Abstract
The Informatics Core of the Alzheimer’s Diseases Neuroimaging Initiative (ADNI) has coordinated data integration and dissemination for a continually growing and complex dataset in which both data contributors and recipients span institutions, scientific disciplines and geographic boundaries. This article provides an update on the accomplishments and future plans.
Keywords: ADNI, Informatics, Alzheimer’s disease, Data management
Introduction
Informatics and data management solutions in multi-site efforts are essential for success. There are many aspects to this including, data ingestion, meta-data tagging, provenance, logging, sophisticated query, data interrogation, tightly coupled analytics and visualization, data format flexibility, data distribution and transmission, data integrity and safety and data use policies.
The degree to which high quality data, efficiently ingested into databases along with comprehensive meta-data and usable provenance determine its value. How easily data can be found and queried by anyone provided access further enhances its usefulness. The ever increasing rate of utilization seen in ADNI and other efforts has been driven largely by the creation and adoption of successful informatics solutions along with the demand for multi-scale, multi-modal, large N data in the investigation of fundamental disease processes [1]; the necessity of applying methodologies and insights from multiple disciplines in order to adequately integrate, query, analyze and interpret the data [2]; and the movement of science in general toward freely and openly available information [3]. We have reached a point in biomedical science where the electronic collection, organization, annotation, storage, and distribution of data are essential activities in most translational discovery processes.
1. Overview
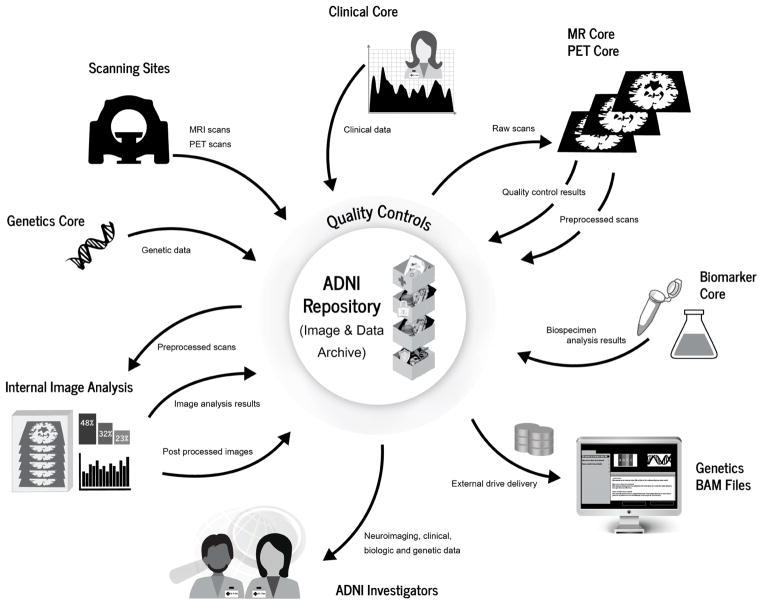
Multidisciplinary scientific collaboration has expedited discovery and understanding of complex problems, with benefits amplified through the application of generous data sharing practices. Although this potential was recognized over a decade ago, only recently has the sociology of scientific communities become accepting and even welcoming of the sharing model building upon participation and contributions across disciplines, institutions, laboratories and investigators [4–6]. The ADNI study unites the expertise and efforts of scientists from multiple disciplines and organizations working toward improving our understanding of Alzheimer’s disease. Eight cores participate in the ongoing conduct of the ADNI study, each employing distinct systems that contribute toward a complex flow of data between systems, individuals and institutions (Figure 1).
Figure 1. ADNI Data Flow.
Data flows from the participating sites and between the ADNI cores is carefully choreographed.
ADNI has set the standard for informatics solutions that facilitate open sharing with a broad data sharing philosophy embodied in the study’s subject consent language, data use agreement, and publication policies. This philosophy has been translated into practice with widespread data sharing and data reuse evident in the number of investigators using ADNI data in their research and the number of ADNI-related publications submitted and published to date. All ADNI data collected and generated through the oversight and efforts of the eight cores are ultimately deposited into the ADNI data repository (Image and Data Archive (IDA), Laboratory of Neuro Imaging, University of Southern California) and data passing quality control checks are made available to the scientific community without embargo, generally within days of collection. The primary goals of the ADNI informatics core are to build and sustain a data repository and information infrastructure that facilitates data integration and sharing for the diverse and growing ADNI scientific community.
The ADNI Informatics Core developed the infrastructure and has played a central role in managing and integrating data collected and analyzed at each of the participating centers, generated by the other ADNI cores and contributed by external investigators since the study began in 2005. The informatics core disseminates ADNI data to a continually growing number of investigators who have written hundreds of scientific papers based upon ADNI data. Research using ADNI data crosses many scientific disciplines, geographic regions and includes computer scientists interested in developing and testing machine learning and classification algorithms, neuroscientists interested in developing and testing models of disease progression, radiologists, geneticists any many others seeking to expand the boundaries of scientific knowledge. ADNI has been a global resource for scientific discovery for a decade. Here we report on the efforts of the informatics core in managing and disseminating these valuable data.
1.1 Background
The ongoing and long-term collection of varied and expanding longitudinal data has produced an accumulation of data that is heterogeneous, complex and large. From the earliest ADNI phase (ADNI 1), different study cohorts (healthy control, mild cognitive impaired (MCI) and Alzheimer’s disease (AD) subjects) followed different protocols. Some subjects underwent FDG-PET scanning with a subset also having Pittsburgh compound B PET scans; a subset of subjects had magnetic resonance (MR) scans conducted on both 1.5 and 3T scanners, while others had MR scans only on 1.5T scanners. In subsequent ADNI phases (ADNI GO and ADNI 2), additional cohorts were added for early MCI (EMCI), late MCI (LMCI) and significant memory concern (SMC) subjects and new clinical assessments were added [7], AV45 PET scans replaced Pittsburgh compound B PET scans [8] and additional vendor-specific MR scanning sequences were added [9–10]. Genome-wide association analysis (GWAS) data collected with Illumina Human610-Quad and Illumina HumanOmniExpress chipsets (Illumina, Inc. San Diego, CA) were added for subsets of ADNI 1 and ADNI GO/2 subjects in 2009 and 2012 respectively, and last year whole genome sequencing of 818 subjects was completed. Throughout the study, image and biospecimen analysts have returned results at different times and for different subsets of subjects. All of these study changes and additions have contributed toward an increasingly valuable but disparate dataset.
As additional data are collected or generated and then deposited into the repository, the resultant size and complexity of the complete data collection offers data management challenges and can be difficult for investigators to comprehend and navigate. Further, many investigators have been working with ADNI data for years and return only periodically to obtain newly deposited data and have specific needs related to ongoing analyses. These issues and the on-going, longitudinal nature of the study contribute to a complicated data sharing landscape. Recent progress to help address data size and complexity have centered on the development of new and improved search functionality, data exploration and data delivery options
2. Methods
Data from examinations and samples collected from subjects at each of the 59 North American ADNI centers are transferred into the ADNI repository in one of two ways. MR and PET imaging data are uploaded directly from the clinical sites using the de-identification and file transfer application within the Image and Data Archive (IDA), usually within 24 hours of scanning. Clinical data entered into the electronic data capture system (EDC) operated by the ADNI clinical core are transferred to the central repository nightly using an IDA file transfer tool that supports incremental updates and full dataset synchronization along with data validation functions.
Data Ingestion
Numerous models of MR and PET scanners are installed at ADNI sites generating images in different file formats and containing variations in the descriptive metadata that comprise the file headers. During the image archiving process the IDA automatically detects file formats and deploys a format-specific de-identification program that strips potentially subject-identifying data from the files before they are transferred to the repository. This is a lightweight applet also that prevents any protected health information (PHI) from being transmitted either across the secure communications link or from inadvertently being deposited into the database. This remote de-identification function greatly improves the flexibility in data distribution also. Once de-identified neuroimaging data are uploaded into the repository, metadata are extracted from the image file headers, used to classify the modality and type of image [11], and inserted into the database to support queries. A validity check of the subject identifier ensures that neuroimaging data contain subject identifiers that match the set of subjects seen at the ADNI centers. Additional validity checks verify that headers are properly populated in accordance with published file format definitions.
Quality Control
Newly uploaded neuroimaging data are queued for the MR and PET imaging cores where they undergo quality assessment. The quarantined data are made available to the MR and PET cores via an applet that downloads in batch all queued scans. Once quality assessment is completed, the results are transmitted back to the repository, usually within a few days of acquisition. As quality assessment results are received, images receiving a passing rating are automatically removed from quarantine, given an “available” status and become immediately accessible to ADNI investigators. Images not receiving a passing rating are given a failed status and remain unavailable. The MR and PET imaging cores also generate corrected preprocessed images that are uploaded into the repository on a daily basis. Preprocessed images are generated only for images that have passed quality assessment [8–9]. MR and PET core image analysts obtain preprocessed images from the repository and provide analysis results on a quarterly basis and these data are also integrated into the repository and shared with ADNI users. The informatics core developed standards and methods for collecting neuroimaging data processing provenance (the processes that an image has undergone) and for reporting image analysis methods. Collection of image processing provenance information and methods, allows external investigators to understand how processed data were generated [12] and how they are linked to primary data, both important aspects of data reuse and replication. Further, provenance metadata provide useful as search criteria for those seeking processed images for their research. Lack of standardized data formats and descriptors often hinders exchange and integration of neuroscience data [13], however, file format translators integrated into the ADNI repository allow investigators to download images in a number of common file formats.
Data Synchronization
The entire clinical database is transferred into the ADNI repository on a daily basis. Biospecimen analysis results generated through the efforts of the biospecimen core and image analysis results produced by MR and PET core analysts are transferred into the repository on a periodic basis as analyses are completed. These non-imaging data are transferred to the repository as comma-separated value (csv) files using an IDA data transfer tool. To help ensure data quality, validation of each transferred file occurs prior to acceptance. After validated data are transferred, a subset of clinical data attributes are integrated into the database and combined with metadata extracted from the images to support searches and exploration of the combined data. Further details about data flows, infrastructure, de-identification and management of these data have been described previously [14].
Data Safety
The quantity of primary and derived clinical, imaging, biochemical and genetic data generated as part of the ADNI study have required a robust and reliable infrastructure for integrating and disseminating ADNI data in a manner that addresses the needs of the ADNI cores and the larger scientific community. To ensure data are long-lived and accessible at all times, a robust computer infrastructure is utilized. Network, server, storage and electrical systems follow high-availability practices. Redundant load balancers distribute requests across multiple servers. Hardware and software failover are built into the design and high availability best practices used to insure the highest up-time. Data are stored in a clustered storage system and replicated database servers are employed. Data and software are backed up on disk and cloned to tape for offsite storage in encrypted form. Backups occur on a regular schedule. Storage nodes retain snapshots of one month’s worth of differential data to offer rapid restoration in the case of a disaster.
Policies
ADNI data are widely shared but remain restricted to investigators who agree to the terms of the ADNI data use agreement and receive the approval of the Data and Publications Committee (DPC). The ADNI repository provides online, web-based interfaces for managing the full range of data access and control activities. These include 1) an online data use application; 2) an online application review system that enables reviewers to quickly and easily review and approve/disapprove application; 3) automated user account creation and notification for approved applicants; 4) online progress report notifications that enable reviewers to notify investigators when a progress report is due and to automatically expire accounts of non-responsive investigators; and, 5) an online manuscript submission system whereby investigators may submit manuscripts to the DPC for review as required under the ADNI data use agreement.
3. Results
3.1 Data dissemination
A scientifically diverse and geographically distributed team of investigators and specialists have come together in the ADNI study to acquire, assess and analyze ADNI data and make it broadly available to the scientific community. The ADNI informatics core provides a crucial infrastructure for managing, integrating and disseminating the ADNI data, supporting efforts of the other ADNI cores and the scientific community at large. Currently, over 200,000 neuroimages comprised of more than 27 million files, 350 clinical, and analysis results datasets containing more than 19,000 data attributes, and dozens of genetic datasets are stored in the ADNI repository and disseminated to ADNI investigators around the globe. Over 6,000 investigators from 80 countries (Figure 2) have requested access to ADNI data and submitted more than 750 manuscripts for review by the ADNI Data and Publication Committee (DPC). Scientific papers utilizing ADNI data now exceeds 600 [15].
Figure 2. Distribution of ADNI data use applicants.

Investigators requesting access to the ADNI data are located in 80 countries and represent research institutions, pharmaceutical, biotech, government, and other sectors.
Many of the typical legal and ethical hurdles to sharing data [16] have been overcome by ensuring subject consents are consistent and allow broad data sharing, developing coherent rules and guidelines for data use, providing a streamlined method for requesting data access and supplying a broad base of support and documentation such that data are accurately described for reuse. To foster meaningful comparison of MR analysis methods, members of the ADNI MR, biostatistics and informatics cores worked together to identify and create ten standardized collections of preprocessed MR images which are available as shared image collections within the repository. In a paper [17], image analysts were encouraged to utilize the complete sets in their analysis. Over 7 million neuroimages and clinical datasets have been downloaded from the ADNI repository (Figure 3).
Figure 3. Download activity and data use application 2006 – 2014.
The number of investigators requesting access to ADNI data and the quantity of data downloads has increased consistently and sharply.
In 2012, the Alzheimer’s Association and the Brin-Wojcicki Foundation provided funds for whole genome sequencing (WGS) of 818 ADNI participants. The resultant quantity of genetics data, close to 200 terabytes, surpasses the ability for delivery over the Internet. The SNPs and Indels data that resulted from this effort may be downloaded from the ADNI repository, however, WGS Researchers interested in obtaining large genetics files must request data through an online application form that provides information about the production of the WGS data and describes how to receive the data on disks. Twenty-six applications for WGS data have been received and over 2.5 Petabytes of WGS have been distributed to requesters.
Support is offered through a comprehensive and informative public ADNI website (http://adni.loni.usc.edu/) that provides study and data access information and documents, news and user support. In addition to a FAQ section and a searchable data dictionary the site offers an “Ask the Experts” section that allows investigators to submit questions to one or more of the ADNI cores. Both the questions and answers submitted through “Ask the Experts” are added to the searchable knowledgebase for the edification of the ADNI community. On average, the ADNI website receives 195,000 page views per year from visitors originating in 139 countries.
3.2 Recent developments
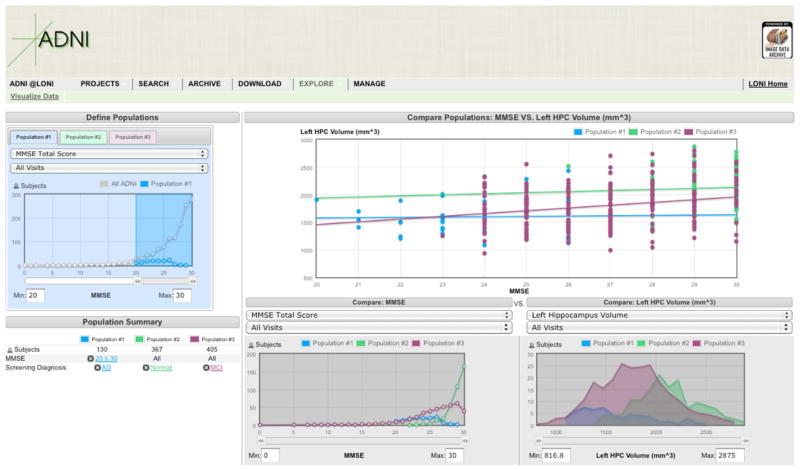
Information discovery is enhanced through interactive visual tools that help investigators explore complex data [18]. The increasing complexity and growth of the ADNI data have emphasized the need for powerful, visual interfaces for data discovery and exploration that enable investigators to fully survey and explore the entirety of the pooled ADNI data, compare cohorts and select data that meet specific criteria, are new or have changed over time. The ADNI informatics core has made improvements to search interfaces that allow users to obtain data with more specificity, to request only data that is new or changed since a specified date and to download only data that the user has not previously downloaded. Investigators may also conduct key-word searches of the non-imaging study data and search the ADNI data dictionary online. A new visual data exploration interface was deployed that allows investigators to interactively compare user-defined subject cohorts (Figure 4). This interface displays, for each data attribute, the distribution of data within each cohort and across the entire study allowing investigators to easily see the range of values and patterns among and between defined cohorts. Currently a subset of ADNI data attributes is incorporated into the visual exploration search interface.
Figure 4. Visual Explorer.
Subject cohorts are defined using the left portion of the visual explorer interface. The right portion displays plots comparing selected attributes.
3.2 Linked efforts
Data from several linked efforts, the Australian Imaging, Biomarker & Lifestyle Flagship Study of Ageing (AIBL), the Study of Brain Aging in Vietnam War Veterans (DOD ADNI) and the ADNI DepressionProject (ADNI-D) are also managed, integrated and disseminated from the IDA repository with supporting usage materials provided through the ADNI website. AIBL follows the same imaging and clinical protocols as ADNI, however while the imaging data are disseminated from the IDA repository, clinical data are available only from the AIBL website. DOD ADNI incorporates a subset of ADNI subjects for its amyloid PET imaging and significant overlap exists between the ADNI, DOD ADNI and ADNI-D clinical protocols. Investigators may obtain access to all four studies from the ADNI website and may search and download data simultaneously from all four studies. More recently, ADNI has partnered with the Global Alzheimer’s Association Interactive Network (GAAIN), a federated data infrastructure providing cohort discovery for dozens of Alzheimer’s and dementia studies from around the world [19–20]. Participation in global initiatives serves to promote ADNI’s contributions in new settings with the potential for reaching unanticipated results and reach broader communities.
4. Discussion
After ten years the informatics core of ADNI has learned a number of lessons. As the science moves forward, the technology, the data, the scientific questions all change. Even the policies, under which we operate, have been modified and tested to accommodate new developments and requirements. The amount of data, the number of users and the sophistication required to support them all have increased.
Users want the system to remember what they did last time, they want to repeat the query the same way without having to re-create all the filters, they want to know how and what data has changed since they searched or downloaded last time. Efficiency is paramount and the query and delivery systems must be continually updated to keep pace with expectations. Users can demand considerable resources, they may repeat downloads, request duplicate accounts and expect considerable hand holding as new users. Sometimes they are less than careful when reading instructions, FAQs (frequently asked questions) news items or forums as they interact with the site. While early in the history of ADNI, there were many novice computer users asking very basic questions, after ten years the level of sophistication and experience among users has increased significantly.
The data use policies of ADNI have been critical in insuring the integrity and value of the data. We developed a policy that when introduced was somewhat unusual and even controversial. Anyone with almost any kind of research or educational goals can have the data BUT the data must be distributed from the ADNI site. In other words we specifically (and emphatically) disallowed redistribution. The intent was to insure that ADNI data was ADNI data, unadulterated and original. Furthermore, we wanted to be able to track how it was being used and to insure those who collected the data and organized the project received credit. We wanted to deliver to the funders, usage statistics that accurately reflected the scientific contributions of the project and their support. A summary list of some of lessons learned includes;
The infrastructure must provide flexible methods for data description and relationships among various metadata characteristics (e.g., provenance);
Subject consent forms must be clear in what and how data are to be used, stored and distributed;
The database must be well organized, algorithmically agile and the user access interface is easy to navigate;
Derived data (pre-processed and post processed data) should be linked to the raw data and easily queriable with comprehensive provenance
Duties and responsibilities of stakeholders, individuals and institutions, must be clearly and precisely specified;
Curation systems governing quality control, data validation, authentication, and authorization must be efficient;
Policies should ensure that requesting investigators give proper attribution to the project and original collectors of the data.
Ancillary Uses
One of the more interesting and yet unanticipated uses of the ADNI data and informatics systems was as a test bed for the development of new tools and competition in their accuracy and sensitivity. In the Organization for Human Brain Mapping 19th annual meeting in 2013, a hackathon was conducted that involved ADNI data to enable those at the competition to test their algorithmic submissions. In another example of this ‘gold standard’ approach to scientific competition, SAGE Bionetworks formed a competition to measure the ability to predict MCI conversion to AD using a specially defined collection of ADNI data. Each participant had to apply for access to the ADNI data in the usual way, but all had access to a uniform collection of data. These examples illustrate how a well-defined collection of high quality data can accelerate the pace of discovery in an environment that is both cooperative and competitive. Coordinating with research communities to provide uniform data and data access for these kinds of efforts promotes a highly collaborative environment and increases the impact of the data.
5. Future directions
We continue working toward a more interactive environment for data discovery that tackles the complexity and expansion of collected data including new imaging, genetic and phenotype data. Central to our plans are improving our automated imaging classification methods and development of a new visual exploration search interface that incorporates the full complement of ADNI data.
We have developed methods for automatically labeling sub-types of MRI and PET scans to better support searches. For example different types of MR scans are tagged in the system as functional MR, diffusion MR or MR angiography based on attributes present in the image files and subtypes of structural MR tagged as T1-weighted, T2-weighted and PD-weighted [11]. PET scans sharing the same radioisotope in image files are relabeled to indicate the radiopharmaceutical used (18F-FDG, 18F-AV45) in a similar way. We plan to refine and expand our image classification capabilities to improve fidelity and automatically identify additional MR and PET subtypes.
Well-designed visualizations of complex data can extend the cognitive ability of the viewer allowing information to be comprehended faster than through reading numbers or text alone [21]. Combined with advances in technology, we can manipulate enormous datasets through interactive visualizations to seek patterns and meaning previously unmanageable [18, 22]. Building on our current visual exploration search interface, we plan to develop a more powerful and expanded visual search interface that allows investigators to search across the entirety of the ADNI dataset, refining and building the datasets that meet their research needs through interactive, visual exploration and filtering of the data.
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; ; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This study was also supported by the National Institutes of Health grants 5P41 EB015922-16 and 1U54EB020406-01).
Footnotes
Conflicts of Interests
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Johansen-Berg H. Human connectomics–what will the future demand? Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toga AW. The clinical value of large neuroimaging data sets in Alzheimer’s disease. Neuroimaging clinics of North America. 2012;22(1):107. doi: 10.1016/j.nic.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross JS, Krumholz HM. Ushering in a New Era of Open Science Through Data Sharing The Wall Must Come Down Open Science Through Data Sharing. JAMA. 2013;309(13):1355–6. doi: 10.1001/jama.2013.1299. [DOI] [PubMed] [Google Scholar]
- 4.Koslow SH. Opinion: sharing primary data: a threat or asset to discovery? Nat Rev Neurosci. 2002;3:311–3. doi: 10.1038/nrn787. [DOI] [PubMed] [Google Scholar]
- 5.Sonnenwald DH. Scientific collaboration. Ann Rev Info Sci Tech. 2007;41:643–681. [Google Scholar]
- 6.Amari S, et al. Neuroinformatics: the integration of shared databases and tools towards integrative neuroscience. J Integr Neurosci. 2002;1(2):117–28. doi: 10.1142/s0219635202000128. [DOI] [PubMed] [Google Scholar]
- 7.Aisen PS, Petersen RC, Donohue MC, Gamst A, Raman R, Thomas RG Alzheimer’s Disease Neuroimaging Initiative, . Clinical Core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimer’s & Dementia. 2010;6(3):239–246. doi: 10.1016/j.jalz.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s & Dementia. 2010;6(3):221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Weiner MW. The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27(4):685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jack CR, Bernstein MA, Borowski BJ, Gunter JL, Fox NC, Thompson PM Alzheimer’s Disease Neuroimaging Initiative, . Update on the magnetic resonance imaging core of the Alzheimer’s disease neuroimaging initiative. Alzheimer’s & Dementia. 2010;6(3):212–220. doi: 10.1016/j.jalz.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neu SC, Crawford KL, Toga AW. Practical management of heterogeneous neuroimaging metadata by global neuroimaging data repositories. Frontiers in Neuroinformatics. 2012:6. doi: 10.3389/fninf.2012.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacKenzie Graham AM, Van Horn JD, Woods RP, Crawford KL, Toga AW. Provenance in neuroimaging. Neuroimage. 2008;42:178–195. doi: 10.1016/j.neuroimage.2008.04.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotter MR, Setzu A, Slim FJ, Van Rooijen N, Franklin RJ. Macrophage depletion impairs oligodendrocyte remyelination following lysolecithin-induced demyelination. Glia. 2001;35(3):204–212. doi: 10.1002/glia.1085. [DOI] [PubMed] [Google Scholar]
- 14.Toga AW, Crawford KL Alzheimer’s Disease Neuroimaging Initiative, . The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s & Dementia. 2010;6(3):247–256. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Trojanowski JQ. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimer’s & Dementia. 2013;9(5):e111–e194. doi: 10.1016/j.jalz.2013.05.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kulynych J. Legal and ethical issues in neuroimaging research: human subjects protection, medical privacy, and the public communication of research results. Brain and cognition. 2002;50(3):345–357. doi: 10.1016/s0278-2626(02)00518-3. [DOI] [PubMed] [Google Scholar]
- 17.Wyman BT, Harvey DJ, Crawford KL, Bernstein MA, Carmichael O, Cole PE Alzheimer’s Disease Neuroimaging Initiative. Standardization of analysis sets for reporting results from ADNI MRI data. Alzheimer’s & Dementia. 2013;9(3):332–337. doi: 10.1016/j.jalz.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson CR, Moorhead R, Munzner T, Pfister H, Rheingans P, Yoo TS. NIH-NSF Visualization Research Challenges Report. IEEE Press; 2006. [DOI] [PubMed] [Google Scholar]
- 19.Toga AW, Neu SC, Bhatt P, Crawford KL, Ashish N. The Global Alzheimer’s Association Interactive Network. Alzheimer’s & Dementia. 2015 doi: 10.1016/j.jalz.2015.06.1896. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neu SC, Crawford KL, Toga AW. Sharing Data in the Global Alzheimer’s Association Interactive Network. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.05.082. (Submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Card SK, Mackinlay JD, Shneiderman B. Readings in Information Visualization: Using Vision to Think. San Francisco: Morgan Kaufmann Publishers; 1999. [Google Scholar]
- 22.Bertin J. Semiology of graphics. 1. Redlands: University of Wisconsin Press; 1983. [Google Scholar]