Abstract
Subclinical atherosclerosis cannot be predicted and novel therapeutic targets are needed. The molecular anatomy of healthy and atherosclerotic tissue is pursued to identify ongoing molecular changes in atherosclerosis development. Mass Spectrometry Imaging (MSI) accounts with the unique advantage of analyzing proteins and metabolites (lipids) while preserving their original localization; thus two dimensional maps can be obtained. Main molecular alterations were investigated in a rabbit model in response to early development of atherosclerosis. Aortic arterial layers (intima and media) and calcified regions were investigated in detail by MALDI-MSI and proteins and lipids specifically defining those areas of interest were identified. These data further complement main findings previously published in J Proteomics (M. Martin-Lorenzo et al., J. Proteomics. (In press); M. Martin-Lorenzo et al., J. Proteomics 108 (2014) 465–468.) [1,2].
Specifications table
| Subject area | Biology |
| More specific subject area | Cardiovascular disease, MSI development and application to arterial tissue |
| Type of data | Table and figure |
| How data was acquired | MALDI-MSI, FTICR |
| Data format | Analyzed |
| Experimental factors | Specific and careful tissue treatment was applied as previously published [1] |
| Experimental features | |
| Data source location | LUMC (Leiden, The Netherlands), IIS-Fundación Jiménez Díaz (Madrid, Spain) |
| Data accessibility |
Value of the data
-
•
A novel unexplored ex vivo imaging approach in cardiovascular disease;
-
•
30 µm high spatial resolution is applied to investigate atherosclerosis tissue layers;
-
•
This is the first time specific protein localization and alteration in response to atherosclerosis is shown by MALDI-MSI;
-
•
TMSB4X up-regulation in atherosclerosis is firstly identified at its original location.
1. Data, experimental design, materials and methods
1.1. Data
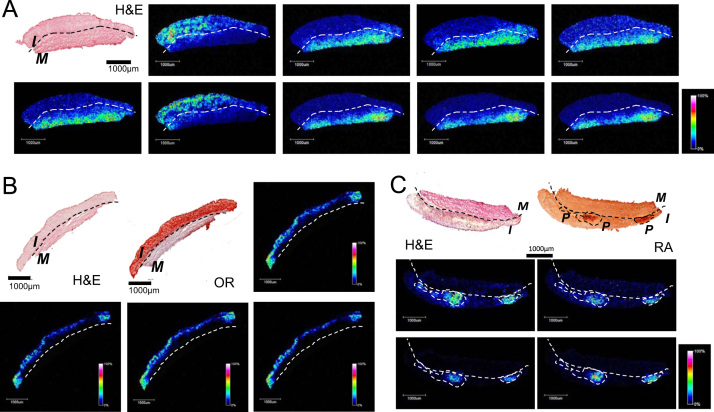
Specific molecular features (m/z values) were identified by MALDI-MSI, corresponding to proteins and lipids specifically defining intima, media or calcified regions in atherosclerotic rabbit aorta (Fig. 1). m/z values with specific location, and fold change in response to atherosclerosis early development are compiled in Table 1. Tentative identification was performed and is also shown.
Fig. 1.
Representative MALDI-MSI images for proteins (A) and lipids (B, C) in rabbit aorta. Intima (I) and media (M) layers and calcified regions (P) in the intima are defined by specific m/z values. Characterization of samples is made according to histology: H&E, Oil-Red (OR) and Red Alizarin (RA).
Table 1.
MALDI-MSI m/z values with specific localization in the intima or media layer are shown (left column): xp means specifically located in the calcified region of the intima layer. Comparison between healthy and atherosclerotic tissues is also included (right column):↑increased in atherosclerosis;↓decreased in atherosclerosis; P: pathologic (atherosclerotic) tissue; C: control (healthy) tissue. Bold numbers show statistical significance (p Value <0.05, Mann–Whitney test). Identification was performed by FT-ICR measurements, MaTisse database, MSiMass list database and literature [12,13].
|
Arterial localization |
Atherosclerosis |
Molecule |
|||||
|---|---|---|---|---|---|---|---|
| m/z | Media | Intima | p-Value | Trend | Fold change (P/C) | p-Value | |
| Proteins | |||||||
| 3011 | x | 0.0108 | ↑ | 1.67 | 0.0022 | SEL1L, IQGAP1, GANAB, NCSTN, UGDH, CYBA, YWHAG, MIF, EIF2S3, SYNM, ITGA5, NDUFS7, COL12A1, VASN, EEF1A1, MYBPC1, HBA1-2, ENO1, UBA1, CA3, MUC5B | |
| 3553 | x | 0.0022 | ↓ | 0.64 | 0.0152 | NSF, PSMC4, ACTB, MYL2, PKM2, HSPD1 | |
| 3569 | x | 0.0022 | ↓ | 0.67 | 0.0303 | DHRS7, ACTB, MYL2, PKM2, ERP44, S100A6 | |
| 4597 | x | 0.0022 | ↓ | 0.92 | 0.4589 | – | |
| 4614 | x | 0.0022 | ↓ | 0.93 | 0.6494 | HBB | |
| 4762 | x | 0.0303 | ↑ | 3.00 | 0.0022 | TMSB4X | |
| 4778 | x | 0.0303 | ↑ | 2.07 | 0.0022 | – | |
| 5620 | x | 0.0022 | ↓ | 0.58 | 0.0087 | – | |
| 6182 | x | 0.0022 | ↓ | 0.49 | 0.0022 | – | |
| 6199 | x | 0.0022 | ↓ | 0.57 | 0.0152 | – | |
| Lipids | |||||||
| 255 | x | 0.0152 | ↑ | 4.98 | 0.0022 | SFA | |
| 518 | x | 0.0022 | ↑ | 8.74 | 0.0022 | Lysolipids | |
| 520 | x | 0.0260 | ↑ | 4.58 | 0.0260 | Lysolipids | |
| 522 | x | 0.0022 | ↑ | 5.64 | 0.0022 | LPC (0:0/18:1), lysolipids | |
| 535 | xP | 0.0381 | ↑ | 4.21 | 0.0381 | – | |
| 536 | xP | 0.3524 | ↑ | 1.42 | 0.1714 | – | |
| 568 | xP | 0.1714 | ↑ | 3.57 | 0.0667 | – | |
| 675 | xP | 0.0667 | ↑ | 6.84 | 0.0190 | PA | |
| 676 | xP | 0.1143 | ↑ | 4.61 | 0.0381 | PA+PG | |
| 691 | xP | 0.0667 | ↑ | 4.43 | 0.0381 | SM+PA+PE−Cer | |
| 722 | xP | 0.1143 | ↑ | 4.76 | 0.1143 | PC+PE | |
| 800 | x | 0.0022 | ↑ | 3.74 | 0.0022 | SM | |
| 864 | x | 0.0087 | ↑ | 9.77 | 0.0022 | PG | |
| 865 | x | 0.0087 | ↑ | 6.54 | 0.0022 | PI | |
| 866 | x | 0.0260 | ↑ | 1.03 | 0.0022 | PC | |
| 891 | x | 0.0931 | ↑ | 6.52 | 0.0022 | Glc−GP+PI | |
| 893 | x | 0.0433 | ↑ | 6.18 | 0.0411 | PS | |
| 895 | xP | 0.3874 | ↑ | 1.51 | 0.1320 | TG | |
1.2. Experimental design
A rabbit model of atherosclerosis was developed as previously published [3] to investigate molecular alterations in arterial tissue in response to atherosclerosis. High-spatial-resolution MALDI-MSI was applied to comparatively analyze histologically-based arterial regions of interest from control and atherosclerotic aortas.
1.3. Materials and methods
The ascending aortic section of each animal was dissected, snap frozen in liquid nitrogen without any fixation and stored at −80 °C [4,5].Three different MALDI-MSI protocols were applied for the detection of proteins [2], lipids [6] and metabolites [7,8]. Public libraries of MALDI-MSI data, MSiMass list database [9] and MaTisse [10] were used to assign identity of the most significantly altered protein molecular feature using a mass tolerance of ±3 Da [11]. Lipid molecular identification was performed by using exact mass measurements, peak peaking and spatial filtering combined with Lipidsmap database using a tolerance of ≤0.005 Da, as previously published [12,13]. For comparison between control and atherosclerotic tissue, a random selection of the whole spectra sets from these regions were then imported into ClinProTools 3.0 (Bruker Daltonik) where they underwent smoothing, baseline subtraction, mass spectral alignment and normalization. Mann–Whitney non-parametric tests were performed using GraphPad Prism software.
Acknowledgments
This work is financially supported by the Cyttron II project “Imaging Mass Spectrometry”, ISCIII (PI11/01401, PI13/01873, CP09/00229) and IDCSalud (Grant number 3371/002). MML is funded by Fundación Conchita Rabago and gratefully acknowledges the travel funding supplied by SePROT and the COST Action BM1104 for the Short Term Scientific Missions to LUMC. BB and RC are funded by the Marie Curie Actions of the European Union (BB no. 331866, SITH FP7-PEOPLE-2012-IEF, RC no. 303344, ENIGMAS FP7-PEOPLE-2011-IEF).
These results are lined up with the Spanish initiative on the Human Proteome Project (SpHPP).
References
- 1.Martin-Lorenzo M., Balluff B., Maroto A.S., Carreira R.J., van Zeijl R.J.M., Gonzalez-Calero L., de la Cuesta F., Barderas M.G., Lopez-Almodovar L.F., Padial L.R., McDonnell L.A., Vivanco F., Alvarez-Llamas G. J. Proteomics. 2015 doi: 10.1016/j.jprot.2015.06.005. (In press) [DOI] [PubMed] [Google Scholar]
- 2.Martin-Lorenzo M., Balluff B., Sanz-Maroto A., van Zeijl R.J., Vivanco F., Alvarez-Llamas G. J. Proteomics. 2014;108:465–468. doi: 10.1016/j.jprot.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Martin-Lorenzo M., Zubiri I., Maroto A., Gonzalez-Calero L., Posada-Ayala M., de la Cuesta F. KLK1 and ZG16B proteins and arginine-proline metabolism identified as novel targets to monitor atherosclerosis, acute coronary syndrome and recovery. Metabolomics. 2014 doi: 10.1007/s11306-014-0761-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chughtai K., Heeren R.M. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 2010;110(5):3237–3277. doi: 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meding S., Walch A. MALDI imaging mass spectrometry for direct tissue analysis. Methods Mol. Biol. 2013;931:537–546. doi: 10.1007/978-1-62703-056-4_29. [DOI] [PubMed] [Google Scholar]
- 6.Willems S.M., van R.A., van Z.R., Deelder A.M., McDonnell L.A., Hogendoorn P.C. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumour type and grade, and reveals biochemical intratumour heterogeneity. J. Pathol. 2010;222(4):400–409. doi: 10.1002/path.2771. [DOI] [PubMed] [Google Scholar]
- 7.Jones E.A., Shyti R., van Zeijl R.J., van Heiningen S.H., Ferrari M.D., Deelder A.M. Imaging mass spectrometry to visualize biomolecule distributions in mouse brain tissue following hemispheric cortical spreading depression. J Proteomics. 2012;75(16):5027–5035. doi: 10.1016/j.jprot.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Dekker T.J., Jones E.A., Corver W.E., van Zeijl R.J., Deelder A.M., Tollenaar R.A. Towards imaging metabolic pathways in tissues. Anal. Bioanal. Chem. 2014 doi: 10.1007/s00216-014-8305-7. [DOI] [PubMed] [Google Scholar]
- 9.McDonnell L.A., Walch A., Stoeckli M., Corthals G.L. MSiMass list: a public database of identifications for protein MALDI-MS imaging. J. Proteome Res. 2013 doi: 10.1021/pr400620y. [DOI] [PubMed] [Google Scholar]
- 10.Maier S.K., Hahne H., Gholami A.M., Balluff B., Meding S., Schoene C. Comprehensive identification of proteins from MALDI imaging. Mol. Cell Proteomics. 2013;12(10):2901–2910. doi: 10.1074/mcp.M113.027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balluff B., Rauser S., Meding S., Elsner M., Schone C., Feuchtinger A. MALDI imaging identifies prognostic seven-protein signature of novel tissue markers in intestinal-type gastric cancer. Am. J. Pathol. 2011;179(6):2720–2729. doi: 10.1016/j.ajpath.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chughtai K., Jiang L., Greenwood T.R., Glunde K., Heeren R.M. Mass spectrometry images acylcarnitines, phosphatidylcholines, and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res. 2013;54(2):333–344. doi: 10.1194/jlr.M027961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schiller J., Zschornig O., Petkovic M., Muller M., Arnhold J., Arnold K. Lipid analysis of human HDL and LDL by MALDI-TOF mass spectrometry and (31)P NMR. J. Lipid Res. 2001;42(9):1501–1508. [PubMed] [Google Scholar]