAGT CAAAGTGACGGCTCTGGTAGTCCT |
| TIMP2 | TIMP2 | mouse | GACTCCCCCTCAGACTCTCCCTAC CATATTGATACCACCGCACAGGAA |
| TIMP3 | TIMP3 | mouse | CACATCAAGGTGCCATTCAGGTAG GTTCTCTCCTCCTCAACCCAAACA |
| NOS2 | nitric oxide synthase-2, iNOS | mouse | CTCATGACATCGACCAGAAGCGT TATATTGCTGTGGCTCCCATGTTG |
| IL-6 | interleukin-6 | mouse | GTTCTCTGGGAAATCGTGGA TTCTGCAAGTGCATCATCGT |
| CCL2 | chemokine (C-C motif) ligand-2, MCP1 | rabbit | GCTTGCCCAGCCAGATGCCGTGAA GGTTGGCAATGGCATCCTGGACCC |
| NF-κB1 | nuclear factor of kappa light chain polypeptide gene enhancer in B-cells 1 | mouse | GGAGGCATGTTCGGTAGTGG CCCTGCGTTGGATTTCGTG |
| ARG1 | arginase-1 | mouse | AGTCTGGCAGTTGGAAGCATCTCT TTCCTTCAGGAGAAAGGACACAGG |
| ARG2 | arginase-2 | mouse | ACAGCCAGACTAGCACTGGATGTC CGAATGCCTTGCAACTCTGTAATG |
| Chi3l3 | chitinase-like 3, Ym1 | mouse | CAGGTCTGGCAATTCTTCTG GTCTTGCTCATGTGTGTAAGTG |
| CD206 | mannose receptor, MRC1 | mouse | CCATTTATCATTCCCTCAGCAAGC AAATGTCACTGGGGTTCCATCACT |
| RND3 | member Rho GTPase family | human | GGGACACTTCGGGTTCTCCTTACT TGGACAAAATTCCTGGATTTCACC |
| SARAF | store-operated calcium entry-associated regulatory factor, Tmem66 | mouse | GGCTTTAAGTCGGAGTTCACAGGA TCGAGTCTGCATTAGAGGATGCAC |
| CMPK2 | cytidine monophosphate (UMP-CMP) kinase 2 | mouse | GTTTCCTCGGTGTAGGAGCTGTGT CTCGAAGCTGACTTCACATGCAAT |
| MYH11 | smooth muscle myosin heavy chain v1&2 | mouse | ATGAGGTGGTCGTGGAGTTG GCCTGAGAAGTATCGCTCCC |
| MAPK14 | mitogen-activated protein kinase 14 | mouse | ATAAGAGGATCACAGCAGCCCAAG GACAGAACAGAAACCAGGTGCTCA |
| PICALM | picalm | mouse | CCATTCCAAGCTTAAACCCTTTCC AGGCCACTGTTGGTTTGAGAAGTC |
| GZMA | granzyme a | mouse | TGCTGCCCACTGTAACGTG GGTAGGTGAAGGATAGCCACAT |
| GZMB | granzyme b | mouse | TGCTGCTAAAGCTGAAGAGTAAG CGTGTTTGAGTATTTGCCCAT |
| NPC1 | Niemann Pick type 1 | mouse | GCTGTGAGCTGTGGTCTGC CTCACTCGGCTTCCTTTGGTA |
| NPC2 | Niemann Pick type 2 | mouse | AGGACTGCGGCTCTAAGGT AGGCTCAGGAATAGGGAAGGG |
| CCR7 | C-C chemokine receptor type-7 | mouse | TGTACGAGTCGGTGTGCTTC GGTAGGTATCCGTCATGGTCTTG |
| VDR | vitamin D receptor | mouse | ACCCTGGTGACTTTGACCG GGCAATCTCCATTGAAGGGG |
| CD44 | CD44 | mouse | GTCTTCTTCCGGCTCTCCATGTAA ATCTCACATCCAATGGGACAAGGT |
Immunocytochemistry and (immuno)histochemistry
After isolation, FCMs and NFMs were adhered to coverslips to allow identification of cell type (general leukocyte (Giemsa staining), macrophage or SMC/fibroblast; antibodies as listed in Table 2) after use of a mouse-on-mouse immunodetection kit (BMK-2212, Vector, USA) as necessary. Lipid content was identified after staining with Oil-Red-O (Sigma) and nuclei were counterstained with haematoxylin. Paraffin embedded sections of formalin-fixed sponges or brachiocephalic arteries from ApoE null mice were examined for expression of the various proteins (antibodies listed in Table 2), as were coverslips from RAW or bone marrow-derived monocyte (BMDM) cultures (S1 File). Specimens were visualised using light or fluorescent microscopy after labelling primary antibodies or controls with the appropriate secondary antibodies and substrate/fluorophore systems (DAB (Sigma), streptavidin-594 (Molecular Probes, Life Technologies). Nuclei were stained with haematoxylin (light microscopy) or DAPI (fluorescent; Prolong-Gold anti-fade medium + DAPI, Invitrogen). Irrelevant antibodies or appropriate sera were used in negative controls.
Table 2. Primary and secondary antibodies used in this study.
| Antibody | Clone/Cat # | Type | Supplier |
|---|---|---|---|
| Primary antibodies/controls | |||
| mouse monocyte/macrophage | MOMA2 /MBS530837 | Rt_Mab | Biosource International, USA |
| Human α/γ-SM actin | HHF35 /M0635 | Mm_Mab | Dako, UK |
| human smooth muscle myosin heavy chain (SM-1, SM-2) | hSM-V /M7786 | Mm_Mab | Sigma, USA |
| human biglycan | - /ab58562 | Gt_Pab | Abcam, USA |
| mouse CTGF | - /ab6992 | Rb_Pab | Abcam |
| human LXRα (NR1H3) | - /LS-B3526 | Rb_Pab | Lifespan Biosciences, USA |
| human cFOS | - /ab7963 | Rb_Pab | Abcam |
| human phospho-SMAD2 (ser465/467) | 138D4 /3108 | Rb_Mab | Cell Signalling Technology (New England Biolabs, UK) |
| human total SMAD2 | D43B4 /5339 | Rb_Mab | Cell Signalling Technology |
| human phospho-SMAD3 (ser423/425) | C25A9 /9520 | Rb_Mab | Cell Signalling Technology |
| human total SMAD3 | C67H9 /9523 | Rb_Mab | Cell Signalling Technology |
| rabbit GAPDH | 6C5 /MAB374 | Mm_Mab | Chemicon (Millipore) |
| normal rabbit IgG control | - /X0936 | Dako | |
| - I5006 | Sigma | ||
| normal rat IgG control | - /6-001-A | R&D Systems | |
| normal mouse IgG1 control | - /X0931 | Dako | |
| normal mouse IgG2b control | - /X0944 | Dako | |
| goat serum | - /X0907 | Dako | |
| Secondary antibodies | |||
| Mouse Ig | - /P0260 | Rb_Pab | Dako |
| - /B7264 | Gt_Pab | Sigma | |
| Rat Ig | - /E0468 | Rb_Pab | Dako |
| - /80-9520 | Mm_Pab | Zymed (Invitrogen) | |
| Rabbit Ig | - /P0448 | Gt_Pab | Dako |
| - /A0545 | Gt_Pab | Sigma | |
| - /7074 | Gt_Pab | Cell Signalling Technology | |
| Goat Ig | - /E0466 | Rb_Pab | Dako |
| - /P0449 | Rb_Pab | Dako |
Statistics
Statistical analysis of the microarrays was performed using the 'R' Bioconductor Lumi and Limma packages, applying the linear models and empirical Bayes methods included in the package [25]. Variance stabilizing transformation was performed using the Robust Spline Normalization (RSN) algorithm [26] with unsupervised analysis methods such as Principal Component Analysis and Hierarchical Clustering used for initial data exploration. Statistical analysis of differential expression consisted of a univariate model to detect individual genes that are significantly different in abundance between the conditions [25]. P-values were adjusted for multiple comparisons, using the False Discovery Rate (FDR) Benjamini-Hochberg method. Differential expression was classified as significant (P<0.01 after FDR correction for multiple testing) or suggestive (P<0.01 unadjusted). The significance of the association between the data set and the canonical pathway was measured by either a ratio of the number of molecules from the data set that map to the pathway divided by the total number of molecules that map to the canonical pathway is displayed, or by using a right-tailed Fisher’s exact test, to calculate a P-value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
For the RT-qPCR, data differences between the two groups were tested for significance with Student’s t-test, using a logarithmic transformation or the Mann-Whitney U-test if the data were not normally distributed, using GraphPad Instat (USA). P-values of less than 0.05 were considered statistically significant. Values are expressed as mean ± standard deviation.
Results
Yield and purity of cells isolated from subcutaneous granulomas
The yield of cells purified by floatation and adhesion from sponges implanted for 4 weeks into fat-fed ApoE null mice was 4.6 ± 2.7 x 106. This was comparable to the 6.3 ± 2.4 x 106 cells isolated by adhesion only from control non-fat-fed wild-type mice after 4 weeks, which is similar to values previously reported after 7 days [27]. Cells from the ApoE null and control mice were 96 ± 5.8% or 87 ± 10% MOMA2 positive, respectively, and only 0.86 ± 1.2% or 1.4 ± 1.8% α/γ-SM actin positive, respectively. Data from RT-qPCR confirmed that the cell preparations contained low levels of SM myosin heavy chain (SM1/SM2) (averaging less than 2.5 copies/ng RNA), implying they have little SMC contamination. Whereas almost all of the cells in the preparations from the ApoE null mice had lipid-rich (Oil-Red-O positive) inclusions, there were none in cell preparations from control mice (Fig 1). Hence cells isolated from the fat-fed ApoE null mice were indeed foam cell macrophages (FCMs), whereas those from the control mice were non-foamy macrophages (NFMs).
Fig 1. Lipid droplets were found only in macrophages from mice fed a high-fat diet.
Oil-Red-O staining for lipid (orange-red) in A) FCMs and B) NFMs isolated from sponges. Nuclei appear purple (haematoxylin). Magnification x 400.
Genes and pathways indicated from the array
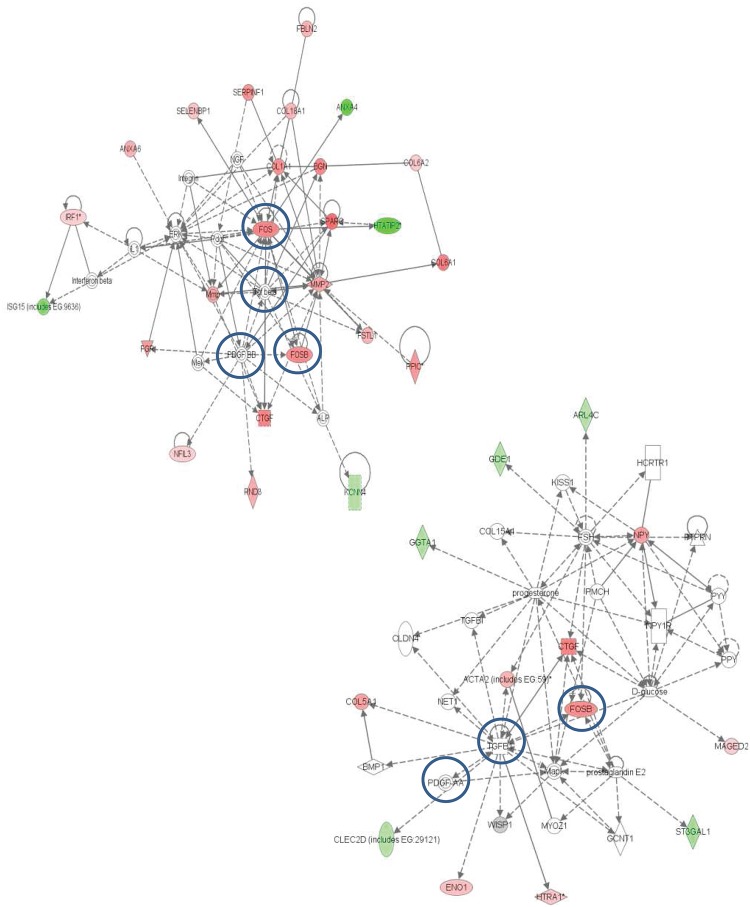
RNA from FCMs and NFMs (n = 4) was subjected to microarray analysis on the Illumina platform (GEO accession number GSE70126). Using an adjusted P-value of <0.01, we found only 29 differentially expressed genes. However, using a less stringent unadjusted P-value <0.01, a total of 749 genes appeared to be differentially expressed, with 369 up-regulated and 380 down-regulated. A detailed list can be found in S1 Table. Canonical signalling analysis revealed that the most enriched pathway in FCMs compared to NFMs was that related to LXR and its obligate partner, retinoid acid receptor (RXR) (Fig 2), confirming the results of a previous study comparing peritoneal FCMs and NFMs from LDL receptor null mice [23]. The second most over-enriched pathway was that previously associated with hepatic fibrosis (Fig 2). Interestingly, functions found to be enriched and/or up-regulated in FCM by Ingenuity Functional Pathway analysis (Table 3) also implicated connective tissue development as well as cell regulation, differentiation and proliferation.
Fig 2. Canonical pathway analysis.

The canonical pathways most enriched in FCMs were anti-inflammatory (LXR) and pro-fibrotic (derived from array data). Blue bars indicate significance, orange line indicates ratio. (Ingenuity Systems Inc).
Table 3. Functions enriched/regulated in FCMs by Ingenuity Functional Pathway analysis.
| Rank | Functions enriched/regulated in FCMs | Score |
|---|---|---|
| 1 | Dermatological diseases and conditions, Connective tissue development and function, Tissue morphology | 52 |
| 2 | Cancer, Cellular growth and proliferation, Nervous system development and function | 30 |
| 3 | Cardiovascular disease, Cellular growth and proliferation, Haematological system development and function | 28 |
| 4 | Cancer, Cellular movement, Cell death | 23 |
| 5 | Cell signalling, Molecular transport, Vitamin and mineral metabolism | 23 |
| 6 | Cellular growth and proliferation, Cancer, Cell death | 13 |
The 2 most statistically significant networks generated from the array data (Fig 3) pointed to a regulatory node around platelet derived growth factor (PDGF) and transforming growth factor-β (TGFβ), which have been previously implicated in tissue fibrosis [28]. Network analysis also suggested that FOS and FosB, which are components of the activator protein-1 (AP-1) transcription factor complex that is known to be downstream of PDGF and TGFβ activation [29], might be major contributors to the differences between FCMs and NFMs (Fig 3). The network analysis focused our attention on extracellular matrix protein and pro-fibrotic genes, including several collagen polypeptides, decorin, biglycan and bone morphogenic protein 1 (BMP1, a pro-collagen convertase), that were strongly over expressed in FCMs relative to NFMs. We also further investigated expression of connective tissue growth factor (CTGF), which is a major contributor to fibrosis; and proteases, including cathepsins and matrix metalloproteinases (MMPs), which have been implicated in plaque rupture [30].
Fig 3. Network maps of genes differentially regulated in FCM and NFM array (top 2 networks).
Molecules are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). Continuous lines represent direct interactions, while indirect ones are represented by interrupted lines. The intensity of the node colour indicates the degree of up- (red) or down- (green) regulation. Colour of node indicates the presence (grey) or absence (white) of a given gene in the study. No change in M1/M2 markers were observed, but there was a regulatory node around PDGF and TGFβ (which were absent from the array) (Ingenuity Systems Inc).
Comparison of array and RT-qPCR measurements of differentially expressed mRNAs
Despite the low stringency and hence potentially high false discovery rate of our initial analysis, most of the genes found to be differentially expressed in the array (Fig 4A) were validated by RT-qPCR, often with increased ratios (Fig 4B and Table 4). In addition, several candidate genes related to the identified pathways were found to be differentially expressed by RT-qPCR on greater numbers of preparations (Fig 4B and Table 4).
Fig 4. Genes differentially regulated in FCM and NFM.
A) array; B) RT-qPCR). Cyan = LXR-related genes, red = extracellular matrix structural proteins, yellow = fibrosis-related signalling and associated molecules, purple/blue = degradative enzymes/inhibitors, green = genes associated with M1 and M2 polarisation, peach = miscellaneous genes. Genes not present on the Illumina chip or not qPCR verified are left blank. *P<0.05.
Table 4. Levels of mRNAs by RT-qPCR in FCMs and NFMs (n = 5–7).
| Gene | FCM/NFM | FCMs (copies/ng RNA) | FCM SD | NFMs (copies/ng RNA) | NFM SD | P-value |
|---|---|---|---|---|---|---|
| 36B4 | 1.39 | 3564 | 683.1 | 2558 | 228.0 | 0.3434 |
| RXRβ | 1.98 | 5903 | 3100 | 2982 | 1173 | 0.0303 |
| RXRα | 2.08 | 167.1 | 59.10 | 80.25 | 32.69 | 0.0170 |
| LXRα | 4.10 | 23.37 | 18.822 | 5.703 | 2.298 | 0.0177 |
| LXRβ | 2.69 | 2.000 | 1.219 | 0.743 | 0.108 | 0.0478 |
| Srebp1 | 2.13 | 1332 | 712.8 | 624.3 | 174.7 | 0.0303 |
| FASN | 6.90 | 57.23 | 60.56 | 8.297 | 4.795 | 0.0182 |
| MSR1 | 17.5 | 37074 | 45725 | 2116 | 1002 | 0.0038 |
| CD36 | 2.47 | 1912 | 1890 | 774.8 | 880.3 | 0.4318 |
| SCARB1 | 2.92 | 150.7 | 117.6 | 51.51 | 25.46 | 0.1061 |
| ABCA1 | 1.48 | 44043 | 27951 | 29749 | 14210 | 0.3222 |
| PPARγ | 1.83 | 248580 | 95420 | 136141 | 30771 | >0.9999 |
| Col1α1 | 17.8 | 18252 | 14723 | 1024 | 708.9 | 0.0016 |
| Col4α1 | 11.1 | 347.1 | 304.9 | 31.39 | 18.82 | 0.0041 |
| Col6α1 | 19.7 | 1045 | 1194 | 52.95 | 41.89 | 0.0078 |
| Col8α1 | 84.4 | 3.529 | 4.392 | 0.0418 | 0.0494 | 0.0380 |
| BGN | 9.08 | 22551 | 21086 | 2484 | 1678 | 0.0185 |
| DCN | 8.30 | 9964 | 9985 | 1201 | 795 | 0.0482 |
| hCTGF | 63.5 | 3.951 | 2.933 | 0.0623 | 0.0637 | 0.0001 |
| BMP1 | 11.4 | 544680 | 473325 | 47913 | 20134 | 0.0081 |
| THBS1 | 0.29 | 23317 | 7268 | 81297 | 37646 | 0.0051 |
| TGFβ1 | 1.19 | 37.65 | 20.84 | 31.52 | 14.56 | 0.5855 |
| FOS | 13.7 | 29675 | 21365 | 2171 | 1416 | 0.0038 |
| FosB | 8.02 | 128.4 | 77.37 | 16.00 | 5.779 | 0.0001 |
| JUN | 3.09 | 24095 | 11387 | 7793 | 7144 | 0.0185 |
| JunB | 1.95 | 186454 | 63475 | 95703 | 51901 | 0.0255 |
| SMAD2 | 0.87 | 946 | 585 | 1091 | 350 | 0.6325 |
| SMAD3 | 0.82 | 132.5 | 100.0 | 161.1 | 105.1 | 0.6421 |
| ACVR1 | 1.21 | 84.40 | 52.27 | 69.62 | 25.40 | 0.5750 |
| TGFBR1 | 0.70 | 972 | 382 | 1392 | 1174 | 0.3101 |
| CTSC | 6.60 | 4003 | 2215 | 606.5 | 246.2 | 0.0033 |
| CTSE | 13.4 | 22.72 | 10.60 | 1.6993 | 1.1338 | <0.0001 |
| CSTB | 0.72 | 27327 | 11840 | 38125 | 14139 | 0.1805 |
| CSTF | 0.82 | 235.1 | 127.5 | 285.1 | 136.3 | 0.5296 |
| LYZ | 8.84 | 105455 | 82637 | 11936 | 14926 | 0.0177 |
| MMP2 | 9.82 | 654.4 | 500.6 | 66.62 | 39.62 | 0.0103 |
| MMP9 | 0.94 | 799.2 | 562.8 | 854.6 | 823.8 | 0.8920 |
| MMP12 | 1.07 | 13415 | 8590 | 12553 | 9439 | 0.8804 |
| MMP13 | 0.20 | 2330 | 1452 | 11368 | 4487 | 0.0007 |
| MMP14 | 0.75 | 35000 | 12138 | 46875 | 25061 | 0.4901 |
| MMP23 | 6.98 | 5109 | 3772 | 732.0 | 388.9 | 0.0080 |
| TIMP1 | 0.67 | 2037 | 1393 | 3023 | 1707 | 0.2955 |
| TIMP2 | 3.23 | 692.0 | 637.5 | 214.5 | 148.5 | 0.2677 |
| TIMP3 | 3.81 | 19.70 | 18.27 | 5.165 | 5.289 | 0.2677 |
| NOS2 | 1.27 | 13143 | 8979 | 10331 | 5791 | 0.5550 |
| IL6 | 0.33 | 443.6 | 467.4 | 1333 | 1419 | 0.2126 |
| CCL2 | 2.15 | 3.290 | 2.290 | 1.530 | 0.850 | 0.1061 |
| NF-κB1 | 0.41 | 978.9 | 405.6 | 2378 | 1248 | 0.0219 |
| ARG1 | 1.71 | 72449 | 55730 | 42307 | 45624 | 0.3862 |
| ARG2 | 0.63 | 6887 | 4015 | 10866 | 6745 | 0.2270 |
| YM1 | 0.63 | 1198 | 610.1 | 1905 | 891 | 0.1315 |
| CD206 | 2.22 | 34553 | 14498 | 15565 | 5164 | 0.5934 |
| RND3 | 8.43 | 104.32 | 54.87 | 12.38 | 6.575 | 0.0004 |
| SARAF | 2.88 | 996.1 | 710.6 | 345.3 | 147.0 | 0.0194 |
| Cmpk2 | 0.25 | 1265 | 895 | 5010 | 2654 | 0.0129 |
LXR-related genes
Genes associated with the LXR pathway that was identified by canonical signalling analysis were of special interest, given the previous data comparing peritoneal FCMs and NFMs from LDL receptor null mice [23]. RXRβ was increased 1.2-fold in the array (P = 0.0019, Fig 4A) and this was confirmed by RT-qPCR (2-fold, P = 0.0303) with over 1000 copies/ng RNA in FCMs (Fig 4B and Table 4). RXRα was also over expressed in FCMs (P = 0.0170) but these cells only had 160 copies/ng RNA. LXRα and LXRβ were over expressed in FCM (P = 0.0177 and P = 0.0478, respectively), although both had less than 25 copies/ng RNA. Looking at downstream genes, sterol regulatory element binding protein 1 (Srebp1) was increased in FCMs (P = 0.0303) and had over 1000 copies/ng RNA, whereas fatty acid synthase (FASN), also increased in FCMs (P = 0.0182), had less than 60 copies/ng RNA. Scavenger receptor class F member 2 (SCARF2) was 2-fold elevated in FCMs in the array (P = 0.0081) and macrophage scavenger receptor 1 (MSR1, scavenger receptor A1 (SR-A1)) was increased 17.5-fold in FCMs by RT-qPCR (P = 0.0038). The mRNAs for scavenger receptors CD36 and scavenger receptor class B, member 1 (SCARB1, SR-B1) (both associated with TGFβ [28,31–34]), although abundant in FCMs, were not significantly different from those in NFMs. There were no significant differences between FCMs and NFMs in ATP binding cassette transporter isoform A1 (ABCA1) or peroxisome proliferator-activated receptor-γ (PPARγ expression, although both FCMs and NFMs had very high expression levels of these genes (130000–250000 copies/ng RNA), as previously reported [10].
Extracellular matrix structural proteins
Probably the most unexpected finding from our experiments was that several collagen polypeptide genes, as well as core proteins of the proteoglycans, biglycan and decorin, and the matricellular protein CTGF, were 2 to 6.5-fold up-regulated in FCM compared with NFM in the array (Fig 4A). All of these genes are known targets for induction by PDGF and/or TGFβ [29,35]. The large increases in mRNAs for collagens 1, 4 and 6 were confirmed by RT-qPCR (11–20 fold increase in FCM, Fig 4B and Table 4). Collagen1α1 was the most abundant, with 18000 copies/ng RNA in FCMs and 1000 in NFMs (P = 0.0016, Table 4). FCMs had more than 63-fold more mRNA for CTGF than did NFM (P = 0.0001), also confirming the array result. The mRNAs for biglycan and decorin core proteins were also significantly increased and highly abundant, with 23000 or 100000 copies in FCM and only 2900 or 1200 in NFM, respectively (biglycan P = 0.0185, decorin P = 0.0482). The expression of BMP1, which is known to be up-regulated by TFGβ and potentially involved in the regulation of TGFβ activation [36], was considerably increased in FCM, with 540000 and 48000 copies/ng RNA in FCMs and NFMs, respectively (P = 0.0081; Fig 4B and Table 4). By contrast, thrombospondin 1 (THBS1), which is known to bind and activate TGFβ [36] was significantly reduced in FCM (P = 0.0051; Fig 4B), although both FCMs and NFMs had abundant THBS1 mRNA (23000 and 81000 copies/ng, respectively, Table 4).
Fibrosis-related signalling and associated molecules
Levels of TGFβ1 mRNA were similar in FCMs and NFMs (Fig 4B). However, the PDGF and TGFβ related activator protein-1 (AP-1) family members FOS and FosB were over expressed over 5-fold, whereas JUN and JunB were 1.5 and 2.4 fold higher, respectively, in FCMs compared to NFMs in the array (Fig 4A). Data from RT-qPCR (Fig 4B) confirmed that FCMs expressed more FOS, JUN and JunB than NFMs, with several thousand copies of each per ng RNA (Table 4). The level of FosB expression was lower than the other signalling molecules, but still over-expressed in FCMs compared with NFMs, with 130 copies/ng RNA in FCM and only 16 copies/ng RNA in NFM (P = 0.0001; Fig 4B). By contrast with the increases in AP-1 factors, signalling factors SMAD 2 and 3 that are activated by TGF family members by phosphorylation appeared to be down-regulated in FCMs in the array, as were the TGFβ receptors activin A receptor type 1 (ACVR1, ALK2) and TGFβ Receptor 1 (TGFBR1, ALK5) (Fig 4A). However, none of these small fold changes could be confirmed by RT-qPCR (Fig 4B).
Degradative enzymes
The most differentially expressed gene in the array was cathepsin C (CTSC), with a 20-fold change between the FCM and the NFM (P = 0.0000, Fig 4A). This difference was confirmed by RT-qPCR (Fig 4B) with 4000 and 600 copies in FCMs and NFMs, respectively (Table 4). Cathepsin E (CTSE) was also over expressed in FCMs (Fig 4A and 4B), although there were few copies of mRNA in either type of macrophage. In contrast, cystatins E and F were either not changed or slightly decreased in FCMs (Fig 4A and 4B and Table 4). Lysozyme (LYZ) was over-expressed (4-fold) in FCMs in the array (Fig 4A) and RT-qPCR (Fig 4B), where FCMs and NFMs contained 105000 and 12000 lysozyme copies/ng RNA, respectively (P = 0.0177, Table 4). Turning to the MMPs, we found that the gelatinase, MMP2, and the cysteine-rich MMP, MMP23, were over expressed in FCMs (650 or 5100 copies in FCM and only 67 or 730 in NFM, respectively). In contrast, for those MMPs that are related to M1 activation [24], there was no change in MMP9 (Table 4) and the abundant M1-related collagenase, MMP13, was down-regulated in FCMs (2.9-fold, P = 0.0302 in the array and 4.9 fold P = 0.0007 by RT-qPCR). The tissue inhibitor of MMPs, TIMP-2, appeared to be increased in FCMs compared with NFMs in the array, although this could not be confirmed using RT-qPCR (Fig 4A and 4B and Table 4).
Genes associated with M1 and M2 polarization
From the array, there were no differences in genes associated with M1 macrophage activation, such as inducible nitric oxide synthase (NOS2), IL-6, MMP9 or chemokine (C-C motif) ligand 2 (CCL2, MCP1). However, as mentioned above, MMP13 was decreased, and in the NF-κB pathway, which is implicated in M1 activation [37,38], NF-κB1 was also down-regulated in FCMs in the array (Fig 4A). Data from RT-qPCR (Fig 4B), confirmed that NOS2 expression was high in both FCM and NFM, 13000 and 10000 copies/ng RNA, respectively (P = 0.5550). CCL2 levels were also not significantly different in FCMs and NFMs (P = 0.1061), nor was MMP9 (Table 4). NF-κB1 was lower in FCM than in NFM, with 980 and 2400 copies/ng RNA in FCMs and NFMs, respectively (P = 0.0219, Fig 4B and Table 4), confirming the array data. There were no differences in M2 marker genes, Arg1, Ym1 and the mannose receptor (CD206) in the array (Fig 4A). By RT-qPCR, Arg1 expression in both FCMs and NFMs was high, with 72450 and 42300 copies/ng RNA, respectively (P = 0.3862). Although Arg2 levels (suggesting either M1 or M2c activation) were lower in the FCM, the differences did not reach significance (FCM 6900, NFM 11000 copies/ng RNA, respectively, P = 0.2270). Both FCMs and NFMs expressed the M2 markers Ym1 (1000–2000 copies/ng RNA) and CD206 (15000–35000 copies/ng RNA) at similar levels. Together these results indicate that FCMs and NFMs display similar levels of established markers for either M1 or M2 polarization.
Miscellaneous genes
Other genes that were confirmed as differentially regulated in the two set of macrophages were Rho family GTPase 3 (RND3), which regulates the organisation of the actin cytoskeleton in macrophages and other cells [39] and store-operated calcium entry-associated regulatory factor (SARAF, formerly Tmem66); both were elevated in FCMs (P = 0.0004 and P = 0.0194, respectively). Cytidine monophosphate (UMP-CMP) kinase 2 (Cmpk2), which is involved in the terminal differentiation of monocytic cells [40], was lower in FCMs (P = 0.0129, Fig 4B). Further details of the differences in the differentially expressed genes that were validated using RT-qPCR can be found in Table 4.
Immunohistochemical validation of selected findings from the array
Given that the two pathways most increased at the mRNA levels in FCMs compared to NFMs were the LXR and hepatic fibrosis pathways (Fig 2), we used immunohistochemistry to validate these findings in sections of subcutaneous sponges from ApoE null mice. We also took the opportunity, where possible, to compare plaque FCMs and adventitial NFMs in the same atherosclerotic plaques from the brachiocephalic artery of ApoE null mice.
LXRα
LXRα protein was identified in 52 ± 26% of cells (n = 5) in sections of sponges from ApoE null mice (Fig 5A) and in 53 ± 23% of the cells in plaques from brachiocephalic arteries from ApoE null mice (Fig 5B, 5B’ and 5C,). Cells with nuclear (pink) and cytoplasmic (red) staining were found throughout the plaque. Cells containing LXRα in their cytoplasm were most often found near the lumen, while those cells closest to the internal elastic lamina, deep within the plaque, tended to have less cytoplasmic LXRα (Fig 5B). Little cytoplasmic (and even less nuclear) staining was found in adventitial cells (Fig 5C), and no staining at all in IgG control sections (Fig 5D and 5E).
Fig 5. LXRα in sponges and arteries from fat-fed ApoE null mice.
LXRα is present in the cytoplasm (red) and/or nucleus (pink, arrows) of A) FCMs in sections from a subcutaneous sponge, or; B) cells in the plaque of a brachiocephalic artery. B’ higher magnification of plaque in B. C) LXRα is occasionally present in the cytoplasm of the adventitial cells that are close to the media; D) sponge section negative control (only the sponge spicule is red); E) negative control in a section from the same plaque as B. Blue = nuclei (DAPI), green = autofluorescence. Magnification x 400 A-E, x 1000 B’.
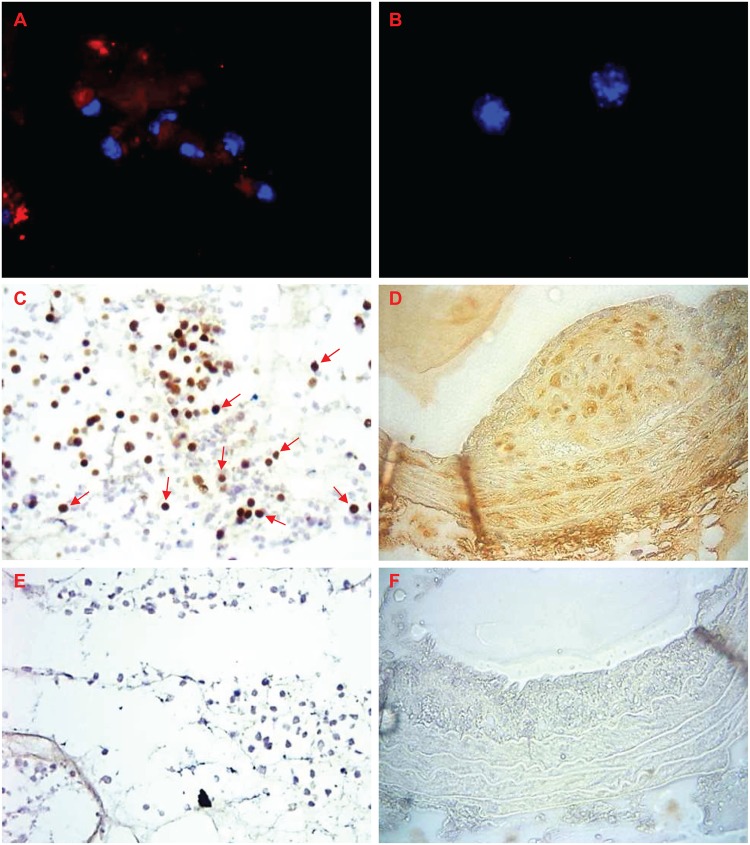
CTGF
Most FCMs but very few NFMs isolated from sponges stained for CTGF (Fig 6A and 6B; red staining), which supports the mRNA data. Of cells in sponge sections from fat-fed ApoE null mice, 49 ± 22% (n = 10) stained for CTGF (arrows in Fig 6C). FCMs in brachiocephalic artery plaque sections also had strong staining for CTGF (Fig 6D). SMC in the media also stained for CTGF, as did the extracellular matrix of the adventitia, which obscured any staining by adventitial cells. Approximately 90–95% (n = 6–7) of cells in the plaque, media and adventitia stained for CTGF protein, intracellularly or in the surrounding matrix. This data therefore confirmed the over expression of CTGF in sponge and plaque FCMs, and suggested that there might be reduced staining from NFMs, at least in in the sponges.
Fig 6. CTGF in sponges and arteries from mice.
CTGF (red) is present in A) in FCMs, but not B) NFMs isolated from sponges. Blue = nuclei (DAPI). CTGF (brown) is present in C) FCMs in sponge sections or; D) throughout the plaque, media and adventitia of a brachiocephalic artery from a fat-fed ApoE null mouse. E) sponge section negative control; F) negative control in a section from a brachiocephalic artery plaque. Magnification x 200 (C, E), x 400 (A, B, D, F).
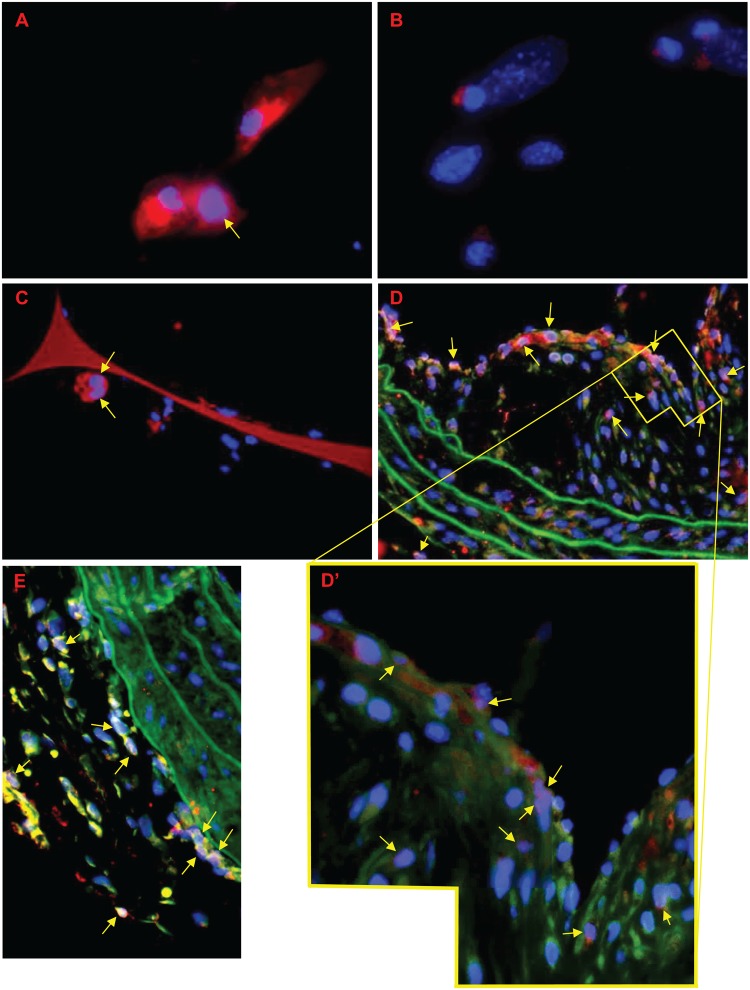
cFOS
FCMs isolated from sponges had prominent cFOS immunostaining in their cytoplasm (red), with a third of cells also expressing cFOS in their nucleus (pink) (Fig 7A). Staining was less pronounced in isolated NFMs (Fig 7B), and was only observed in a few nuclei. Staining for cFOS was also observed of FCMs in subcutaneous sponge granulomas (Fig 7C), with 42 ± 22% of cells (n = 5) having nuclear cFOS staining (pink), and many having cFOS staining (red) in their cytoplasm. 43–44% of cells within the BCA plaques (n = 2) also expressed nuclear (pink) cFOS, with many also having cytoplasmic cFOS staining (Fig 7D and 7D’). A similar proportion, 45–55%, of adventitial cells (from their shape possibly fibroblasts) also had nuclear cFOS staining (Fig 7D and 7E). Overall, almost half of all cells in the plaque, adventitia or sponge sections had nuclear cFOS staining. Clearly, at least some plaque FCMs were cFOS positive, which confirms the array and RT-qPCR findings.
Fig 7. cFOS in sponge and artery macrophages from mice.
cFOS is present in the cytoplasm (red) or nucleus (pink, arrows) in isolated macrophages from A) mice fed a high-fat diet (FCMs) or B) a normal diet (NFMs). cFOS was also observed in the cytoplasm (red, orange, yellow) and/or nucleus (pink, arrows) of cells in sections from C) a subcutaneous sponge granuloma or D, E) a brachiocephalic artery from a fat-fed ApoE null mouse. D’) higher magnification of plaque in D. Blue = nuclei (DAPI), green = autofluorescence. Magnification x 400 A-E, x 1000 D’. See Fig 5C and 5D for negative control staining.
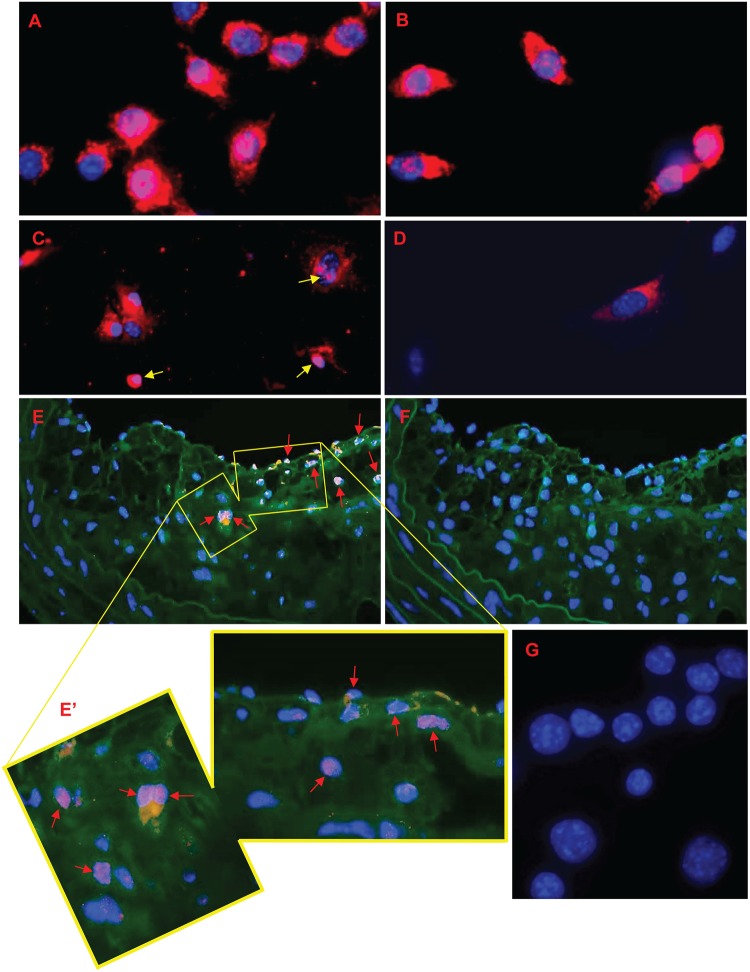
Role of TGFβ1 and activation of SMAD2 signalling in FCMs
Members of the TGF family signal through phosphorylation and nuclear translocation of SMADs, especially SMAD2. Hence we hypothesised that FCMs in sponges and plaques might contain increased levels of nuclear pSMAD2 detectable by immunohisto/cytochemistry. As a positive control, we first showed that TFGβ1 quickly stimulated SMAD2 and SMAD3 phosphorylation and translocation to the nucleus by 45 minutes in mouse RAW cells (Fig 8A and 8B and Table B in S1 File). Staining for nuclear pSMAD2 (pink) was found in 50 ± 12% of isolated FCMs; many also had cytoplasmic staining (red). By contrast, nuclear pSMAD2 staining was detected in only 10 ± 10% of NFM (n = 3, P = 0.0110, Fig 8C and 8D). Furthermore, 74–76% of the FCMs in brachiocephalic artery plaques had pSMAD2 present in their nuclei (pink), with many also having pSMAD2 in their cytoplasm (orange) (Fig 8E and 8E’). No staining was observed in an IgG control section from the same plaque or in IgG control RAW cells (Fig 8F and 8G). This provides strong evidence for SMAD2 signalling in FCMs in sponges and plaques.
Fig 8. Phosphorylated SMAD proteins in RAW 264.7 cells and sponge or artery macrophages from mice.
RAW cells were exposed to 10 ng/ml TGFβ1 for 45 minutes. A) phospho-SMAD2 and B) phospho-SMAD3 were present in the cytoplasm and nucleus of many cells. pSMAD2 was also found in C) the cytoplasm and nucleus (arrows) from isolated FCMs, but only in the D) cytoplasm of NFMs. E) pSMAD is present in the cytoplasm and nuclei (arrows) of plaques in a brachiocephalic artery a fat-fed ApoE null mouse. E’) higher magnification of plaque in E. F) negative control in a section from the same plaque. G) RAW negative control staining (rabbit IgG). Red/orange = cytoplasmic staining, pink = nuclear staining, blue = nuclei (DAPI), green = autofluorescence. A, B, E’, G magnification x 1000, C, D, E, F magnification x 400.
Discussion
Main findings
Our experiments document novel differences between the transcriptomes of FCMs and NFMs obtained from subcutaneous sponge-induced granulomas in living ApoE null mice. The most significant changes were in the LXR/RXR pathway, which concurs with a study of peritoneal FCMs and NFMs in LDL receptor null mice [23]. We also observed significant increases in fibrosis-related gene expression in FCMs, including several collagen polypeptides and proteoglycan core proteins. Furthermore, we observed elevated levels of FOS and JUN transcription factors that are associated with pro-fibrotic actions [41] and phosphorylation of the SMAD2 transcription factor that mediates actions of TGF family members [42]. FCMs from subcutaneous sponges from ApoE null mice were polarized neither towards the M1 (classically activated) nor M2 (alternatively activated) phenotypes. Our results demonstrate that FCMs formed in vivo adopt a pro-fibrotic phenotype.
LXR/RXR pathway
LXRs act by heterodimer formation with RXRs and subsequent binding of the complex with LXR response elements in target genes. They can also inhibit expression of other genes by antagonising the activity of transcription factors or by preventing the release of co-repressor complexes from target-gene promoters [43]. We found that FCMs had increased expression of LXRα and LXRβ, as well of their binding partners RXRα and RXRβ, as well as several down-stream mediators (e.g. Srepb1, FASN). Similar observations were reported previously in peritoneal macrophages isolated from fat-fed LDL receptor null mice [23]; and loading of mouse BMDM with acetylated low density lipoprotein (LDL) in vitro also upregulates several LXR-related genes [44]. On the other hand, loading with oxidised- (ox-) LDL for 24 hours down-regulates MSR1, FASN and SCARB1 but upregulates CD36 and ABCA1, most likely by modulating transcription by ATF3 [45]. Such disparities dependent on both the type of lipids used and the phenotypic state of the mouse macrophages before loading have been previously reviewed [46].
The pro-fibrotic response of FCMs
In addition to lipid-related genes, we found that the mRNAs of many extracellular matrix proteins were up-regulated in FCMs. These included mRNAs for collagens 1, 4, 5, 6, 8, 16 and 18 as well as the pro-collagen convertase, BMP-1. Furthermore, mRNAs for the core proteins of biglycan, decorin and versican, which are proteoglycans that bind to collagen and promote matrix assembly and maturation, were elevated in FCMs (Table 4 and S1 Table). All of these matrix proteins are present at high levels within plaques [47–49] but have been thought of as products of SMCs or fibroblasts [35]. Our new data suggests that FCMs contribute to their own surrounding extracellular matrix. Interestingly, FCMs recovered from the peritoneum of LDL receptor null mice also overexpress collagen 1α2, collagen 3α1, collagen 6α1 and decorin by 2–4 fold compared to NFMs (Supplementary Table IC of [23]). Hence the pro-fibrotic transformation we observed in cells from subcutaneous granulomas in ApoE mice is replicated in the LDL receptor null background and at another site. On the other hand, several transcriptomic studies of mouse BMDM loaded with lipids for short periods in vitro did not note any changes in fibrosis-related genes [44,45,50], which suggests that such changes may evolve slowly or depend on the local microenvironment in vivo.
Collagens 1 and 3 are believed to be important in stabilizing plaques against rupture [51]. In contrast, collagen 8 coincides with active remodelling, migration and increased production of some MMPs [52,53]. Biglycan and versican are known to trap lipid within the matrix and hence perpetuate lesion progression leading ultimately to instability [48,49,54]. The impact of fibrotic transformation of FCMs could therefore be beneficial for stability through collagen synthesis but promote plaque progression through proteoglycans.
Canonical pathway analysis and network maps suggested PDGF and TGFβ as a regulatory node in the gene expression changes in FCMs compared to NFMs (Fig 3). At first sight this seems counter-intuitive, because TGFβ has been shown to reduce foam cell formation in vitro by decreasing scavenger receptor mRNA expression and oxidised LDL uptake [4]. However, much previous literature establishes that TGFβ can stimulate collagen synthesis in the plaques [29,35], as well as during the development of restenosis [55]. Proteolytic activation of latent TGFβ, rather than increased expression, is often responsible for increased TGFβ activity [29]. Consistent with this, we found that TGFβ1 mRNA was not increased in FCMs compared with NFMs; nor was the TGFβ1 receptor, ALK5. However, MMP2 was increased; and BMP1, which can also activate TGFβ, was up-regulated 11-fold in FCMs compared with NFMs. CTGF (CCN2), a multi-functional growth factor that is up-regulated by TGFβ via AP-1 and SMADs [28,56,57] was also increased in FCMs. CTGF may act synergistically with TGFβ [28,56] and can promote monocyte migration into the atherosclerotic plaque [58]. To validate our differential gene expression data in vivo, we showed that CTGF staining in both FCM- and SMC-rich areas of mouse plaques (Fig 6D). CTGF staining has also been reported in human atherosclerotic plaques, particularly near areas associated with large numbers of macrophages e.g. the shoulder region and surrounding the lipid core [58], and alveolar macrophages examined in vivo have been shown to express CTGF [59]. On the other hand, thrombospondin-1, which is also implicated in the binding and activation of TGFβ [31], was significantly decreased in FCMs.
TGFβ signalling is mediated by phosphorylation of SMADs, and by up-regulation of FOS, JUN and LXR transcription factors [28,34,36,55,60,61]. Consistent with this, we found that although the mRNA expression of SMADs 2 and 3 were not elevated in FCMs, levels of phosphorylated, nuclear localised SMAD2 were increased in granuloma FCMs relative to NFMs and in plaque FCMs, suggesting that TGFβ1 signalling was occurring. SMAD2 staining has been previously associated with FCMs in fibro-fatty lesions [60]. FCMs also had elevated expression of FosB, JUN and JunB. Interestingly, other factors that are affected by the presence of FosB, such as the extracellular matrix protein tenascin C [57], were also significantly up-regulated in our FCMs (S1 Table). We also confirmed that mouse FCMs overexpressed cFOS, with protein expression often found in the nucleus.
Proteinases
Cathepsins C and E were overexpressed in in vivo generated FCMs in our study and also in mouse BMDM loaded with acetylated LDL in vitro [44]. However, these are acidic proteases, and only cathepsins with activity at neutral pH have been directly implicated in destabilization of plaques [62,63]. We found increased expression of MMP2, which promotes SMC migration and proliferation [64]. MMP2 is also known to activate latent TGFβ [65] and release TGFβ from extracellular matrix stores [55], which could further contribute to a pro-fibrotic action. MMP9, which also promotes migration of SMC [64], was not changed in our study, despite data from peritoneal FCMs showing a decrease [23]. Instead, we observed decreased expression of MMP13, which is the main collagenase of mouse atherosclerosis [66]. On the other hand, the expression of MMP23, which has not been studied in the context of atherosclerosis, was increased.
Polarization towards M1 or M2 phenotypes
Peritoneal FCMs appeared to be polarized away from M1 compared to NFMs in LDL receptor null mice [23]. Characteristic M1 genes, including IL1β and MMP9, were down-regulated and, moreover, peritoneal FCMs were resistant to the M1 polarizing effects of added toll-like receptor ligands ex vivo, in part because accumulation of desmosterol led to ligation of LXR, which stabilized the co-repressor complex, NcoR [23]. In contrast with these results, we did not observe a significant decrease in M1 marker genes, including MMP9, NOS2, CCL2, IL-6 and ARG2, in granuloma FCMs compared with NFMs, although MMP13 and NF-κB1 levels, which are also M1-related genes [24,37,38], were decreased. There could be several reasons for this discrepancy, including different diets and background strains in the two studies and the metabolic consequences of ApoE compared with LDL receptor knockout. Another difference is that we took FCMs from subcutaneous granulomas rather than the peritoneum, and the cells therefore experienced a different inflammatory environment in vivo. NFMs from the peritoneum are known to be strongly polarized towards M2 [67], whereas, given the foreign body reaction in granulomas, it is not surprising that we measured considerable levels of M1 markers in both NFMs and FCMs (Table 4). Our previous work on subcutaneous granulomas in lipid- or chow-fed, wild-type rabbits also demonstrated the presence of M1 markers, but these were greater in FCMs than NFMs. FCMs had greater activation of NF-κB and up-regulation of the NF-κB dependent genes, MMP1 and MMP3; they also had decreased expression of arginase-1 and increased nitrite production compared with NFMs [21,22], confirming polarization away from M2. In the only available study of human plaque cells examined ex vivo, cells including FCMs also expressed M1 genes, and this was dependent on toll-like receptor-2 (TLR2) stimulation [68]. Hence M1/M2 polarisation appears to depend crucially on the microenvironment from which the FCMs are obtained. FCMs appear polarised towards M1 when extracted from human plaques [68] and rabbit granulomas [21,22], towards M2 when extracted from the peritoneum of LDL receptor null mice [23] or neither M1 nor M2 when extracted from mouse subcutaneous granulomas in this study. Consistent with this, FCMs bearing M1 markers, M2 markers or neither are detected in mouse plaques by histology [69] and FCMs observed histologically in human plaques also demonstrate a wide variety of phenotypes [9]. Many FCMs appear to be M1, based on nuclear NF-κB localisation and expression of marker genes such as NOS2 and COX2 [8,19]. Laser capture dissected plaque macrophages also show up-regulation of several M1-related genes, including NOS2, Arg2, TLR2 and IL1r1 [70]. However, FCMs bearing M2 markers such as CD206 and PPARγ [19] are also present, although they tend to be less foamy, probably thanks to up-regulation of reverse transporters [13]. In plaques, FCMs may also ingest particles other than LDLs, including bacteria, apoptotic bodies and cholesterol crystals that can cause additional polarization towards M1 [71,72], or take up haem and become deactivated [16].
A similar disparity of findings has been obtained in in vitro studies, depending on the type of lipid used (e.g. minimally compared to extensively oxidised LDL, acetylated LDL, oxidised phospholipids or cholesterol, cholesterol crystals) and the phenotypic state of the mouse macrophages before loading (reviewed by Adamson and Leitinger [46]). For example, ox-LDL loading of M2 macrophages generates a pro-inflammatory state [50], but oxidised phospholipid treatment leads to a distinct, anti-oxidant state [69]. Lipid ligands of PPARγ can prime macrophages towards an anti-inflammatory state [13]. In agreement with this diversity of responses, one study of ox-LDL loaded human monocyte derived macrophages observed overexpression of M1 genes thanks to toll-like receptor activation [20] but another found little effect on M1 or M2 markers, MMP-14 or TIMP-3, at least at the mRNA level [73], consistent with our present findings.
Limitations of our study
Since FCMs and NFMs were both harvested from subcutaneous sponges in our study, the foreign body response to the sponge itself could not obscure the transcriptomic differences, including up-regulation of pro-fibrotic genes, which we observed in FCMs. However, we do not yet know how significantly these contribute to plaque progression and stability in man. Supporting the relevance of our present data in mice, formation of human FCMs in vitro has been previously shown to up-regulate expression of many of the same genes, including CTSC, LXR, Cmpk2, and fibrotic genes [14,17,18,40,74]. Studies in human and mouse plaques, including our own reported here, also corroborate the findings from isolated FCMs studied ex vivo, but these have so far relied almost entirely on immunohistochemistry. In the future, it would be desirable to confirm them with other methods such as transcriptomics of laser capture dissected plaque cells. However, this will not be an easy task, owing to the limited amounts and quality of extracted RNA, the difficulties of extracting FCMs and NFMs from the same microenvironments, and also of distinguishing macrophages from smooth muscle cells based on CD68 staining [6].
In addition, although our differential gene analysis implicated pro-fibrotic signalling pathways related to PDGF and TGFβ, their precise roles will require further verification. Macrophage-selective knockout of individual pro-fibrotic mediators and their receptors would shed additional light but they are clearly beyond the present scope of this study. The translational potential of our studies also needs further consideration. It will be interesting to investigate the roles of the pro-fibrotic genes we identified in mice, using bio-bank and genetic approaches in man. From a treatment perspective, selective oestrogen receptor modulators such as tamoxifen that are known to increase active TGFβ levels in patients appear to stabilize plaques and reduce acute coronary syndromes [75,76]. Based on our findings, further approaches, including targeting production of FOS and JUN transcription factors might represent alternative strategies.
Implications for plaque rupture
Our novel data show conclusively that FCMs overexpress mRNAs for collagen and other matrix proteins that would tend to stabilize plaques. At first sight, this conclusion is paradoxical and needs to be reconciled with the more commonly-held view that FCMs overexpress matrix-degrading enzymes and therefore promote collagen degradation and plaque rupture. However, it is worth remembering that arterial fatty streaks and other xanthomas that contain few VSMCs or fibroblasts neither rupture nor cause thrombosis. Our conclusion is, therefore, that formation of FCMs is intrinsically pro-fibrotic and this may be necessary to stabilize early lesions. FCMs in more advanced plaques, for example at the vulnerable shoulder regions of plaques, are exposed to locally-acting inflammatory stimuli, which initiate other transcriptional programmes that tip the balance from collagen synthesis to degradation and therefore promote plaque rupture.
Supporting Information
(DOCX)
(XLS)
Acknowledgments
The authors would like to express their appreciation of expert assistance from Drs Elaine Mo Hayes, Stephen White, Jason Johnson, Graciela Sala-Newby, Ray Bush and Chris Jackson, and Mrs Laura Bevan.
Data Availability
All relevant data are available via the Gene Expression Omnibus (GEO) under accession number GSE70126.
Funding Statement
This work was supported by the British Heart Foundation and the NIHR Bristol Cardiovascular Biomedical Research Unit (ACT and ACN). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Ross R. Atherosclerosis-an inflammatory disease. N Engl J Med 1999;340: 115–126. [DOI] [PubMed] [Google Scholar]
- 2. Thomas AH, Edelman ER, Stultz CM. Collagen fragments modulate innate immunity. Exp Biol Med 2007;232: 406–411. [PubMed] [Google Scholar]
- 3. Ley K, Miller YI, Hedrick CC. Monocyte and macrophage dynamics during atherogenesis. Arterioscler Thromb Vasc Biol 2011;31: 1506–1516. 10.1161/ATVBAHA.110.221127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McLaren JE, Michael DR, Ashlin TG, Ramji DP. Cytokines, macrophage lipid metabolism and foam cells: Implications for cardiovascular disease therapy. Prog Lipid Res 2011;50: 331–347. 10.1016/j.plipres.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 5. Cookson FB. The origin of foam cells in atherosclerosis. Br J Exp Pathol 1971;52: 62–69. [PMC free article] [PubMed] [Google Scholar]
- 6. Allahverdian S, Chehroudi AC, McManus BM, Abraham T, Francis GA. Contribution of intimal smooth muscle cells to cholesterol accumulation and macrophage-like cells in human atherosclerosis. Circulation 2014;129: 1551–1559. 10.1161/CIRCULATIONAHA.113.005015 [DOI] [PubMed] [Google Scholar]
- 7. Galkina E, Ley K. Immune and inflammatory mechanisms of atherosclerosis. Annu Rev Immunol 2009;27: 165–197. 10.1146/annurev.immunol.021908.132620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang WC, Sala-Newby GB, Susana A, Johnson JL, Newby AC. Classical macrophage activation up-regulates several matrix metalloproteinases through mitogen activated protein kinases and nuclear factor-κB. PLoS ONE 2012;7: e42507 10.1371/journal.pone.0042507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stoger JL, Gijbels MJ, van der Velden S, Manca M, van der Loos CM, Biessen EA, et al. Distribution of macrophage polarization markers in human atherosclerosis. Atherosclerosis 2012;225: 461–468. 10.1016/j.atherosclerosis.2012.09.013 [DOI] [PubMed] [Google Scholar]
- 10. Shashkin P, Dragulev B, Ley K. Macrophage differentiation to foam cells. Curr Pharmaceut Design 2005;11: 3061–3072. [DOI] [PubMed] [Google Scholar]
- 11. Suzuki M, Becker L, Pritchard DK, Gharib SA, Wijsman E, Bammler TK, et al. Cholesterol accumulation regulates expression of macrophage proteins implicated in proteolysis and complement activation. Arterioscler Thromb Vasc Biol 2012;32: 2910–2918. 10.1161/ATVBAHA.112.300383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li ZY, Howarth S, Trivedi RA, U-King-Im JM, Graves MJ, Brown A, et al. Stress analysis of carotid plaque rupture based on in vivo high resolution MRI. J Biomech 2006;39: 2611–2622. [DOI] [PubMed] [Google Scholar]
- 13. Chinetti-Gbaguidi G, Baron M, Bouhlel MA, Vanhoutte J, Copin C, Sebti Y, et al. Human atherosclerotic plaque alternative macrophages display low cholesterol handling but high phagocytosis because of distinct activities of the PPARγ and LXRα pathways. Circ Res 2011;108: 985–995. 10.1161/CIRCRESAHA.110.233775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolfs IM, Donners MM, de Winther MP. Differentiation factors and cytokines in the atherosclerotic plaque micro-environment as a trigger for macrophage polarisation. Thromb Haemost 2011;106: 763–771. 10.1160/TH11-05-0320 [DOI] [PubMed] [Google Scholar]
- 15. Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TR, Reedquist KA, et al. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J Immunol Methods 2012;375: 196–206. 10.1016/j.jim.2011.10.013 [DOI] [PubMed] [Google Scholar]
- 16. Boyle JJ. Heme and haemoglobin direct macrophage Mhem phenotype and counter foam cell formation in areas of intraplaque haemorrhage. Curr Opin Lipidol 2012;23: 453–461. [DOI] [PubMed] [Google Scholar]
- 17. Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med 2003;9: 213–219. [DOI] [PubMed] [Google Scholar]
- 18. Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. J Immunol 2006;177: 7303–7311. [DOI] [PubMed] [Google Scholar]
- 19. Bouhlel MA, Derudas B, Rigamonti E, Dievart R, Brozek J, Haulon S, et al. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metabol 2007;6: 137–143. [DOI] [PubMed] [Google Scholar]
- 20. Lundberg AM, Yan ZQ. Innate immune recognition receptors and damage-associated molecular patterns in plaque inflammation. Curr Opin Lipidol 2011;22: 343–349. [DOI] [PubMed] [Google Scholar]
- 21. Chase AJ, Bond M, Crook MF, Newby AC. Role of nuclear factor-κB activation in metalloproteinase-1, -3, and -9 secretion by human macrophages in vitro and rabbit foam cells produced in vivo. Arterioscler Thromb Vasc Biol 2002;22: 765–771. [DOI] [PubMed] [Google Scholar]
- 22. Thomas AC, Sala-Newby GB, Ismail Y, Johnson JL, Pasterkamp G, Newby AC. Genomics of foam cells and nonfoamy macrophages from rabbits identifies arginase-I as a differential regulator of nitric oxide production. Arterioscler Thromb Vasc Biol 2007;27: 571–577. [DOI] [PubMed] [Google Scholar]
- 23. Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell 2012;151: 138–152. 10.1016/j.cell.2012.06.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hayes EM, Tsaousi A, Di Gregoli K, Jenkinson SR, Bond AR, Johnson JL, et al. Classical and alternative activation and metalloproteinase expression occurs in foam cell macrophages in male and female ApoE null mice in the absence of T- and B-lymphocytes. Front Immunol 2014;5: 537 10.3389/fimmu.2014.00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3: Article3 [DOI] [PubMed] [Google Scholar]
- 26. Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res 2008;36: e11 10.1093/nar/gkm1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE. The phenotype of murine wound macrophages. J Leukoc Biol 2010;87: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leask A, Abraham DJ. TGF-β signaling and the fibrotic response. FASEB J 2004;18: 816–827. [DOI] [PubMed] [Google Scholar]
- 29. Border WA, Noble NA. Transforming growth factor β in tissue fibrosis. N Engl J Med 1994;331: 1286–1292. [DOI] [PubMed] [Google Scholar]
- 30. Newby AC. Proteinases and plaque rupture: Unblocking the road to translation. Curr Opin Lipidol 2014;25: 358–366. [DOI] [PubMed] [Google Scholar]
- 31. Yehualaeshet T, O'Connor R, Green-Johnson J, Mai S, Silverstein R, Murphy-Ullrich JE, et al. Activation of rat alveolar macrophage-derived latent transforming growth factor β-1 by plasmin requires interaction with thrombospondin-1 and its cell surface receptor, CD36. Am J Pathol 1999;155: 841–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Draude G, Lorenz RL. TGF-β1 downregulates CD36 and scavenger receptor A but upregulates LOX-1 in human macrophages. Am J Physiol Heart Circ Physiol 2000;278: H1042–H1048. [DOI] [PubMed] [Google Scholar]
- 33. Zuckerman SH, Panousis C, Evans G. TGF-β reduced binding of high-density lipoproteins in murine macrophages and macrophage-derived foam cells. Atherosclerosis 2001;155: 79–85. [DOI] [PubMed] [Google Scholar]
- 34. Hu YW, Wang Q, Ma X, Li XX, Liu XH, Xiao J, et al. TGF-β1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor α signaling pathway in THP-1 macrophage-derived foam cells. J Atheroscler Thromb 2010;17: 493–502. [DOI] [PubMed] [Google Scholar]
- 35. Rekhter MD. Collagen synthesis in atherosclerosis: Too much and not enough. Cardiovasc Res 1999;41: 376–384. [DOI] [PubMed] [Google Scholar]
- 36. ten Dijke P, Arthur HM. Extracellular control of TGFβ signalling in vascular development and disease. Nat Rev Mol Cell Biol 2007;8: 857–869. [DOI] [PubMed] [Google Scholar]
- 37. de Winther MP, Kanters E, Kraal G, Hofker MH. Nuclear factor κB signaling in atherogenesis. Arterioscler Thromb Vasc Biol 2005;25: 904–914. [DOI] [PubMed] [Google Scholar]
- 38. Martinez FO. Regulators of macrophage activation. Eur J Immunol 2011;41: 1531–1534. 10.1002/eji.201141670 [DOI] [PubMed] [Google Scholar]
- 39. Guasch RM, Scambler P, Jones GE, Ridley AJ. RhoE regulates actin cytoskeleton organization and cell migration. Mol Cell Biol 1998;18: 4761–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu Y, Johansson M, Karlsson A. Human UMP-CMP kinase 2, a novel nucleoside monophosphate kinase localized in mitochondria. J Biol Chem 2008;283: 1563–1571. [DOI] [PubMed] [Google Scholar]
- 41. Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol 1997;9: 240–246. [DOI] [PubMed] [Google Scholar]
- 42. Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003;425: 577–584. [DOI] [PubMed] [Google Scholar]
- 43. Pourcet B, Feig JE, Vengrenyuk Y, Hobbs AJ, Kepka-Lenhart D, Garabedian MJ, et al. LXRα regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ Res 2011;109: 492–501. 10.1161/CIRCRESAHA.111.241810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berisha SZ, Hsu J, Robinet P, Smith JD. Transcriptome analysis of genes regulated by cholesterol loading in two strains of mouse macrophages associates lysosome pathway and ER stress response with atherosclerosis susceptibility. PLoS ONE 2013;8: e65003 10.1371/journal.pone.0065003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gold ES, Ramsey SA, Sartain MJ, Selinummi J, Podolsky I, Rodriguez DJ, et al. ATF3 protects against atherosclerosis by suppressing 25-hydroxycholesterol-induced lipid body formation. J Exp Med 2012;209: 807–817. 10.1084/jem.20111202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Adamson S, Leitinger N. Phenotypic modulation of macrophages in response to plaque lipids. Curr Opin Lipidol 2011;22: 335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Merrilees M, Beaumont B, Scott L, Hermanutz V, Fennessy P. Effect of TGF-β1 antisense S-oligonucleotide on synthesis and accumulation of matrix proteoglycans in balloon catheter-injured neointima of rabbit carotid arteries. J Vasc Res 2000;37: 50–60. [DOI] [PubMed] [Google Scholar]
- 48. Wight TN, Merrilees MJ. Proteoglycans in atherosclerosis and restenosis: Key roles for versican. Circ Res 2004;94: 1158–1167. [DOI] [PubMed] [Google Scholar]
- 49. Merrilees MJ, Beaumont BW, Braun KR, Thomas AC, Kang I, Hinek A, et al. Neointima formed by arterial smooth muscle cells expressing versican variant V3 is resistant to lipid and macrophage accumulation. Arterioscler Thromb Vasc Biol 2011;31: 1309–1316. 10.1161/ATVBAHA.111.225573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Tits LJH, Stienstra R, van Lent PL, Netea MG, Joosten LAB, Stalenhoef AFH. Oxidized LDL enhances pro-inflammatory responses of alternatively activated M2 macrophages: A crucial role for Kruppel-like factor 2. Atherosclerosis 2011;214: 345–349. 10.1016/j.atherosclerosis.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 51. Libby P. Collagenases and cracks in the plaque. J Clin Invest 2013;123: 3201–3203. 10.1172/JCI67526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Weitkamp B, Cullen P, Plenz G, Robenek H, Rauterberg J. Human macrophages synthesize type VIII collagen in vitro and in the atherosclerotic plaque. FASEB J 1999;13: 1445–1457. [DOI] [PubMed] [Google Scholar]
- 53. Adiguzel E, Ahmad PJ, Franco C, Bendeck MP. Collagens in the progression and complications of atherosclerosis. Vasc Med 2009;14: 73–89. 10.1177/1358863X08094801 [DOI] [PubMed] [Google Scholar]
- 54. Bobik A, Agrotis A, Kanellakis P, Dilley R, Krushinsky A, Smirnov V, et al. Distinct patterns of transforming growth factor-β isoform and receptor expression in human atherosclerotic lesions. Colocalization implicates TGF-β in fibrofatty lesion development. Circulation 1999;99: 2883–2891. [DOI] [PubMed] [Google Scholar]
- 55. Suwanabol PA, Kent KC, Liu B. TGF-β and restenosis revisited: A Smad link. J Surg Res 2011;167: 287–297. 10.1016/j.jss.2010.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Moritani NH, Kubota S, Eguchi T, Fukunaga T, Yamashiro T, Takano-Yamamoto T, et al. Interaction of AP-1 and the ctgf gene: A possible driver of chondrocyte hypertrophy in growth cartilage. J Bone Miner Metab 2003;21: 205–210. [DOI] [PubMed] [Google Scholar]
- 57. Ramachandran A, Gong EM, Pelton K, Ranpura SA, Mulone M, Seth A, et al. FosB regulates stretch-induced expression of extracellular matrix proteins in smooth muscle. Am J Pathol 2011;179: 2977–2989. 10.1016/j.ajpath.2011.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cicha I, Yilmaz A, Klein M, Raithel D, Brigstock DR, Daniel WG, et al. Connective tissue growth factor is overexpressed in complicated atherosclerotic plaques and induces mononuclear cell chemotaxis in vitro. Arterioscler Thromb Vasc Biol 2005;25: 1008–1013. [DOI] [PubMed] [Google Scholar]
- 59. Xifteri A, Drakopanagiotakis F, Tzouvelekis A, Tsiambas E, Karameris A, Tsakanika K, et al. Expression of CTGF and TNFa in alveolar macrophages of patients with idiopathic pulmonary fibrosis before and after treatment with azathioprine or interferon-γ-1β. Pneumon 2011;24: 149–156. [Google Scholar]
- 60. Kalinina N, Agrotis A, Antropova Y, Ilyinskaya O, Smirnov V, Tararak E, et al. Smad expression in human atherosclerotic lesions. Evidence for impaired TGF-β/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol 2004;24: 1391–1396. [DOI] [PubMed] [Google Scholar]
- 61. Chen G, Chen X, Sukumar A, Gao B, Curley J, Schnaper HW, et al. TGFβ receptor I transactivation mediates stretch-induced Pak1 activation and CTGF upregulation in mesangial cells. J Cell Sci 2013;126: 3697–3712. 10.1242/jcs.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Rodgers KJ, Watkins DJ, Miller AL, Chan PY, Karanam S, Brissette WH, et al. Destabilizing role of cathepsin S in murine atherosclerotic plaques. Arterioscler Thromb Vasc Biol 2006;26: 851–856. [DOI] [PubMed] [Google Scholar]
- 63. Liu J, Sukhova GK, Sun JS, Xu WH, Libby P, Shi GP. Lysosomal cysteine proteases in atherosclerosis. Arterioscler Thromb Vasc Biol 2004;24: 1359–1366. [DOI] [PubMed] [Google Scholar]
- 64. Galis ZS, Khatri JJ. Matrix metalloproteinases in vascular remodeling and atherogenesis: The good, the bad, and the ugly. Circ Res 2002;90: 251–262. [PubMed] [Google Scholar]
- 65. Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev 2000;14: 163–176. [PMC free article] [PubMed] [Google Scholar]
- 66. Quillard T, Araujo HA, Franck G, Tesmenitsky Y, Libby P. Matrix metalloproteinase-13 predominates over matrix metalloproteinase-8 as the functional interstitial collagenase in mouse atheromata. Arterioscler Thromb Vasc Biol 2014;34: 1179–1186. 10.1161/ATVBAHA.114.303326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xu W, Schlagwein N, Roos A, van den Berg TK, Daha MR, van Kooten C. Human peritoneal macrophages show functional characteristics of M-CSF-driven anti-inflammatory type 2 macrophages. Eur J Immunol 2007;37: 1594–1599. [DOI] [PubMed] [Google Scholar]
- 68. Monaco C, Gregan SM, Navin TJ, Foxwell BM, Davies AH, Feldmann M. Toll-like receptor-2 mediates inflammation and matrix degradation in human atherosclerosis. Circulation 2009;120: 2462–2469. 10.1161/CIRCULATIONAHA.109.851881 [DOI] [PubMed] [Google Scholar]
- 69. Kadl A, Meher AK, Sharma PR, Lee MY, Doran AC, Johnstone SR, et al. Identification of a novel macrophage phenotype that develops in response to atherogenic phospholipids via Nrf2. Circ Res 2010;107: 737–746. 10.1161/CIRCRESAHA.109.215715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tuomisto TT, Korkeela A, Rutanen J, Viita H, Brasen JH, Riekkinen MS, et al. Gene expression in macrophage-rich inflammatory cell infiltrates in human atherosclerotic lesions as studied by laser microdissection and DNA array: Overexpression of HMG-CoA reductase, colony stimulating factor receptors, CD11A/CD18 integrins, and interleukin receptors. Arterioscler Thromb Vasc Biol 2003;23: 2235–2240. [DOI] [PubMed] [Google Scholar]
- 71. Duewell P, Kono H, Rayner KJ, Sirois CM, Vladimer G, Bauernfeind FG, et al. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature 2010;464: 1357–1361. 10.1038/nature08938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rajamaki K, Lappalainen J, Oorni K, Valimaki E, Matikainen S, Kovanen PT, et al. Cholesterol crystals activate the NLRP3 inflammasome in human macrophages: A novel link between cholesterol metabolism and inflammation. PLoS ONE 2010;5: e11765 10.1371/journal.pone.0011765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Johnson JL, Jenkins N, Huang WC, Di Gregoli K, Sala-Newby G, Scholtes V, et al. Relationship of MMP-14 and TIMP-3 expression with macrophage activation and human atherosclerotic plaque vulnerability. Mediators Inflamm 2014;2014: Article ID 276457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wilson HM, Chettibi S, Jobin C, Walbaum D, Rees AJ, Kluth DC. Inhibition of macrophage nuclear factor-κB leads to a dominant anti-inflammatory phenotype that attenuates glomerular inflammation in vivo. Am J Pathol 2005;167: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Grainger DJ, Metcalfe JC. Tamoxifen: Teaching an old drug new tricks? Nat Med 1996;2: 381–385. [DOI] [PubMed] [Google Scholar]
- 76. Hackshaw A, Roughton M, Forsyth S, Monson K, Reczko K, Sainsbury R, et al. Long-term benefits of 5 years of tamoxifen: 10-year follow-up of a large randomized trial in women at least 50 years of age with early breast cancer. J Clin Oncol 2011;29: 1657–1663. 10.1200/JCO.2010.32.2933 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(XLS)
Data Availability Statement
All relevant data are available via the Gene Expression Omnibus (GEO) under accession number GSE70126.