Abstract
Proteome changes in the longissimus thoracis bovine muscle in response to pre-slaughter stress were assessed on the basis of two-dimensional electrophoresis (2-DE) data. In this study, the bootstrap resampling statistical technique and a new measure of relative change of the volume of 2-DE protein spots are shown to be more efficient than commonly used statistics to reliably quantify changes in protein abundance in stress response. The data are supplied in this article and are related to “Tackling proteome changes in the longissimus thoracis bovine muscle in response to pre-slaughter stress” by Franco et al. [1].
Keywords: Bovine muscle proteome, Stress-dependent proteome changes, Two-dimensional electrophoresis, Bootstrap resampling method, Relative and fold change measures, Quantitative proteomics
Specifications table
| Subject area | Biology |
| More specific subject area | Meat science, bovine muscle proteome |
| Type of data | Table, figure |
| How data was acquired | 2-DE gel images stained for total protein with SYPRO Ruby stain were acquired using a Gel Doc XR+system. Image analysis of digitalized gels was performed through PDQuest Advanced software |
| Data format | Analyzed output data |
| Experimental factors | 2-DE was performed using total protein extracts from normal meat and DFD meat of animals affected by pre-slaughter stress |
| Experimental features | 2-DE gels (1st dimension pH 4-7 gradient; 2nd dimension 12% SDS-PAGE) were obtained, protein spots were excised and peptides were analyzed after trypsin digestion by LC-MS/MS and MALDI-TOF/TOF MS |
| Data source location | Lugo, Spain |
| Data accessibility | Data are with this article and provided in Supplementary materials directly with this article |
Value of the data
-
•
Proteome changes in bovine muscle in response to pre-slaughter stress using 2-DE data.
-
•
Application of the bootstrap statistical technique for testing proteome changes in cattle.
-
•
Illustration of the efficiency of “relative change” as a new measure in quantitative proteomics.
1. Experimental design
Valuable information about the proteome changes in the longissimus thoracis (LT) bovine muscle in response to pre-slaughter stress (PSS) was obtained from 2-DE data. The occurrence of Dark, Firm and Dry (DFD) meat was used as indicator of animals affected by PSS. A total of four biological replicates of control (normal or non-DFD) and DFD meats from male calves of the Rubia Gallega breed (Spain) were used in this study. DFD and control samples were selected from 76 male calves after evaluation of meat quality parameters that differentiate both types of meat [1]. 2-DE protein spots with statistically significant changes in protein abundance between control and DFD samples were identified by mass spectrometry (MS).
2. Materials and methods
2.1. Meat sample preparation, protein extraction and quantification
Meat samples from the LT bovine muscle were excised from the left half of each carcass at 24 h post-mortem. A 2.5 cm thick steak was taken at the fifth rib and packed under vacuum conditions at the abattoir and subsequently transported to laboratory under refrigerated conditions. Meat samples were then lyophilized under optimal conditions [2] and frozen at −80 °C until proteome analysis. Lyophilization is a cheap, practical and safe alternative for the storage of samples prior to protein extraction, separation and quantification by 2-DE [2].
Lyophilized beef powder (50 mg) was resuspended in 1.5 mL of lysis buffer (7 M urea; 2 M thiourea; 4% CHAPS; 10 mM DTT; and 2% pharmalyte pH 3-10, GE Healthcare) for 2 h at 25 °C. An aliquot of 250 µL was lysed using a Sonifier 250 (Branson) by cycling. Protein purification and extraction from crude cell lysates were carried out with the Clean-Up kit (GE Healthcare) as described in manufacturer׳s indications. The proteins were then resuspended in 250 µL of lysis buffer. Protein quantification was assessed for each extraction using the CB-X protein assay kit (G-Biosciences) according to manufacturer׳s recommendations. The BSA protein standard was used to get a calibration curve.
2.2. Two- dimensional electrophoresis
2-DE was carried out from 350 μg of total protein extract dissolved in lysis and rehydration (7 M urea, 2 M thiourea, 4% CHAPS, 0.002% bromophenol blue) buffers. Protein extracts were loaded into 24-cm-long ReadyStrip IPG Strips (Bio-Rad Laboratories) with linear pH gradient of 4-7, together with 0.6% DTT and 1% IPG buffer (Bio-Rad Laboratories). First dimensional isoelectric focusing (IEF) was performed using a PROTEAN IEF cell system (Bio-Rad Laboratories). Gels were initially rehydrated for 12 h at 50 V and rapid voltage ramping was subsequently applied to reach a total of 70 kVh. After IEF, strips were equilibrated for 15 min at room temperature in the equilibration solution I (50 mM Tris pH 8.8, 6 M urea, 2% SDS, 30% glycerol and 1% DTT) and then with equilibration solution II (50 mM Tris pH 8.8, 6 M urea, 2% SDS, 30% glycerol and 4% iodoacetamide) under the same conditions. Second dimension electrophoresis was run on 12% SDS-PAGE gels using an Ettan DALTsix vertical slab gel system (GE, Healthcare) and Tris-glycine-SDS (50 mM Tris, 384 mM glycine and 0.2% SDS) as electrode buffer. Gels were run at a constant current of 32 mA at 25 °C.
2.3. Image analysis
2-DE gel images stained for total protein with SYPRO Ruby stain (Lonza) were acquired using a Gel Doc XR+ system (Bio-Rad Laboratories). Image analysis of digitalized gels was performed through PDQuest Advanced software v. 8.0.1 (Bio-Rad Laboratories). 2-DE gels were matched across biological replicates and volume of each spot was quantitatively determined after background subtraction and normalization using total density of validated spots across all replicate gels. The pI and Mr of spots were determined from their position on the IEF-strips and standard molecular mass markers ranging from 15 to 200 kDa (Fermentas), respectively.
Sample lyophilization, protein extraction and 2-DE methods used in the study resulted in good quality and highly reproducible 2-DE gel images (Supplementary Fig. 1).
2.4. Mass spectrometry analysis
A total of ten 2-DE protein spots showed statistically significant differential abundance in muscle conversion to DFD meat [1]. Differentially abundant protein spots were excised and peptides were analyzed after trypsin digestion by LC-MS/MS and MALDI-TOF/TOF MS as described in Franco et al. [1]. The proteins identified by MS were: myosin light chain 3 (MYL3), myosin light chain 6B (MYL6B), myosin regulatory light chain 2 (MYL2), troponin C type 2 (TNNC2), beta-galactoside alpha-2,6-syalyltransferase 1 (ST6GAL1), ATP synthase subunit beta (ATP5B), triosephosphate isomerase (TPI1), cofilin-2 (CFL2) and two fast skeletal myosin regulatory light chain 2 isoforms (MYLPF and MYLPF-1) (Supplementary Tables 1 and 2).
2.5. Statistical analysis
Statistically significant differential abundance between control and DFD groups was assessed by bootstrap resampling. Bootstrapping was used to calculate 95% and 99% non-parametric confidence intervals (CIs) for the means of the observed spot volumes in DFD and control biological replicates by the bias-corrected percentile method [3]. For each set of N (=4) estimates of spot volume, 2000 bootstrap samples of size N were drawn with replacement following a Monte Carlo algorithm. Random numbers were obtained using the standard multiplicative linear congruential generator implemented by Schrage [4], which uses the multiplier 16807 and prime modulus 231−1 to generate very long sequences of “pseudo-random” numbers that have the appearance of randomness. Bootstrap CIs (95 and 99%) were then constructed from distribution of 2000 bootstrap mean replications. The bias was corrected from the proportion of bootstrap mean replications less than the original estimate of the mean using the theoretical normal distribution [3]. The bootstrapped CIs were calculated with the software DIANA [6].
The bootstrap-based statistical approach offers clear advantages in comparison with commonly used tests for testing differential protein abundance between samples: Student׳s t-test and Mann–Whitney U-test [3,5]. The bootstrap algorithm uses the empirical distribution for the sample mean. In contrast, the t-test is based on the t-distribution (small samples) or normal distribution (large samples), which are parametric and theoretical distributions that are generally only approached by the sample data. On the other hand, the Mann–Whitney is a non-parametric test but it cannot provide estimates of the magnitude of any difference because the observed data values are replaced by their ranks.
Biased volume distributions of 2-DE protein spots have a marked negative effect on the efficiency of the Student׳s t-test and the Mann–Whitney U-test. Bias of mean volume (×10−2) averaged across biological replicates for each 2-DE protein spot with significant differential abundance between control and DFD samples is shown in Table 1. The bias for each spot was estimated as the percentage of 2000 bootstrap mean replications with mean volume less than the observed mean volume that deviates from expected percentage of 50% for unbiased distributions. It can be seen that empirical distributions of mean volumes were highly biased for an important number of the identified proteins in PSS response, with biases up to 50% (Table 1). It is noteworthy that the sampling distribution of the means of random samples of any distribution will approach the normal distribution when the sample size is sufficiently large, according with the central limit theorem [5]. However, small replicate numbers are commonly used in proteomic studies.
Table 1.
Biases of mean volumes for differentially abundant protein spots between control and DFD meat samples.
| Protein (Abbrev.) | Control meat |
DFD meat |
||
|---|---|---|---|---|
| Mean volume | Bias | Mean volume | Bias | |
| (±SE, standard error) | (%) | (±SE, standard error) | (%) | |
| MYL3 | 2.16±0.46 | 0.6 | 0.00 | N/A |
| MYL6B | 13.6±8.41 | 17.2 | 2.40±0.71 | 10.6 |
| MYL2 | 18.7±3.24 | 6.2 | 7.81±1.83 | 9.4 |
| TNNC2 | 2.27±0.94 | 8.0 | 0.00 | N/A |
| ST6GAL1 | 0.28±0.28 | 49.2 | 5.60±3.31 | 51.0 |
| ATP5B | 0.62±0.19 | 11.2 | 2.35±0.49 | 7.6 |
| TPI1 | 3.03±0.12 | 6.0 | 6.83±2.61 | 10.2 |
| CFL2 | 0.00 | N/A | 4.72±1.34 | 11.4 |
| MYLPF | 0.00 | N/A | 18.1±6.15 | 1.0 |
| MYLPF-1 | 0.00 | N/A | 38.1±14.5 | 4.6 |
2.6. Measures to quantify changes in protein abundance
The commonly used measure of “fold change” (FC) was applied to quantify volume changes of 2-DE protein spots from control to DFD meat as follows:
where and are the volume means for each spot across replicate gels in DFD and control samples, respectively. FC-values less than one were represented as their negative reciprocal. In this study, a new measure is used for determination of quantitative changes in protein abundance between groups. The measure named “relative change” (RC) is defined as follows:
where is a measure of the differential volume between samples, and is the maximum observed value of over spots.
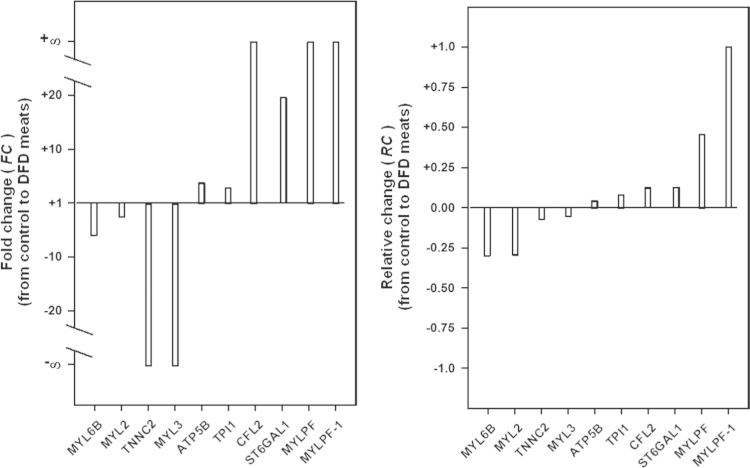
The RC measure provides more genuine information than FC to assess differential protein abundance between groups. By way of illustration, changes in protein abundance between control and DFD meat samples as measured by FC and RC statistics are shown in Fig. 1. It can be seen that both measures provide very discrepant estimates of the strength of change in protein abundance from control to DFD samples. However, RC outperforms FC to assess volume changes because FC ranges from − to + while RC ranges between −1.0 and +1.0 and takes a value of zero when there is no volume change between groups. Note that the FC measure gives values of −/+ for qualitative changes in protein abundance (i.e., presence or absence of a given spot in only one of the sample groups) when the absence of a given spot can be due merely to the occurrence of protein amounts undetectable by 2-DE. In contrast, RC allows us the joint analysis of spots with qualitative and quantitative changes under the same range of variation. Overall, the RC measure enables to assess more exactly the relative strength (weak, moderate or strong) of change in protein abundance between sample groups.
Fig. 1.
Fold change (FC) and relative change (RC) in the volume of 2-DE protein spots.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.04.011.
Supplementary materials
Supplementary material
References
- 1.Franco D., Mato A., Salgado F.J., López-Pedrouso M., Carrera M., Bravo S., Parrado M., Gallardo J.M., Zapata C. Tackling proteome changes in the longissimus thoracis bovine muscle in response to pre-slaughter stress. J. Proteomics. 2015;122:73–85. doi: 10.1016/j.jprot.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 2.Carpentier S.C., Dens K., van den Houwe I., Swennen R., Panis B. Lyophilization, a practical way to store and transport tissues prior to protein extraction for 2-DE analysis? Pract. Proteomics. 2007;1:64–69. doi: 10.1002/pmic.200700529. [DOI] [PubMed] [Google Scholar]
- 3.B. Efron, The Jackknife, the Bootstrap and Other Resampling Plans, CBMS-NSF Regional Conference Series in Applied Mathematics No. 38, Society for Industrial and Applied Mathematics, Philadelphia, 1982.
- 4.Schrage L. A more portable Fortran random number generator. ACM Trans. Math. Softw. 1979;5:132–138. [Google Scholar]
- 5.Sokal R.R., Rohlf F.J. Biometry. fourth ed. W.H. Freeman and Company; New York: 2012. [Google Scholar]
- 6.C. Zapata, DIANA: software for analysis of linkage disequilibrium, unpublished.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material