Abstract
The data provides information in support of the research article, “Differential Cysteine Labeling and Global Label-Free Proteomics Reveals an Altered Metabolic State in Skeletal Muscle Aging”, Journal of Proteome Research, 2014, 13 (11), 2008–21 [1]. Raw data is available from ProteomeXchange [2] with identifier PDX001054. The proteome of gastrocnemius muscle from adult and old mice was analyzed by global label-free proteomics and the relative quantification of specific reduced and reversibly oxidized Cysteine (Cys) residues was performed using Skyline [3]. Briefly, reduced Cysteine (Cys) containing peptides was alkylated using N-ethylmalemide (d0-NEM). Samples were desalted and reversibly oxidized Cys residues were reduced using tris(2-carboxyethyl)phosphine (TCEP) and the newly formed reduced Cys residues were labeled with heavy NEM( d5-NEM). Label-free analysis of the global proteome of adult (n=5) and old (n=4) gastrocnemius muscles was performed using Peaks7™ mass spectrometry data analysis software [4]. Relative quantification of Cys containing peptides that were identified as reduced (d(0) NEM labeled) and reversibly oxidized d(5)–NEM labeled was performed using the intensity of their precursor ions in Skyline. Results indicate that muscles from old mice show reduced redox flexibility particularly in proteins involved in the generation of precursor metabolites and energy metabolism, indicating a loss in the flexibility of the redox energy response.
Keywords: Redox proteomics, Skeletal muscle, Aging, Oxidative stress
Specifications Table
| Subject area | Biology |
|---|---|
| More specific subject area | Redox proteomics and aging |
| Type of data | Excel sheets, figures |
| How data was acquired | QExactive mass spectrometer coupled with ultimate 3000 RSLC nano system (Thermo Scientific). |
| Data format | Raw and processed data. |
| Experimental factors | Initial blocking of free thiols with d(0)NEM, reduction of reversibly oxidized Cys residues and labeling of newly reduced residues with d(5) NEM. |
| Experimental features | Label-free analysis of global proteome and relative quantification of oxidation state of redox sensitive Cysteine residues. |
| Data source location | Liverpool, United Kingdom |
| Data accessibility | Data is available through ProteomeXchange with identifier PDX001054 (http://www.ebi.ac.uk/pride/archive/projects/PXD001054). |
Value of the data
-
•
A number of metabolic and muscle diseases are associated with aberrant redox regulation. The combination of label free analysis and redox proteomic data provides additional information on the functional proteome.
-
•
This is the first article to our knowledge that combines differential Cys labeling with global label-free analysis of the proteome.
-
•
Data provides valuable information on the changes that occur in the skeletal muscle proteome with aging.
-
•
Skeletal muscle aging is an ideal model for redox proteomics [5] and our findings suggest that skeletal muscle aging is associated with a decrease in redox flexibility.
1. Data, experimental design, materials and methods
1.1. Sample preparation
Adult (12 months) and aged (25 months) C57BL/6 male mice were purchased from Charles River and housed in the Specific Pathogen-Free (SPF) Facility at the University of Liverpool for at least 2 weeks before use. All experiments were performed in accordance with United Kingdom Home Office guidelines under the United Kingdom Animals (Scientific Procedures) Act 1986. Animals were sacrificed by cervical dislocation and gastrocnemius muscles were immediately dissected. A gastrocnemius muscle from each mouse was placed immediately in a thiol blocking buffer containing (25 mM d(0) NEM, 50 mM ammonium bicarbonate, pH 8) for redox proteomic analysis. Briefly protein extracts for redox analysis were prepared in the presence of thiol blocking buffer containing d(0) NEM under anaerobic conditions. Homogenized protein lysates were cleared by centrifugation at 15,000g for 10 min at 4 °C and protein concentrations were calculated by Bradford assay (BioRad) using BSA as a standard. Protein extracts for redox analysis were desalted using Zeba spin desalting columns (Thermo) and protein concentrations were re-calculated as before. 200 µg in 160 µl of 25 mM ammonium bicarbonate of the desalted protein extract was denatured by addition of 10 μL of 1% w/v RapiGest™ (Waters, Manchester, UK) in 25 mM ammonium bicarbonate followed by incubation at 80 °C for 10 min. Reversibly oxidized Cys residues were reduced by the addition of 10 µl of 100 mM TCEP and incubated at 60 °C for 10 min. Newly reduced Cys residues were subsequently alkylated with 10 µl of 200 mM d(5) NEM and incubated at room temperature for 30 min. Trypsin (Sigma, Poole, UK) was reconstituted in 50 mM acetic acid and 2 μg added to the samples followed by incubation overnight at 37 °C. The digestion was terminated and RapiGest™ removed by acidification (3 μL of TFA and incubation at 37 °C for 45 min) and centrifugation (15,000 × g for 15 min).
1.2. Sample analysis
The data-dependent label-free analysis was performed using an Ultimate 3000 RSLC™ nano system coupled to a QExactive™ mass spectrometer (Thermo Scientific). The sample (5 µL corresponding to 200 ng of protein) was loaded onto the trapping column (Thermo Scientific, PepMap100, C18, 75 μm×20 mm), using partial loop injection, for 7 min at a flow rate of 4 μL/min with 0.1% (v/v) TFA. The sample was resolved on the analytical column (Easy-Spray C18 75 µm×500 mm×2 µm column) using a gradient of 97% A (0.1% formic acid) 3% B (99.9% ACN 0.1% formic acid) to 60% A 40% B over 120 min at a flow rate of 300 nL/min. Data dependent acquisition consisted of a 70,000 resolution full-scan MS scan (AGC set to 106 ions with a maximum fill time of 250 ms). The 10 most abundant peaks were selected for MS/MS using a 17,000 resolution scan (AGC set to 5×104 ions with a maximum fill time of 250 ms) with an ion selection window of 3 m/z and a normalized collision energy of 30. To avoid repeated selection of peptides for MS/MS the program used a 30 s dynamic exclusion window.
1.3. Data processing and quantification
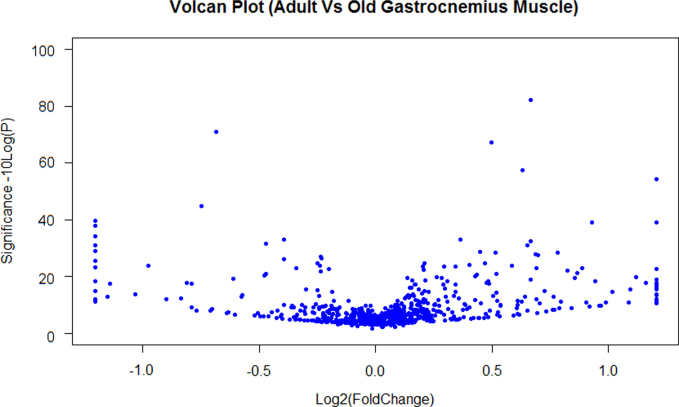
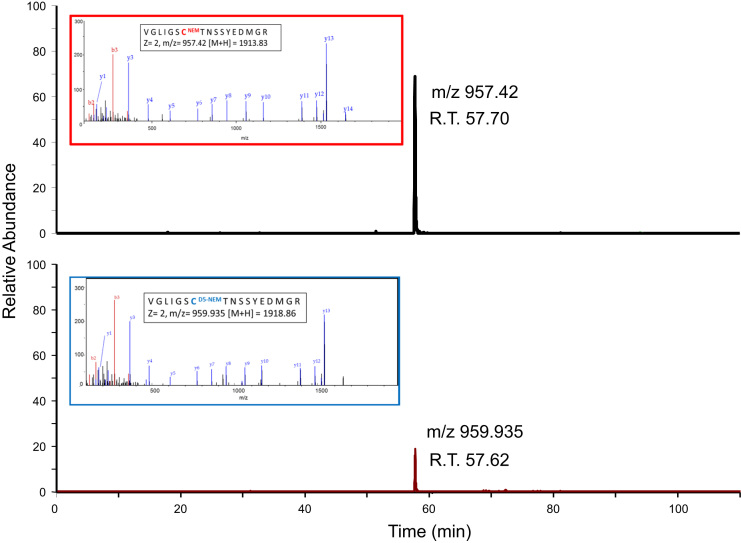
Raw spectra were converted to mascot generated files (mgf) using Proteome Discoverer software (Thermo Scientific). The resulting mgf files were searched against Uniprot Mouse database sequence database (12/05/2012, 16376 sequences) using an in-house Mascot server (Matrix Science, London, UK). Search parameters used were: peptide mass tolerances, 10 ppm; fragment mass tolerance, 0.01 Da, 1+, 2+ and 3+ ions; missed cleavages, 1; instrument type, ESI-TRAP. Variable modifications included were: d(0) NEM, d(5) NEM, mono-, di- and tri-oxidation of Cys residues and oxidation of methionine. Label-free relative quantification software Peaks7™ was used to analyze RAW data files against the same mouse protein database used for identifications with Mascot. Proteins were considered significantly changed between adult and aged samples using a -10logP score of 20 (equivalent to a p value of 0.01), a fold change ≥1.5, using a quality value of 0.8 and FDR set to 1%. Supplementary file 1 contains additional information on the search and result parameters used including a Volcano plot for peptides, the distribution of feature vector ratio by quality, distribution of feature vector ratio by intensity, retention time shift distribution and M/Z Shift distribution. A volcano plot of the expression of proteomic data is provided in Fig. 1 and a list of proteins identified and quantified is included in Supplementary Table 1. An example of a peptide containing a redox sensitive Cys residue (Cys385 from aconitase) is presented in Fig. 2. Analysis of redox peptides identified with both d(0) NEM and d(5) NEM was performed using Skyline and the relative quantification of the reversible oxidation state of Cys residues (reduced:reversibly oxidized) was calculated from the intensity of precursor ions and is included in Supplementary Table 2.
Fig. 1.
Volcano plot showing distribution and log 2 fold change of proteins identified from Gastrocnemius muscle from adult and old mice.
Fig. 2.
Representative extracted ion chromatogram (XIC) and fragmentation of the double charged Aconitase peptide VGILSCTNNSSYEDMGR (containing Cys385) from a single analysis. The peptide containing the Cys residue labeled with d(0) NEM corresponds to the reduced form of the peptide and the d(5) NEM labeled peptide corresponds to reversibly oxidized Cys residue. The ratio of the intensity of the redox state of the Cys residue can be estimated from the intensity of the precursor ions and can be combined with complimentary data on the protein׳s abundance.
Acknowledgments
BMcD is supported by the Wellcome Trust ISSF Fund and the University of Liverpool (097826/Z/11/Z).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.06.012.
Supporting information
Supplementary data Supplementary File 1. MS search and results parameters.
Supplementary data Supplementary Table 1. List of proteins identified and quantified using Peaks7 analysis software.
Supplementary data Supplementary Table 2. Relative quantification of the redox state of reduced and reversibly oxidized Cys residues.
References
- 1.McDonagh B., Sakellariou G.K., Smith N.T., Brownridge P., Jackson M.J. Differential Cysteine labelling and global lable-free proteomics reveals an altered metabolic state in skeletal muscle aging. J. Proteome Res. 2014;13:2008–2021. doi: 10.1021/pr5006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vizcaíno J.A., Deutsch E.W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J.A., Sun Z., Farrah T., Bandeira N., Binz P.A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R.J., Kraus H.J., Albar J.P., Martinez-Bartolomé S., Apweiler R., Omenn G.S., Martens L., Jones A.R., Hermjakob H. ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;30:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang J., Xin L., Shan B., Chen W., Xie M., Yuen D., Zhang W., Zhang Z., Lajoie G., Ma B. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics. 2011;11 doi: 10.1074/mcp.M111.010587. M111.010587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schilling B., Rardin M.J., MacLean B.X., Zawadzka A.M., Frewen B.E., Cusack M.P., Sorensen D.J., Bereman M.S., Jing E., Wu C.C., Verdin E., Kahn C.R., Maccoss M.J., Gibson B.W. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McDonagh B., Sakellariou G.K., Jackson M.J. Application of redox proteomics to skeletal muscle aging and exercise. Biochem. Soc. Trans. 2014;42:965–970. doi: 10.1042/BST20140085. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data Supplementary File 1. MS search and results parameters.
Supplementary data Supplementary Table 1. List of proteins identified and quantified using Peaks7 analysis software.
Supplementary data Supplementary Table 2. Relative quantification of the redox state of reduced and reversibly oxidized Cys residues.