Abstract
Many studies have reported the association between the matrix metalloproteinase (MMP) polymorphisms and lung cancer susceptibility, but the results were inconclusive. We conducted a meta-analysis, using a comprehensive strategy based on the logistic regression and a model-free approach, to derive a more precise estimation of the relationship between MMP1, MMP2, MMP9 and MMP13 polymorphisms with lung cancer risk. A total of 22 case-control studies including 8202 cases and 7578 controls were included in this meta-analysis. For MMP1-1607 1G/2G, increased lung cancer risk was found among Asians in additive model(OR = 1.34, 95%CI:1.18-1.53) and with model-free approach(ORG = 1.41, 95%CI:1.21-1.65). For MMP2-1306 C/T and -735 C/T, based on the model-free approach, a significantly reduced risk was found in Asians(MMP2-1306 C/T:ORG = 0.49,95%CI:0.42-0.57; MMP2-735 C/T: ORG = 0.71, 95%CI:0.61-0.84). For MMP9-1562 C/T, a significantly increased risk was found among Asians(OR = 2.73, 95%CI:1.74-4.27) with model-free approach. For MMP13-77A/G, there was no association between this polymorphism and lung cancer risk in the recessive model(OR = 1.02, 95%CI:0.83-1.26) and with the model-free approach(ORG = 0.95, 95%CI:0.76-1.17). Therefore, this meta-analysis suggests that the MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T polymorphisms were risk factors for lung cancer among Asians, while MMP13 -77A/G polymorphism was not associated with lung cancer risk.
Lung cancer is the leading cause of cancer-related death worldwide and responsible for approximately 1.3 million deaths each year1. Despite the great progress made in several areas of oncology, the prognosis of lung cancer remains dismal2. The exact cause and mechanism of lung cancer are still under investigation. Epidemiological studies have demonstrated tobacco smoking as well as exposure to environmental tobacco smoke in healthy non-tobacco users as the major risk factor for lung cancer3. However, not all smokers develop lung cancer and a fraction of life long non-smokers will die from lung cancer indicating that genetic factors may play a significant role in determining the susceptibility to lung cancer3,4.
The matrix metalloproteases (MMPs) are zinc-dependent endopeptidases that degrade the extracellular matrix collagens and belong to a larger family of proteases known as the metzincin superfamily. 5,6 Matrix metalloproteinase-1 (MMP-1) may degrade a broad range of substrates including the interstitial types I, II, III collagens as well as casein and contribute to tumor growth and spread by altering the cellular microenvironment to favor tumor formation. 5,6,7,8 Among secreted MMPs, MMP-2 and MMP-9 are known to play a major role in cancer invasion and metastasis development by their ability to degrade type IV collagen9. Furthermore, overexpression of MMP13 has been related to more aggressive tumors and poor prognosis in lung cancer10,11.
Polymorphisms in the regulatory regions of MMPs have been associated with changes in the expression level of these genes in different human cancer12,13,14. Up to now, many molecular epidemiological studies have investigated the association between the MMPs polymorphism and lung cancer risk15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31. However, the results remain controversial and ambiguous. Several meta-analysis have been performed to assess MMPs polymorphism in lung cancer, but these analyses are mainly based on traditional approaches, which would lead to multiple comparisons or erroneous mode specification without prior biological evidence. Therefore, we conducted this meta-analysis based 22 case-control studies by a comprehensive statistical strategy of a logistic regression and a model-free approach32,33.
Materials and Methods
Search Strategy
We searched for relevant studies up to May 2014 through the PubMed, Embase, Wanfang (http://www.wanfangdata.com.cn), China National Knowledge Infrastructure Platform (CNKI; http://www.cnki.net) database with the following terms and their combinations: “lung cancer/carcinoma”, “polymorphism/variant”, and “metalloproteinase/MMP”. We tried to identify potentially relevant studies from the whole reference lists by orderly reviewing title, abstract and full text.
Selection criteria
The inclusion criteria were as follows: a) case-control studies focused on the association of MMP1, MMP2, MMP9 or MMP13 polymorphism and lung cancer; b) genotype and allele data available. Studies were excluded for following reasons: a) unpublished papers, reviews and duplication of publications; b) data unavailable for calculating genotype or allele frequencies; c) no control population. Additionally, investigations of departure from Hardy-Weinberg equilibrium (HWE) was excluded from the final analysis. If more than one article was published using the same case series, we selected the study with the largest sample size.
Data extraction
All the available data were extracted from each study by two investigators (H X L and X Y L) independently according to the inclusion criteria listed above. For each study, we recorded the first author, year of publication, country of origin, ethnicity, the method of genotyping, the number of cases and controls and genotype distributions in cases and controls.
Statistical analysis
Hardy-Weinberg equilibrium was examined by chi-square goodness-of-fit test (P > 0.05) using gene frequencies of the healthy individuals. Metagen (http://bioinformatics.biol.uoa.gr/~pbagos/metagen/) was used by selecting the genetic model. Two parameters, θ2 and θ3, were calculated using the formula: log it (πij) = αi + θ2zi2 + θ3zi3 and OR AB/AA = exp (θ2), OR BB/AA = exp (θ3); where αi is the indicator of study-specific fixed-effect; θ2 and θ3 are dummy variables of genotypes AB and BB. The appropriate genetic model was identified using the following criteria:(i) No association: θ2 = θ3; (ii) Dominant model: θ2 = θ3 > 0; (iii) Recessive model: θ2 = 0 and θ3 > 0; (iv) Additive model: 2θ2 = θ3; (v) Co-dominant model: θ3 > θ2 > 0; (vi) Complete overdominant model: θ2 > 0 and θ3 = 0. Finally, once the most appropriate genetic model was identified, the pooled OR with corresponding 95% confidence interval (95% CI) was estimated in logistic regression model. Additionally, Zintzaras reported a novel method to calculate the generalized odds ratio (ORG) based on a genetic model-free approach, which may overcome the short-comings of multiple model testing or erroneous model specification33. Thus, the ORG calculations were also performed.
The heterogeneity of the studies was assessed using the Cochran’s Q test (considered significant for P < 0.10) and was quantified by the I2 statistic. Both fixed effects (Mantel-Haenszel) and random effects (Der Simonian and Laird) models were used to combine the data. Relative influence of each study on the pooled estimate was assessed by omitting one study at a time for sensitivity analysis. Publication bias was evaluated by visual inspection of symmetry of Begg’s funnel plot and assessment of Egger’s test (P < 0.05 was regarded as representative of statistical significance). Statistical analyses were done in ORGGASMA, metan and metagen in STATA software, version 11.0 (STATA Corp., College Station, TX, USA), and all tests were two-sided.
Results
Characteristics of the studies
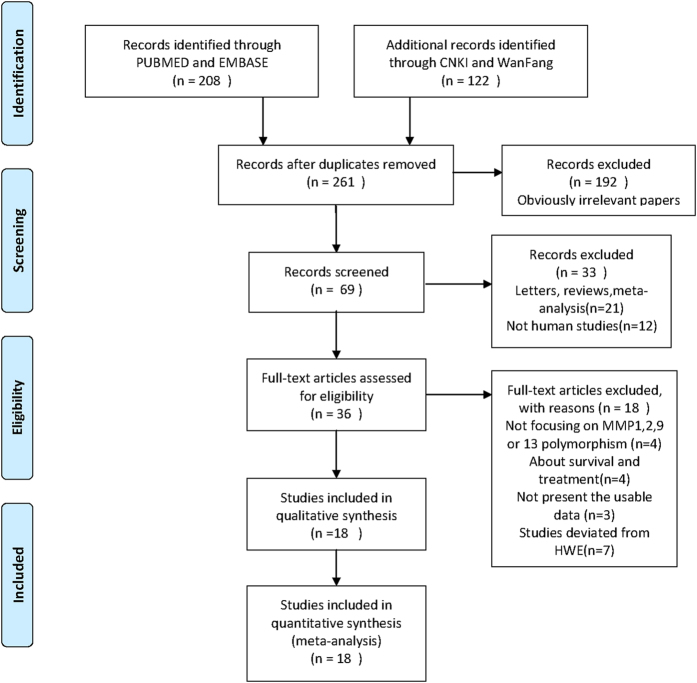
There were 330 papers relevant to the search words. The flow chart of selection of studies and reasons for exclusion is presented in Fig. 1. Overall, 18 publications with 22 case-control studies including 8202 cases and 7578 controls were available for this analysis. Seven studies with 3996 cases and 3507 controls for MMP1-1607 1G/2G polymorphism, five studies with 2004 cases and 1967 controls for MMP2-1306 C/T polymorphism, three studies with 1229 cases and 1303 controls for MMP2-735 C/T polymorphism, four studies with 1202 cases and 1039 controls for MMP9-1562 C/T polymorphism, and three studies with 1221 cases and 1225 controls for MMP13-77A/G polymorphism. Study characteristics are summarized in Table 1. The genotype distributions in the controls of all studies were consistent with HWE.
Figure 1.
Flow diagram of studies identification.
Table 1. Characteristics of studies included in this meta-analysis.
| Author | Year | Country | Ethnicity | Source of control | Genotyping methods | Sample size (case/control) | Case | Control | PHWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMP1 -1607 1G/2G | 1G/1G | 1G/2G | 2G/2G | 1G/1G | 1 G/2 G | 2 G/2 G | |||||||
| Biondi | 2000 | Italy | Caucasian | NA | NA | 29/164 | 7 | 16 | 6 | 42 | 86 | 36 | 0.520 |
| Su | 2006 | USA | Caucasian | PB | Taqman | 2014/1323 | 541 | 1015 | 458 | 367 | 642 | 314 | 0.310 |
| Zhang | 2006 | China | Asian | PB | PCR-RFLP | 150/200 | 32 | 70 | 48 | 60 | 98 | 42 | 0.865 |
| Gonzalez-Arriaga | 2008 | Spain | Caucasian | HB | PCR-RFLP | 501/510 | 128 | 248 | 125 | 119 | 259 | 132 | 0.712 |
| Hart | 2011 | Norway | Caucasian | PB | Taqman | 436/434 | 115 | 207 | 114 | 132 | 198 | 104 | 0.081 |
| Liu | 2011 | China | Asian | PB | PCR-RFLP | 825/825 | 74 | 323 | 428 | 100 | 367 | 358 | 0.691 |
| Fakhoury | 2012 | Lebanon | Caucasian | PB | PCR-RFLP | 41/51 | 5 | 17 | 19 | 7 | 16 | 28 | 0.081 |
| MMP2 -1306 C/T | CC | CT | TT | CC | CT | TT | |||||||
| Yu | 2002 | China | Asian | PB | PCR-DHPLC | 781/852 | 644 | 127 | 10 | 585 | 248 | 19 | 0.220 |
| Zhou | 2005 | China | Asian | PB | PCR-RFLP | 770/777 | 635 | 124 | 11 | 539 | 220 | 18 | 0.421 |
| Rollin | 2007 | France | Caucasian | HB | PCR-RFLP | 90/90 | 60 | 28 | 2 | 60 | 29 | 1 | 0.217 |
| Song | 2007 | China | Asian | PB | PCR | 163/148 | 129 | 32 | 2 | 100 | 44 | 4 | 0.747 |
| Bayramoglu | 2011 | Turkey | Caucasian | HB | PCR-RFLP | 200/100 | 123 | 73 | 4 | 65 | 32 | 3 | 0.692 |
| MMP2 -735 C/T | CC | CT | TT | CC | CT | TT | |||||||
| Zhou | 2005 | China | Asian | PB | PCR-RFLP | 770/777 | 506 | 230 | 34 | 425 | 313 | 39 | 0.052 |
| Rollin | 2007 | France | Caucasian | HB | PCR-RFLP | 89/90 | 69 | 18 | 2 | 67 | 21 | 2 | 0.816 |
| Jia | 2009 | China | Asian | PB | PCR-RFLP | 370/436 | 260 | 96 | 14 | 292 | 123 | 21 | 0.092 |
| MMP9 -1562C/T | CC | CT | TT | CC | CT | TT | |||||||
| Zhang | 2005 | China | Asian | PB | PCR-RFLP | 150/200 | 83 | 60 | 7 | 155 | 42 | 3 | 0.936 |
| Rollin | 2007 | France | Caucasian | HB | PCR-RFLP | 90/90 | 68 | 22 | 0 | 64 | 21 | 5 | 0.085 |
| Bayramoglu | 2009 | Turkey | Caucasian | HB | PCR-RFLP | 200/100 | 150 | 48 | 2 | 67 | 30 | 3 | 0.871 |
| Gonzalez-Arriaga | 2012 | Spain | Caucasian | HB | PCR-RFLP | 762/649 | 581 | 174 | 7 | 483 | 148 | 18 | 0.110 |
| MMP13 -77A/G | AA | AG | GG | AA | AG | GG | |||||||
| Gonzalez-Arriaga | 2008 | Spain | Caucasian | HB | PCR-RFLP | 501/506 | 248 | 208 | 45 | 267 | 197 | 42 | 0.508 |
| Peng | 2010 | China | Asian | PB | PCR-RFLP | 420/419 | 105 | 207 | 108 | 91 | 227 | 101 | 0.085 |
| Wang | 2013 | China | Asian | PB | PCR-RFLP | 300/300 | 85 | 132 | 83 | 55 | 156 | 89 | 0.354 |
PB, Population–based; HB, Hospital–based; PCR-RFLP: Polymerase Chain Reaction-restriction Fragment Length Polymorphism; HWE: Hardy-Weinberg Equilibrium.
Quantitative synthesis
There was a variation in the 2G allele frequency of the MMP1-1607 1G/2G polymorphism among the controls across different ethnicities, ranging from 0.46 to 0.71. For Asian controls, the 2G allele frequency was 0.56, which was slightly higher than that in Caucasian controls (0.53, P = 0.791; Fig. 2A). Another variation was in the T allele frequency of the MMP2-1306 C/T polymorphism among the controls across different ethnicities, ranging from 0.17 to 0.19. For Asian controls, the T allele frequency was 0.17, which was slightly lower than that in Caucasian controls (0.18, P = 0.249; Fig. 2B).
Figure 2.
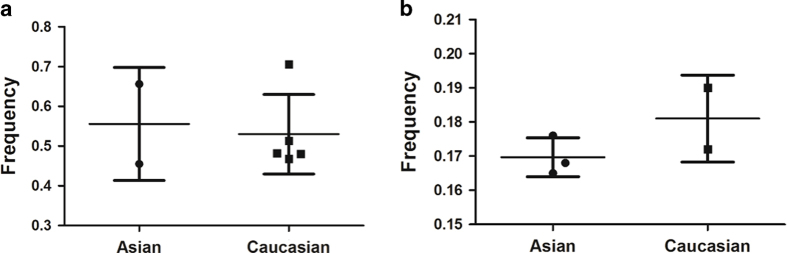
Frequencies of the variant alleles among control subjects stratified by ethnicity. (a) MMP1 -1607 2G allele; (b) MMP2 -1306 T allele.
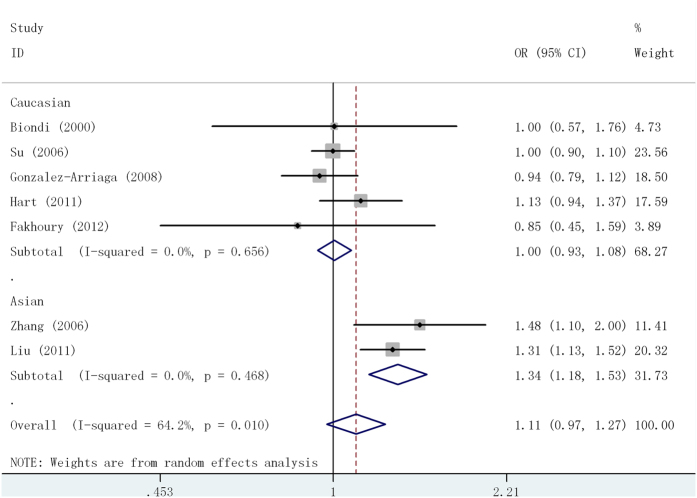
Five common SNPs occurred in MMP1, MMP2, MMP9 and MMP13 sequences were included in the quantitative synthesis, and detail results were shown in Table 2. For the MMP1 -1607 1G/2G polymorphism, the pooled OR1G2G/1G1G and OR2G2G/1G1G were 1.08(95%CI = 0.96-1.21) and 1.16(95%CI = 1.02-1.33), respectively, suggesting an additive model was assessed using traditional method. Overall, no significant association with lung cancer risk was detected for MMP1 -1607 1G/2G polymorphism in additive model and heterogeneity between studies was observed in the overall comparison. In subgroup analysis based on ethnicity, however, the heterogeneity disappeared and a significantly increased risk was found in Asians(OR = 1.34, 95%CI:1.18-1.53) (Fig. 3). Based on the model-free approach, significant result was also produced for MMP1 -1607 1G/2G polymorphism and lung cancer risk among Asians(ORG = 1.41, 95%CI:1.21-1.65). No significant association was found in subgroup analyses based on the source of control and sample size (Table 2).
Table 2. Meta-analysis of MMP1,MMP2,MMP9 and MMP13 polymorphism and lung cancer risk.
| Studycharacteristics | Case/controls | Genetic model | OR(95%CI) | I2 (%) | P forheterogeneity | |
|---|---|---|---|---|---|---|
| MMP1 -1607 1G/2G | ||||||
| Total (N = 7) | 3996/3507 | Additive Model | 1.11(0.97-1.27) | 64.2 | 0.010 | |
| ORG | 1.13(0.96-1.32) | 63.4 | 0.012 | |||
| Ethnicity | Caucasian(N = 5) | 3021/2482 | Additive Model | 1.01(0.93-1.09) | 0 | 0.656 |
| ORG | 1.01(0.92-1.10) | 0 | 0.652 | |||
| Asian(N = 2) | 975/1025 | Additive Model | 1.34(1.18-1.53) | 0 | 0.468 | |
| ORG | 1.41(1.21-1.65) | 0 | 0.473 | |||
| Source of control | PB(N = 5) | 3466/2833 | Additive Model | 1.16(0.99-1.37) | 70.9 | 0.008 |
| ORG | 1.19(0.98-1.44) | 70.2 | 0.009 | |||
| HB(N = 1) | 501/510 | Additive Model | 0.94(0.79-1.12) | — | — | |
| ORG | 0.93(0.75-1.14) | — | — | |||
| NA(N = 1) | 29/164 | Additive Model | 1.00(0.57-1.76) | — | — | |
| ORG | 1.00(0.51-1.97) | — | — | |||
| Sample size | ≥ 500(N = 4) | 3776/3092 | Additive Model | 1.08(0.94-1.25) | 74.4 | 0.008 |
| ORG | 1.10(0.93-1.30) | 73.8 | 0.010 | |||
| 500(N = 3) | 220/415 | Additive Model | 1.18(0.83-1.67) | 39.2 | 0.193 | |
| ORG | 1.30(0.97-1.74) | 42.8 | 0.174 | |||
| MMP2 -1306 C/T | ||||||
| Total (N = 5) | 2004/1967 | No association | ||||
| ORG | 0.64(0.46-0.87) | 73.2 | 0.005 | |||
| Ethnicity | Caucasian(N = 2) | 290/190 | ORG | 1.09(0.74-1.59) | 0 | 0.782 |
| Asian(N = 3) | 1714/1777 | ORG | 0.49(0.42-0.57) | 0 | 0.863 | |
| Source of control | PB(N = 3) | 1714/1777 | ORG | 0.49(0.42-0.57) | 0 | 0.863 |
| HB(N = 2) | 290/190 | ORG | 1.09(0.74-1.59) | 0 | 0.782 | |
| Sample size | ≥ 500(N = 2) | 1551/1629 | ORG | 0.48(0.41-0.57) | 0 | 0.840 |
| 500(N = 3) | 453/338 | ORG | 0.84(0.63-1.15) | 55.9 | 0.104 | |
| MMP2 -735 C/T | ||||||
| Total (N = 3) | 1229/1303 | No association | ||||
| ORG | 0.72(0.62-0.84) | 21.1 | 0.281 | |||
| Ethnicity | Caucasian(N = 1) | 89/90 | ORG | 0.85(0.44-1.67) | — | — |
| Asian(N = 2) | 1140/1213 | ORG | 0.71(0.61-0.84) | 56.3 | 0.130 | |
| Source of control | PB(N = 2) | 1140/1213 | ORG | 0.71(0.61-0.84) | 56.3 | 0.130 |
| HB(N = 1) | 89/90 | ORG | 0.85(0.44-1.67) | — | — | |
| Sample size | ≥ 500(N = 2) | 1140/1213 | ORG | 0.71(0.61-0.84) | 56.3 | 0.130 |
| 500(N = 1) | 89/90 | ORG | 0.85(0.44-1.67) | — | — | |
| MMP9 -1562 C/T | ||||||
| Total (N = 4) | 1202/1039 | Complete overdominant model | 0.84(0.51-1.39) | 79.1 | 0.002 | |
| ORG | 1.07(0.59-1.95) | 87.0 | < 0.001 | |||
| Ethnicity | Caucasian(N = 3) | 1052/839 | Complete overdominant model | 1.04(0.84-1.29) | 0 | 0.568 |
| ORG | 0.84(0.68-1.03) | 0 | 0.600 | |||
| Asian(N = 1) | 150/200 | Complete overdominant model | 0.40(0.25-0.64) | — | — | |
| ORG | 2.73(1.74-4.27) | — | — | |||
| Source of control | PB(N = 1) | 150/200 | Complete overdominant model | 0.40(0.25-0.64) | — | — |
| ORG | 2.73(1.74-4.27) | — | - | |||
| HB(N = 3) | 1052/839 | Complete overdominant model | 1.04(0.84-1.29) | 0 | 0.568 | |
| ORG | 0.84(0.68-1.03) | 0 | 0.600 | |||
| Sample size | ≥ 500(N = 1) | 762/649 | Complete overdominant model | 1.00(0.78-1.28) | — | — |
| ORG | 0.89(0.70-1.13) | — | — | |||
| 500(N = 3) | 440/390 | Complete overdominant model | 0.79(0.36-1.73) | 83.3 | 0.003 | |
| ORG | 1.14(0.44-2.94) | 89.7 | <0.001 | |||
| MMP13 -77 A/G | ||||||
| Total (N = 3) | 1221/1225 | Recessive model | 1.02(0.83-1.26) | 0 | 0.711 | |
| ORG | 0.95(0.76-1.17) | 57.8 | 0.094 | |||
| Ethnicity | Caucasian(N = 1) | 501/506 | Recessive model | 1.09(0.70-1.69) | ||
| ORG | 1.12(0.90-1.41) | |||||
| Asian(N = 2) | 720/719 | Recessive model | 1.01(0.80-1.27) | 0 | 0.446 | |
| ORG | 0.87(0.73-1.04) | 39.6 | 0.198 | |||
| Source of control | PB(N = 2) | 720/719 | Recessive model | 1.01(0.80-1.27) | 0 | 0.446 |
| ORG | 0.87(0.73-1.04) | 39.6 | 0.198 | |||
| HB(N = 1) | 501/506 | Recessive model | 1.09(0.70-1.69) | |||
| ORG | 1.12(0.90-1.41) | |||||
ORG: The Generalized Odds Ratio.
Figure 3.
Odds ratios (OR) and 95% confidence interval (CI) of individual studies and pooled data for the association of MMP1-1607 1G/2G polymorphism and lung cancer risk in additive model.
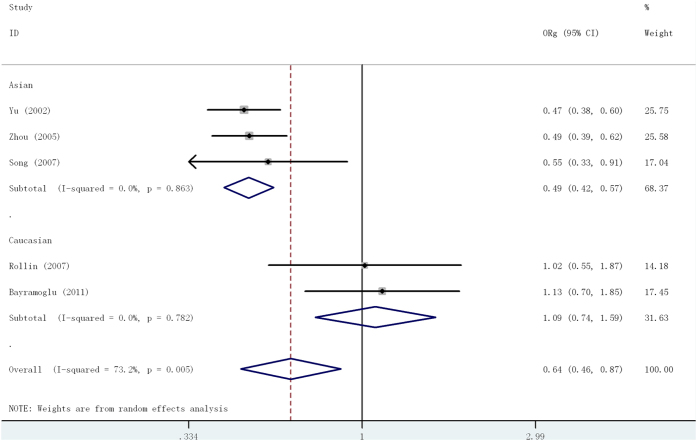
For the MMP2 -1306 C/T polymorphism, the pooled ORCT/CC and ORTT/CC were 0.54(95%CI = 0.47-0.63) and 0.53(95%CI = 0.33-0.85), respectively, suggesting no appropriate genetic model was assessed using traditional method. Based on the model-free approach, significant result was found in the overall comparison (ORG = 0.64, 95%CI:0.46-0.87) and among Asians (ORG = 0.49, 95%CI:0.42-0.57), but not among Caucasians(ORG = 1.09, 95%CI:0.74-1.59) (Fig. 4). Stratified by source of control, a significantly risk was found in the population-based studies, however, no significant association was observed in the hospital-based studies (Table 2). When stratifying by sample size, a significant association was found in sample size ≥ 500 studies (Table 2). No significant heterogeneity between studies was observed in subgroup analyses.
Figure 4.
Forest plot of the generalized odds ratio (ORG) and 95% confidence intervals (CIs) of studies on the association between lung cancer and the MMP2-1306 C/T polymorphism based on model-free approach.
For the MMP2 -735 C/T polymorphism, the pooled ORCT/CC and ORTT/CC were 0.70(95%CI = 0.59-0.83) and 0.75(95%CI = 0.51-1.10), respectively, suggesting no appropriate genetic model was assessed using traditional method. Based on the model-free approach, significant result was found in the overall comparison (ORG = 0.72, 95%CI:0.62-0.84) and among Asians(ORG = 0.71, 95%CI:0.61-0.84), but not among Caucasians(ORG = 0.85, 95%CI:0.44-1.67). Stratified by source of control, a significantly risk was found in the population-based studies, however, no significant association was observed in the hospital-based studies (Table 2). When stratifying by sample size, a significant association was found in sample size ≥ 500 studies (Table 2). No significant heterogeneity between studies was observed in the overall comparisons as well as in subgroup analyses.
For the MMP9 -1562 C/T polymorphism, the pooled ORCT/CC and ORTT/CC were 1.14(95%CI = 0.70-1.87) and 0.46(95%CI = 0.11-2.00), respectively, suggesting a complete overdominant model was assessed using traditional method. Overall, no significant association with lung cancer risk was detected for MMP9 -1562 C/T polymorphism in complete overdominant model and heterogeneity between studies was observed in the overall comparison. In subgroup analysis based on ethnicity, however, a significantly decreased risk was found in Asians(OR = 0.40, 95%CI:0.25-0.64), suggesting homozygotes were at a lesser risk of lung cancer than heterozygotes. Based on the model-free approach, significant result was also found in Asians(ORG = 2.73, 95%CI:1.74-4.27), suggesting lung cancer cases with higher mutational load than healthy individuals have higher risk for lung cancer susceptibility. Stratified by source of control, a significantly risk was found in the population-based studies, however, no significant association was observed in the hospital-based studies (Table 2). When stratifying by sample size, no significant association was found (Table 2). Heterogeneity between studies was observed in the overall comparisons and subgroup analysis based on sample size, but not in subgroup analysis based on ethnicity.
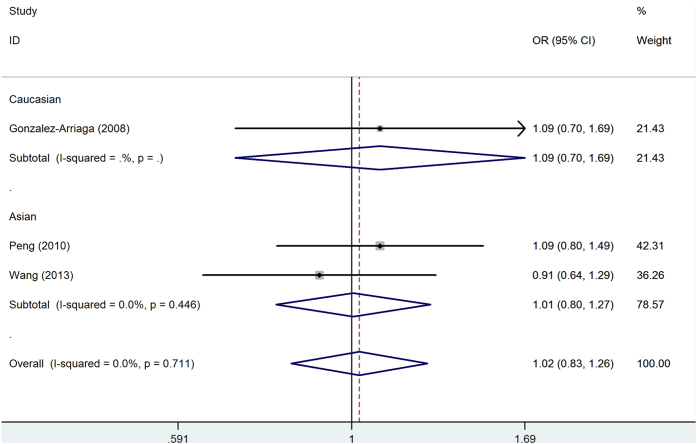
For the MMP13 -77A/G polymorphism, the pooled ORAG/AA and ORGG/AA were 0.99(95%CI = 0.83-1.18) and 1.32(95%CI = 1.03-1.67), respectively, suggesting a recessive model was assessed using traditional method. In recessive model, no significant association with lung cancer risk was detected for MMP13 -77A/G polymorphism in overall comparison and subgroup analysis (Fig. 5). Based on the model-free approach, no significant result was also found in overall comparison and subgroup analysis (Table 2).
Figure 5.
Forest plot of the generalized odds ratio (ORG) and 95% confidence intervals (CIs) of studies on the association between lung cancer and the MMP13-77 A/G polymorphism in recessive model.
Sensitive analysis
Sensitivity analyses were performed to assess the influence of individual dataset on the pooled ORs by sequential removing each eligible study. As seen in Fig. 6, any single study was omitted, while the overall statistical significance does not change, indicating that our results are statistically robust.
Figure 6.
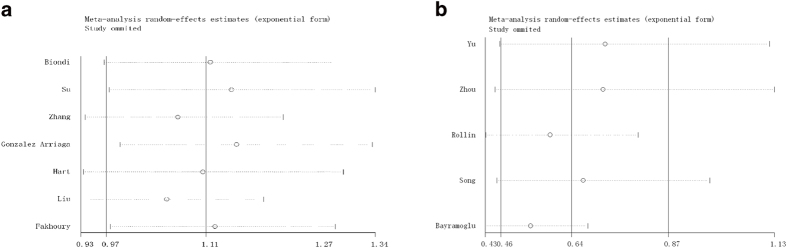
Sensitivity analysis: examining the influence of individual studies to pooled odds ratios (OR). (a) MMP1-1607 1G/2G polymorphism in additive model; (b) MMP2-1306 C/T polymorphism for generalized odds ratio (ORG).
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess publication bias among the literatures. As shown in Fig. 7, there was no evidence of publication bias for MMP1 -1607 1G/2G in additive model (Begg’s test P = 1.000; Egger’s test P = 0.703) and MMP2 -1306 C/T in generalized odds ratio (Begg’s test P = 0.221; Egger’s test P = 0.076).
Figure 7.
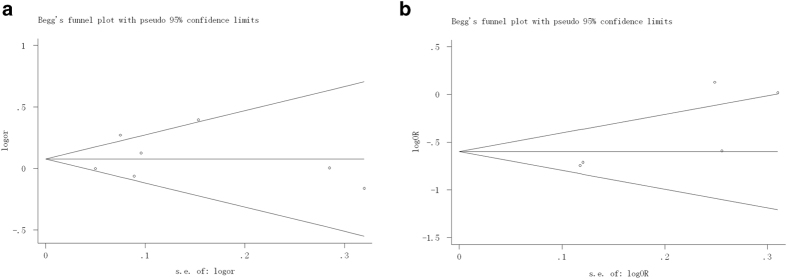
Begg’s funnel plot for publication bias test. Each point represents a separate study for the indicated association. (a) Funnel plot for additive model of MMP1-1607 1G/2G polymorphism; (b) Funnel plot for generalized odds ratio (ORG) of MMP2-1306 C/T polymorphism.
Discussion
Meta-analysis is a powerful statistical tool to resolve the discrepancies across individual studies by integrating existing published data. At present, the majority of meta-analyses of genetic association studies are usually conducted by comparing genotype frequencies between cases and controls under various genetic models. However, these genetic models are not independent, and a priori knowledge or biological justification for model selection is seldom available34,35. Therefore, we performed this meta-analysis about MMP1, 2, 9 and 13 polymorphisms and lung cancer risk by a comprehensive strategy, including logistic regression and model-free approach32,33, to avoid erroneous model specification and multiple model tests with the risk of an inflated Type I error rate.
In the current study, a total of 22 case-control studies with 8202 cases and 7578 controls were included in the meta-analysis12,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31, and the association between MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T and MMP13 -77A/G polymorphisms and lung cancer risk was explored. Our results suggest that MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T polymorphisms were significantly associated with lung cancer risk among Asian population, but there is no association found between MMP13 -77A/G polymorphism and susceptibility to lung cancer.
This finding may be biologically plausible. MMPs play roles in many important physiological and pathological processes including cancer and lung inflammation through degradation of basal membranes and extracellular matrix24,25,36. Expression of MMPs has been linked to a wide range of cancer types including lung cancer and has been reported to be correlated with tumor invasion and poor prognosis24,25,26,27. In recent years, several SNPs (MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T and MMP13 -77A/G) in the promoter region of the MMP genes have been reported26,27,28,31. Functional analyses of these polymorphisms in MMP genes have found their modulatory effect on transcriptional activity, leading to alterations in the gene expression13,14,36,37. For MMP1-1607 1G/2G polymorphism, the promoter with the 2G allele has significantly stronger transcriptional activity compared with the 1G promoter, because the 2G allele creates a transcription factor binding site and increases transcription capacity38. It has been demonstrated that individuals with CC genotype of both MMP2 -735 C/T and -1306 C/T polymorphisms have higher promoter activity and higher MMP-2 enzyme activity compared with those with the TT genotype, and thus may have potentially higher risk of lung cancer39,40. The MMP9 -1562 C to T substitution has been shown to up-regulate the promoter activity and the presence of the -1562T allele has also been found to associated with the decease of the capacity of a putative transcription repressor protein with a subsequent increase in gene expression41. Results from six independent transfection experiments in vitro with MMP13 -77A/G constructs indicated that the constructs with A had about twice as much transcriptional activity as the constructs with G in the same location37. It has been suggested that these SNPs are associated with the development of different human cancer13,14,15,16,42,43.
MMP polymorphisms and lung cancer risk have been investigated by several meta-analyses44,45,46,47. Recently, Hu et al conducted a comprehensive meta-analysis about five MMP polymorphisms and lung cancer susceptibility, and found that the MMP1-1607 1G/2G and MMP2-1306 C/T confer significantly susceptibility to lung cancer, and MMP1-1607’s effect was dependent on ethnicity, consistent with the results of this meta-analysis44. Compared with Hu’s work, we excluded three studies deviating from Hardy-Weinberg equilibrium(HWE)7,48,49, identified more eligible studies21,28,29,30 and performed a detailed analysis by logistic regression and model-free approach. We also found some significant associations that were not observed in Hu’s study, one example of which was that we found the MMP2 -735 C/T decreased lung cancer risk for Asians, whereas no significant result was found for Caucasians. On the other hand, we also analyzed the MMP13 -77A/G polymorphism. Compared with several other meta-analysis about MMP polymorphisms and lung cancer risk reported by Guo XT et al45, Hu J et al 46 and Wang J et al 47, we identified more eligible studies, evaluated more SNPs(MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T and MMP13 -77A/G) and performed analysis by a comprehensive strategy, while they only analyzed single polymorphism and lung cancer risk.
Some heterogeneity factors between studies that could limit the strengths of the meta-analysis should be addressed. First, ethnicity was one of the most important factors that could lead to heterogeneity because of the diverse genetic backgrounds and environmental factors in different ethnicities. Second, the source of the controls was another factor that could lead to heterogeneity. Population-based controls could be more reliable than hospital-based controls because the genotype distributions in hospital-based controls may be deviated from normal. In this study, significant heterogeneity was found in three of the five polymorphisms. For these polymorphisms, the heterogeneity disappeared in subgroup analysis based on ethnicity, suggesting that ethnicity of the studied population are the major source of the heterogeneity.
The current study has some inevitable limitations that should be acknowledged. First, only published studies were included in this meta-analysis, which may have biased our results. Second, there was significant heterogeneity among included studies. Even though we used the random-effect model to calculate pool ORs, the precision of outcome would be affected. Third, our results were based on an unadjusted estimated, a more precise analysis would have been conducted if more detailed individual data were available.
In summary, we concluded that the MMP1-1607 1G/2G, MMP2-1306 C/T, MMP2-735 C/T, MMP9 -1562 C/T polymorphisms were risk factors for lung cancer among Asians, while MMP13 -77A/G polymorphism was not associated with lung cancer risk. However, future well designed large studies, particularly stratified by gene-gene and gene-environment interactions might be necessary to clarify the possible role of the MMP polymorphisms in the susceptibility to lung cancer.
Additional Information
How to cite this article: Li, H. et al. Association of matrix metalloproteinase family gene polymorphisms with lung cancer risk: logistic regression and generalized odds of published data. Sci. Rep. 5, 10056; doi: 10.1038/srep10056 (2015).
Footnotes
Author Contributions H.X.L. and X.L. carried out the experimental studies, participated in data analysis and drafted the manuscript. X.Q. carried out the experimental studies and participated in data analysis. S.H.C. and S.Y.Y. participated in the design of the study and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
References
- Jemal A., et al. Global cancer statistics. C.A. Cancer. J. Clin. 61,69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Sugimura H. & Yang P. Long-term survivorship in lung cancer: a review. Chest. 129,1088–1097 (2006). [DOI] [PubMed] [Google Scholar]
- Hecht S. S. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet. Oncol. 3,461–469 (2002). [DOI] [PubMed] [Google Scholar]
- Adlkofer F. Lung cancer due to passive smoking--a review. Int. Arch. Occup. Environ. Health. 74,231–241 (2001). [DOI] [PubMed] [Google Scholar]
- Vincenti M. P., White L. A., Schroen D. J., Benbow U. & Brinckerhoff C. E. Regulating expression of the gene for matrix metalloproteinase-1 (collagenase): mechanisms that control enzyme activity, transcription, and mRNA stability. Crit. Rev. Eukaryot. Gene. Expr. 6,391–411 (1996). [DOI] [PubMed] [Google Scholar]
- Galateau-Salle, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in bronchial squamous preinvasive lesions. Hum. Pathol. 31,296–305 (2000). [DOI] [PubMed] [Google Scholar]
- Zhu Y., Spitz M. R., Lei L., Mills G. B. & Wu X. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter enhances lung cancer susceptibility. Cancer. Res. 61,7825–7829 (2001). [PubMed] [Google Scholar]
- Egeblad M. & Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2,161–174 (2002). [DOI] [PubMed] [Google Scholar]
- Bjorklund M. & Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim. Biophys. Acta. 1755, 37–69 ( 2005). [DOI] [PubMed] [Google Scholar]
- Gupta G. P., et al. Mediators of vascular remodelling coopted for sequential steps in lung metastasis. Nature. 446, 765–770 (2007). [DOI] [PubMed] [Google Scholar]
- Hsu C. P., Shen G. H. & Ko J. L. Matrix metalloproteinase-13 expres-sion is associated with bone marrow microinvolvement and prognosis in non-small cell lung cancer. Lung. Cancer. 52, 349–357 (2006). [DOI] [PubMed] [Google Scholar]
- Zhou Y., et al. Functional haplotypes in the promoter of matrix metalloproteinase-2 and lung cancer susceptibility. Carcinogenesis. 26,1117–1121 (2005). [DOI] [PubMed] [Google Scholar]
- O-charoenrat P., Leksrisakul P. & Sangruchi S. A functional polymorphism in the matrix metalloproteinase-1 gene promoter is associated with susceptibility and aggressiveness of head and neck cancer. Int. J. Cancer. 118, 2548–2553 (2006). [DOI] [PubMed] [Google Scholar]
- Kader A. K., et al. Matrix metalloproteinase polymorphisms are associated with bladder cancer invasiveness. Clin. Cancer. Res. 13,2614–2620 (2007). [DOI] [PubMed] [Google Scholar]
- Biondi M. L., et al. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin. Chem. 46, 2023–2024 (2000). [PubMed] [Google Scholar]
- González-Arriaga P., et al. Polymorphism+17 C/G in matrix metalloprotease MMP8 decreases lung cancer risk. BMC. Cancer. 8, 378 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart K., et al. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung. Cancer. 71, 123–129 (2011). [DOI] [PubMed] [Google Scholar]
- Liu L., et al. A functional polymorphism (-1607 1G>2G) in the matrix metalloproteinase-1 promoter is associated with development and progression of lung cancer. Cancer. 117, 5172–5181 (2011). [DOI] [PubMed] [Google Scholar]
- Fakhoury H., Noureddine S., Chmaisse H. N., Tamim H. & Makki R. F. MMP1-1607(1G>2G) polymorphism and the risk of lung cancer in Lebanon. Ann. Thorac. Med. 7, 130–132 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Q., Lin H., Zhou Y. A., Wang Y. J. & Cheng Q. S. Association of MMP1-1607(1G>2G)single nucleotide polymorphism with susceptibility to lung cancer in Northwestern Chinese population of Han nationality. [Article in Chinese]. Zhonghua. Yi. Xue. Yi. Chuan. Xue. Za. Zhi. 23, A313–315 (2006). [PubMed] [Google Scholar]
- Jia S. X. & Ding C. M. Association of single nucleotide polymorphisms in the promoter of MMP-2 and TIMP-2 genes with lung cancer. [Article in Chinese] Wanfang Master Thesis Database. (2009) Available at: http://d.wanfangdata.com.cn/Thesis_Y1636837.aspx. (Date of access:10/05/2014).
- Song X. Y., Li L., Zhang L. & Xiong X. Association polymorphisms in the matrix metalloproteinases-2 (MMP-2) gene with non-small cell lung cancer. [Article in Chinese]. Sichuan Zhong Liu Fang Zhi. 20, 257–259 (2007). [Google Scholar]
- Aysegul B., et al. Is a single nucleotide polymorphism a risk factor for lung cancer in the matrixmetalloproteinase-2 promoter? Mol. Biol. Rep. 38, 1469–1474 (2011). [DOI] [PubMed] [Google Scholar]
- Rollin J., et al. Influence of MMP-2 and MMP-9 promoter polymor-phisms on gene expression and clinical outcome of non-small cell lung cancer. Lung. Cancer. 56, 273–280 (2007). [DOI] [PubMed] [Google Scholar]
- Yu C., et al. Correlation between a single nucleotide polymorphism in the matrix metalloproteinase-2 promoter and risk of lung cancer. Cancer. Res. 62, 6430–6433 (2002). [PubMed] [Google Scholar]
- Gonzalez-Arriaga P., et al. Genetic polymorphisms in MMP 2, 9 and 3 genes modify lung cancer risk and survival. BMC. Cancer. 12, 121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayramoglu A., et al. The association of MMP-9 enzyme activity, MMP-9 C1562T polymorphism, and MMP-2 and -9 and TIMP-1,-2, -3, and -4 gene expression in lung cancer. Genet. Test. Mol. Biomarkers. 13, 671–678 (2009). [DOI] [PubMed] [Google Scholar]
- Peng J. C. Association of single nucleotide polymorphisms in the MMP-12 and MMP-13 genes with Lung cancer. [Article in Chinese]. Wanfang Master Thesis Database. (2010) Available at: http://d.wanfangdata.com.cn/Thesis_Y1779924.aspx. (Date of access:11/05/2014)
- Wang W., et al. Association between matrix metalloproteinase 13 gene polymorphism and susceptibility to non-small cell lung cancer. [Article in Chinese]. China Journal of Modern Medicine. 23, 35–39 (2013). [Google Scholar]
- Zhang W. A study of the relationship between lung cancer sensitibity and gene polymorphisms of MMP-1,-9. [Article in Chinese] Wanfang Master Thesis Database. (2005) Available at: http://d.wanfangdata.com.cn/Thesis_Y726994.aspx. (Date of access:11/05/2014)
- Su L., et al. Genotypes and haplotypes of matrix metalloproteinase1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis. 27, 1024–1029 (2006). [DOI] [PubMed] [Google Scholar]
- Bagos P. G. & Nikolopoulos G. K. A method for meta-analysis of case-control genetic association studies using logistic regression. Stat. Appl. Genet. Mol. Biol. 6, 17 (2007). [DOI] [PubMed] [Google Scholar]
- Zintzaras E. The generalized odds ratio as a measure of genetic risk effect in the analysis and meta-analysis of association studies. Stat. Appl. Genet. Mol. Biol. 9, 21 (2010). [DOI] [PubMed] [Google Scholar]
- Kavvoura F. K. & Ioannidis J. P. A. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum. Genet. 123, 1–14 (2008). [DOI] [PubMed] [Google Scholar]
- Salanti G., Sanderson S. & Higgins J. P. T. Obstacles and opportunities in meta-analysis of genetic association studies. Genet. Med. 7, 13–20 (2005). [DOI] [PubMed] [Google Scholar]
- Stetler-Stevenson W. G. The role of matrix metalloproteinases in tumor invasion, metastasis and angiogenesis. Surg. Oncol. Clin. N. Am. 10, 383–392 (2001). [PubMed] [Google Scholar]
- Yoon S., et al. MMP13 promoter polymorphisms is associated with atherosclerosis in the abdominal aorta of young black males. Matrix. Biol. 21, 487–498 (2002). [DOI] [PubMed] [Google Scholar]
- Rutter J. L., et al. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer. Res. 58, 5321–5325 (1998). [PubMed] [Google Scholar]
- Nelson A. R., Fingleton B., Rothenberg M. L., & Matrisian L. M. Matrix metalloproteinases: biologic activity and clinical implications. J. Clin. Oncol. 18, 1135–1135 (2000). [DOI] [PubMed] [Google Scholar]
- Price S. J., Greaves D. R., & Watkins H. Identification of Novel, Functional Genetic Variants in the Human Matrix Metalloproteinase-2 Gene ROLE OF Sp1 IN ALLELE-SPECIFIC TRANSCRIPTIONAL REGULATION. J. Biol. Chem. 276, 7549–7558 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang B., et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 99, 1788–1794 (1999). [DOI] [PubMed] [Google Scholar]
- Li Y., et al. The functional polymorphisms on promoter region of matrix metalloproteinase-12, -13 genes may alter the risk of epithelial ovarian carcinoma in Chinese. Int. J. Gynecol. Cancer. 19, 129–133 (2009). [DOI] [PubMed] [Google Scholar]
- Li Y., et al. Association of functional polymorphisms in MMP genes with gastric cardia adenocarcinoma and esophageal squamous cell carcinoma in high incidence region of North China. Mol. Biol. Rep. 37, 197–205 (2010). [DOI] [PubMed] [Google Scholar]
- Hu C., et al. Current evidence on the relationship between five polymorphisms in the matrix metalloproteinases (MMP) gene and lung cancer risk: a meta-analysis. Gene. 517, 65–71 (2013). [DOI] [PubMed] [Google Scholar]
- Guo X. T., Wang J. F., Zhang L. Y. & Xu G. Q. Quantitative Assessment of the Effects of MMP-2 Polymorphisms on Lung Carcinoma Risk. Asian. Pac. J. Cancer. Prev. 13, 2853–2856 (2012). [DOI] [PubMed] [Google Scholar]
- Hu J., Pan J. & Luo Z. G. MMP1 rs1799750 Single Nucleotide Polymorphism and Lung Cancer Risk: A Meta-analysis. Asian. Pac. J. Cancer. Prev. 13, 5981–5984 (2012). [DOI] [PubMed] [Google Scholar]
- Wang J. & Cai Y. Matrix metalloproteinase 2 polymorphisms and expression in lung cancer: a meta-analysis. Tumour. Biol. 33, 1819–1828 (2012). [DOI] [PubMed] [Google Scholar]
- Fang S., et al. Polymorphisms in the MMP1 and MMP3 promoter and non-small cell lung carcinoma in North China. Carcinogenesis. 26, 481–486 (2005). [DOI] [PubMed] [Google Scholar]
- Wang Y., et al. No association between the C-1562T polymorphism in the promoter of matrix metalloproteinase-9 gene and non-small cell lung carcinoma. Lung. Cancer. 49, 155–161 (2005). [DOI] [PubMed] [Google Scholar]