Abstract
We designed a targeted-array called GOLD (Gain or Loss Detection) Chip consisting of 900 FISH-mapped non-overlapping BAC clones spanning the whole genome to enhance the coverage of 66 unique human genomic regions involved in well known microdeletion/microduplication syndromes. The array has a 10 Mb backbone to guarantee the detection of the aneuploidies, and has an implemented resolution for telomeres, and for regions involved in common genomic diseases. In order to evaluate clinical diagnostic applicability of GOLDChip, analytical validity was carried-out via retrospective analysis of DNA isolated from a series of cytogenetically normal amniocytes and cytogenetically abnormal DNA obtained from cultured amniocytes, peripheral blood and/or cell lines. We recruited 47 DNA samples corresponding to pathologies with significant frequencies (Cri du Chat syndrome, Williams syndrome, Prader Willi/Angelman syndromes, Smith-Magenis syndrome, DiGeorge syndrome, Miller-Dieker syndrome, chromosomes 13, 18 and 21 trisomies). We set up an experimental protocol that allowed to identify chromosomal rearrangements in all the DNA samples analyzed. Our results provide evidence that our targeted BAC array can be used for the identification of the most common microdeletion syndromes and common aneuploidies.
Keywords: aCGH, BAC clones, targeted array, aneuploidies, microdeletions, microduplications
Introduction
Microscopic karyotype analysis has been the gold standard for prenatal diagnosis since the development of chromosome banding techniques in the late 1960s.1 Although highly reliable for identifying aneuploidies as well as large chromosomal rearrangements, this procedure presents some limitations due to the low resolution (5–10 Mb) and to the long average time required to get analysis results.2,3 In order to overcome these limitations, alternative molecular cytogenetic analysis based on FISH (Fluorescence in Situ Hybridization) and QF-PCR (Quantitative Fluorescence Polymerase Chain Reaction) techniques have been applied to prenatal diagnosis for a rapid screening of common aneuploidies.4–7
The major limitation of these methods is that they do not provide a genome wide screening. Consequently, these techniques have been applied to clinical samples in addition to, rather than replacing, conventional chromosomal analysis. The Comparative Genomic Hybridization (CGH) analysis was developed as a genome wide screening strategy for detecting DNA copy number imbalances, but its resolution level continued to be low like microscopic karyotype analysis.8
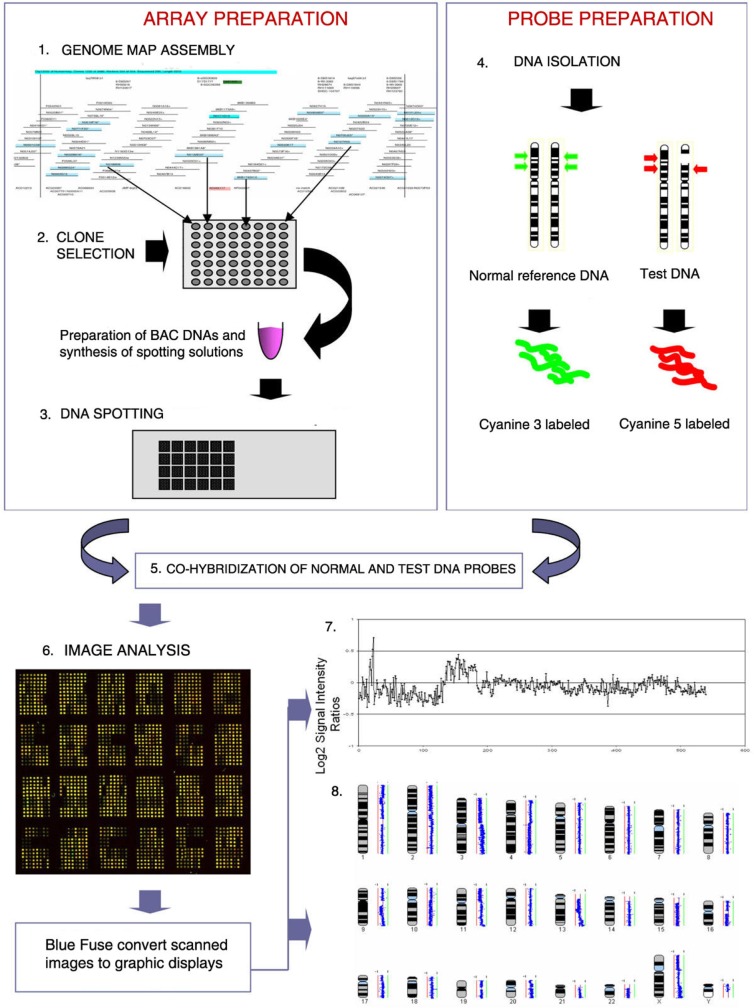
The array-CGH (array-based Comparative Genomic Hybridization) technique is similar in principle to conventional CGH,9,10 but uses arrayed DNA sequences instead of metaphase chromosomes as probes for hybridization, thus providing a direct link between detected aberrations and the physical and genetic maps of the human genome. Array-CGH analysis has a number of significant potential advantages over conventional prenatal testing providing a technique that is not only sensitive and comprehensive, but could be amenable to automation, thus decreasing costs, and the reporting time of results. A classic array-CGH experiment is shown in Figure 1.
Figure 1.
This figure shows the steps in BAC array CGH. (1) BAC clones are selected from a physical map of the genome. (2) DNA samples are extracted from selected BAC clones and their identity is confirmed by DNA fingerprinting or sequence analysis. (3) A multi-step amplification process generates sufficient material from each clone for array spotting. Each clone is spotted in replicate onto a solid support. (4) Reference DNA and test DNA are differentially labeled with cyanine 3 and cyanine 5 respectively. (5) The two labeled products are combined and hybridized onto the spotted slide. (6) Images from hybridized slides are obtained by scanning in two channels. Signal intensity ratios from individual spots can be displayed as a simple plot (7) or by using more complex software such as SeeGH, which can display copy number alterations throughout the whole genome (8).
Whole-genome array-CGH analysis has already been shown to be a useful tool in clinical genetics for detecting cryptic deletions and duplications in patients with mental retardation or learning difficulties, but with apparently normal karyotype. Besides a custom designed microarray can be exploited to analyze specific chromosomal regions. This type of array contains a large number of probes in chromosome regions selected by operator.11–13 Most of these custom arrays have been successfully constructed for all or parts of the human genome and are currently available for research use, but the genome-wide dense arrays would have potential disadvantages in clinical use. More array probes are likely to generate a higher number of false positives, and large arrays are more expensive to fabricate, quality control, and interrogate. Moreover, recent investigations showing significant levels of copy number polymorphism in normal populations14,15 reinforces the desire to test only a limited number of clones, the results of which do not give rise to needless complications in interpretation.
A diagnostically useful microarray must be reliable, must accurately detect the chromosome abnormalities assayed, and must provide interpretable results. Additionally, clinical confidence must be established using microarrays that interrogate regions of known clinical relevance.
Therefore, in the last year targeted arrays have been developed for clinical approach, focusing on medically significant and relatively common chromosomal alterations.16 Shaffer et al17 applied targeted BAC array-CGH for the analysis of subtelomeric and pericentromeric regions and of genomic regions known as critical for microdeletion syndromes, and they reported the identification of abnormalities in a cohort of cases selected for a variety of medical problems including developmental delay, mental retardation, seizures, and various congenital anomalies. In another study, performed on product of conception (POC) samples from spontaneous miscarriages using a low density array, array-CGH analysis was able to detect all abnormalities previously identified by microscopic karyotype analysis, and revealed additional abnormalities in approximately 10% of cases.18,19
The technique therefore holds some promises of combining the speed, sensitivity, and potential for partial automation of a DNA-based test, with the genome screening characteristics of microscopic karyotyping. Although it is becoming accepted that array-CGH will have a place in clinical genetic testing, it is not well established how this will be best applied. Particularly, for prenatal screening when time for further investigation is limited and ambiguous results cause severe anxiety, the ideal array would contain the minimum number of clones that will deliver the required diagnosis.
We designed a new array-CGH microarray called GOLD (Gain or Loss Detection) Chip consisting of 900 non-overlapping BAC (Bacterial Artificial Chromosome) clones spanning the whole genome, concentrating on areas of known clinical significance with dense representation across 66 common microdeletion/microduplications syndromes critical regions (Table 1), and with a lower representation of about one clone per 10 Mb over the remainder of the genome to detect unexpected major chromosome imbalances.
Table 1.
Regions of the genome assayed by the GOLDChip.
| Clinical relevance | Chromosomal region or Karyotype |
|---|---|
| Miller-Dieker lissencephaly syndrome | 17p13.3 |
| Alagille syndrome | 20p12 |
| Muscular dystrophy (Duchenne, Becker) DMD | Xp21.2 |
| ATR-16 | 16pter-p13.3 |
| Nail-Patella syndrome | 9q33.31 |
| Autism, X-linked, susceptibility to, 2, NLGN4 | Xp22.33 |
| Azoospermia factor a (AZFa) | Yq11.2 |
| Azoospermia factor b (AZFb) | Yq11.2 |
| Bruton agammaglobulinemia tyrosine kinase | Xq21.3–q22 |
| Canavan disease (ASPA) | 17pter-p13 |
| Candidate gene for the testis-determining factor (TDF) | Yp11.3 |
| Cat eye syndrome CECR1, CECR5, CECR6 | 22q11 |
| Charcot-Marie-Tooth disease type 1A | 17p11.2 |
| Cornelia de lange syndrome CDLSI | 5p13.1 |
| Cri du chat syndrome | 5p15.2 |
| Dandy-Walker syndrome DWS (ZIC1; ZIC4) | 3q24 |
| DiGeorge syndrome (DGS) | 22q11.2 |
| DiGeorge syndrome critical region 2, | 10p14-p1 |
| Down syndrome 21q22.3 | Xp11.23 |
| Down syndrome critical region, GATA1 | 21q21–21q22.3 |
| Early-onset Alzheimer disease/APP | 21q21 |
| Edwards syndrome | Trisomy 18 |
| Feingold syndrome | 2p24.1 |
| Greig cephalopolysyndactyly syndrome, GLI3 | 7p13 |
| Holoprosencephaly 1 | 21q22.3 |
| Holoprosencephaly 3 | 7q36 |
| Barakat syndrome, GATA3 | 10p |
| Kallmann syndrome 2 (KAL2) | 8p11.2-p11.1 |
| Klinefelter syndrome | XXY |
| Langer giedion type II TRPS2 | 8q24.11–q24.13 |
| Microphthalmia with linear skin defects | Xp22 |
| Beckwith-Wiedemann syndrome | 11p15.5 |
| Brachydactyly-mental retardation syndrome, D2S2338 | 2q37 |
| Neurofibromatosis 1 (NF1) | 17q11.2 |
| Neurofibromatosis 2 (NF2) | 22q12.2 |
| Ovarian dysfunction (FIMIANI; LAPERUTA) | Xq26 |
| Patau syndrome trisomy 13 | Trisomy 13 |
| Pelizaeus-Merzbacher disease | Xq22.2 |
| Potocki-shaffer syndrome | 11p11.2–p12 |
| Prader Willi syndrome/Angelman syndrome | 15q11–q13 |
| Retinoblastoma | 13q14.1–q14.2 |
| Rett syndrome (MECP2) | Xq28 |
| Rieger syndrome type 1 | 4q25 |
| Smith-Magenis syndrome | 17p11.2 |
| Sotos syndrome | 5q35 |
| Split-Hand/Foot Malformation 4 | 3q27 |
| Steroid sulfatase deficiency (STS) | Xp22.31 |
| Synpolydactyly/Syndactyly type II | 2q31–q32 |
| Tuberous sclerosis 2 (TSC2) | 16p13.3 |
| Turner Syndrome | 45, X |
| Van der Woude syndrome | 1q32–q41 |
| Williams syndrome | 7q11.23 |
| Wolf-Hirschhorn candidate 1 (WHSC) | 4p16.3 |
| X-linked lissencephaly | Xq22.3–q23 |
| Leri-Weill syndrome (dischondrosteosis Xp distal deletion SHOX) | Xpter-p22.32 |
| 1p36deletion (monosomy 1p36) | 1p36 |
| 2q37 monosomy | 2q37 |
| Sex reversal deltion 9p | 9p |
| Rubinstein-Taybi | 16p13.3 |
| Saethre-Chotzen Syndrome | 7p21 |
| Sex-determining region, SRY | Yp11.31 |
| Simpson-Golabi-Behmel syndrome | Xq26 |
| Split-Hand/Foot Malformation 3 | 10q24 |
| WAGR syndrome (PAX6) | 11p13 |
| 14q terminal deletion syndrome (van Karnebeek) | 14q |
| Split-Hand/Foot Malformation 5 | 2q31 |
We validated the microarray analyzing 47 cytogenetically abnormal DNA isolated from cultured amniotic fluid, peripheral blood samples and commercial cell lines (Table 2). The selected DNA represents 28 chromosomal abnormalities including common aneuploidies associated with Turner syndrome (45,X), Klinefelter syndrome (47,XXY), Down syndrome (trisomy 21), Edwards syndrome (trisomy 18), Patau syndrome (trisomy 13), and several microdeletions associated, including Wolf-Hirschhorn syndrome (del4p16.3), Cri du Chat syndrome (del5p15.2), Williams syndrome, (del7q11.23), Prader-Willi and Angelman syndromes (del15q11–13), Smith-Magenis syndrome (del17p11.2), DiGeorge syndrome (del22q11.2). Our results provide evidence that GOLDChip can be used for the identification of the most common microdeletions syndromes and common aneuploidies.
Table 2.
Are shown the 28 disease tested in 47 cytogenetically abnormal DNA by GOLDChip (20 cell lines and 27 clinical samples). For each disease are shown OMIM, frequency, the number of sample analyzed, the gene map locus (or Karyotype if possible), and the gain(+) or loss (−) of DNA. ORPHA: data from ORPHANET Journal of Rare Diseases. nr = not reported. * = ECACC (European Collection of Cell Cultures): cell line name AL0053, catalogue number 97091501; ** = ECACC: cell line name AL0021, catalogue number 92102601; *** = ECACC: cell line name AG0847, catalogue number 94032208; **** = ECACC: cell line name AR0189, catalogue number 99012001.
| Disease | OMIM | Frequency | Cell lines | Clinical samples | Total | Gene map locus or Karyotype | Gain/loss |
|---|---|---|---|---|---|---|---|
| Charcot-Marie-Tooth disease type 1A | 118220 | 1/2.500 | 1 | 1 | 17p11.2 | (+) | |
| Cat eye syndrome | 115470 | 1/5.000 | 1 | 1 | 22q11.2 | (−) | |
| Di George (22q12) Syndrome | 188400 | 1/4.000 | 1 | 2 | 3 | 22q11.2 | (−) |
| Di George (10p) syndrome | 601362 | <1/1.000.000 | 1 | 1 | 10p14–p13 | (−) | |
| Down syndrome | 190685 | 1/1.000 | 2 | 2 | 47,XY,+21 | (+) | |
| Williams syndrome | 194050 | 1/10.000 | 1 | 4 | 5 | 7q11.23 | (−) |
| Smith-Magenis syndrome | 182290 | 1/50.000 | 3 | 3 | 17p11.2 | (−) | |
| Patau syndrome | 1660 | nr | 1 | 1 | 47,XX,+13 | (+) | |
| Duchenne syndrome | 310200 | 1/35.000 | 1 | 1 | Xp21.2 | (−) | |
| Miller-Diecker syndrome | 247200 | 1/25.000 | 1 | 1 | 46,XY.ish del(17) (p13.3p13.3) (D17s379−) |
(−) | |
| XYY | 158250 | 1/900 | 1 | 1 | 47,XYY | (+) | |
| Klinefelter’s syndrome (XXY) | 278850 | nr | 2 | 2 | 47,XXY | (+) | |
| Edwards Syndrome | 601161 | 1/6.000 | 1 | 1 | 47,XY,+18 | (+) | |
| Prader Willi/Angelman syndrome | 600161 | 1/20.000 | 2 | 2 | 15q11–q13 | (−) | |
| Ichthyosis-Mental retardation-Kallmann Syndrome | 30870 | 1–9/100.000 | 1 | 1 | 2 | Xp22.3 | (−) |
| Monosomy 1p36 | 607872 | 1/5.000 | 1 | 2 | 3 | 1p36 | (−) |
| Brachydactyly-Mental retardation syndrome | 600430 | >1/1.000.000 | 1 | 2 | 3 | 2q37 | (−) |
| Cri du Chat syndrome | 123450 | 1/50.000 | 1 | 1 | 2 | 5p15.2–>pter | (−) |
| Del 20p | ORPHA1611 | <1/1.000.000 | 1 | 1 | 2 | 20pter* | (−) |
| Wolf-Hirschhorn syndrome (WHS) | 194190 | 1/50.000 | 1 | 1 | 4p16.3–>pter | (−) | |
| Del 6q | ORPHA96151 | <1/1.000.000 | 1 | 1 | 6qter** | (−) | |
| X-linked Ichthyosis | 308100 | 1/6.000 | 1 | 1 | 46,X,del(X) (p22p32) |
(−) | |
| del(3)(p25)->pter | ORPHA1618 | <1/1.000.000 | 1 | 1 | 3p25->pter*** | (−) | |
| Autism/Asperger syndrome | 300497 | 1–5/10.000 | 1 | 1 | 46,X,del(X) (p22.13p22.31) |
(−) | |
| Alagille syndrome | 118450 | 1/100.000 | 1 | 1 | 46,XX,del(20) (p11.23p12.2) |
(−) | |
| Turner syndrome | 158250 | 1/2.500 | 1 | 1 | 45, X**** | (−) | |
| Azoospermia | 415000 | 1–5/10.000 | 1 | 1 | Yq11.2 | (−) | |
| Dandy-Walker syndrome | 220200 | 1/25.000 | 1 | 1 | 3q24 | (−) |
Methods
Array design and production
The targeted-array described in this study was developed using published protocols.20 Briefly, large insert bacterial and plasmid artificial chromosome (BAC and PAC) clones were chosen from the public databases (UCSC, NCBI and Ensembl) to cover each chromosome at a resolution of one clone every 10 Mb. Additional clones were selected for the major common microdeletion syndrome critical regions consulting DECIPHER, OMIM and Orphanet databases, as far as possible covering identified critical regions and microdeletion breakpoints with overlapping clones. Isolated clone DNA was first amplified by degenerate oligonucleotide primed PCR (DOP-PCR), followed by secondary PCR with an amine modified primer. Array clones were spotted in two areas in six replicates onto Aldheyde slides (Genetix).
Degenerate oligonucleotide primed (DOP)-PCR
Degenerate oligonucleotide primed-PCR (DOP-PCR) was performed to amplify target clone DNA using three different PCR primers (DOP 1 primer: CCGACTCGAGNNNNNNCTAGAA; DOP 2 primer: CCGACTCGAGNNNNNNTAGGAG; DOP 3 primer: CCGACTCGAGNNNNNNTTCTAG). PCR was started at 94 °C for 3 min, then cycled first 10 times at 94 °C for 1 min30sec, at 30 °C for 2 min 30 sec and 72 °C for 3 min, then 30 times at 94 °C for 1 min, at 62 °C for 1 min 30 sec and 72 °C for 2 min, finally at 72 °C for 8 min. Gel electrophoresis was carried out as quality control on PCR products. Successfully amplified PCR products, usually 0.2–2 kb in size, were used as template for PCR with 5′ aminolink primer (NH2-GGAAACAGCCCGACTCGAG). The process was started at 95 °C for 10 min, then cycled 34 times at 95 °C for 1 min, at 60 °C for 1 min 30 sec and 72 °C for 7 min, finally at 72 °C for 10 min. PCR products obtained were purified with Wizard SV-96 PCR Clean-up kit (PROMEGA), quantified with Nanodrop and diluted in water to a final concentration of 300 ng/ul. The products were mixed 1:1 with Aldehyde spotting solution (Genetix) and were ready for prints.
Microarray spotting
Amplified DNA was spotted in six replicates onto Aldehyde coated slides (Genetix) using QArray2 arrayer (Genetix). The same sixfold-spot panel was prepared in duplicate as area “up’’ and “down’’ on the same slide. The slides were then pre-treated, denatured, and stored in a desiccator until use.
Array validation
The targeted-array validation was performed by array-CGH analysis of 47 cytogenetically known DNA isolated from 27 cultured amniotic fluid samples, chorionic villus samples and peripheral blood samples and 20 commercial cell lines (Table 2). 20 cytogenetically normal DNA were analyzed as control samples. Dye-reversal array-CGH analysis was performed as described below.
Samples collection
20 cell lines amongst ECACC human genetic collection and 27 cell cultures (Table 2) were selected to represent a broad spectrum of cytogenetic abnormalities including the most common aneuploidies (trisomies of chromosomes 13, 18, and 21, and sex chromosome aneuploidies), with particular emphasis on microdeletion rearrangements and unbalanced structural rearrangements.
DNA was isolated from cultured amniocytes, cultured chorionic villus samples, or postnatal blood specimens for samples previously confirmed by either microscopic karyotype analysis or FISH as carrying chromosomal rearrangements. The results of these investigations were blinded prior to further analysis by array-CGH. Clones exceeding experimental thresholds were identified by Bluefuse Software (BlueGnome).
DNA labelling and array hybridization
Briefly, 600 ng of test DNA were labelled with the cyanine Cy3, and 600 ng of the control DNA with the cyanine Cy5 (CGH 1). In order to conduct a dye-swap experiment, reverse labelling (test DNA with Cy5, and control DNA with Cy3) was also performed (CGH 2). Genomic DNA was labelled with Cy3- or Cy5-dCTP by random prime labelling (BioPrime Genomic labelling System,). After co-precipitation with salmon sperm DNA and human CotI DNA (Roche), labelled probe mixtures of CGH1 and CGH2 were denaturated at 72 °C for 10 min, preannealed at 37 °C for 30 min and then simultaneously applied to area ‘up’ and ‘down’, respectively. Slides were scanned with ScanArray (Perkin-Elmer) and analyzed with Bluefuse software (Bluegnome).
Image acquisition and data analysis
Arrays were scanned using a ScanArray (Perkin-Elmer) and the acquired images were analyzed using Bluefuse software (Bluegnome). Data analysis was performed setting the treashold level to 0,299. Value 0 means no gain or loss of DNA. Values > 0,299 correspond to a DNA duplication, values < 0,299 to a deletion.
Results
We designed a targeted-array called GOLD (Gain or Loss Detection) Chip consisting of 900 FISH-mapped non-overlapping BAC clones spanning the whole genome to enhance the coverage of 66 unique human genomic regions involved in well known microdeletion/microduplication syndromes (Table 1).
We identified multiple clones for each genomic locus. Loci covered by only a single clone may show dosage variation because of the intrinsic technical variability of the procedure or because of polymorphic repetitive sequences inherent to the specific locus. The use of multiple clones provides confidence in the results. All polymorphic clones identified were discarded from the microarray.
In order to evaluate clinical diagnostic applicability of GOLDChip, analytical validity was carried-out via retrospective analysis of DNA isolated from a series of cytogenetically normal amniocytes and cytogenetically abnormal DNA obtained from cultured amniocytes, peripheral blood or cell lines.
We recruited from different centres DNA samples (n = 27) corresponding to pathologies with significant frequence (>1/1000), clinical relevance (Cri du Chat syndrome, Williams syndrome, Prader Willi/Angelman syndromes, Smith-Magenis syndrome, DiGeorge syndrome, Miller-Diecker syndrome, chromosomes 13, 18 and 21 trisomies), and clear known karyotypes. Some pathologies were evaluated on cellular lines commercially available (n = 20).
Table 2 represents the main information of the 47 DNA analyzed (OMIM, frequencies, gene map locus or Karyotype, gain or loss of DNA). We identified all the chromosomal rearrangements previously characterized and excluded false negative clones through comparison of clinical known samples.
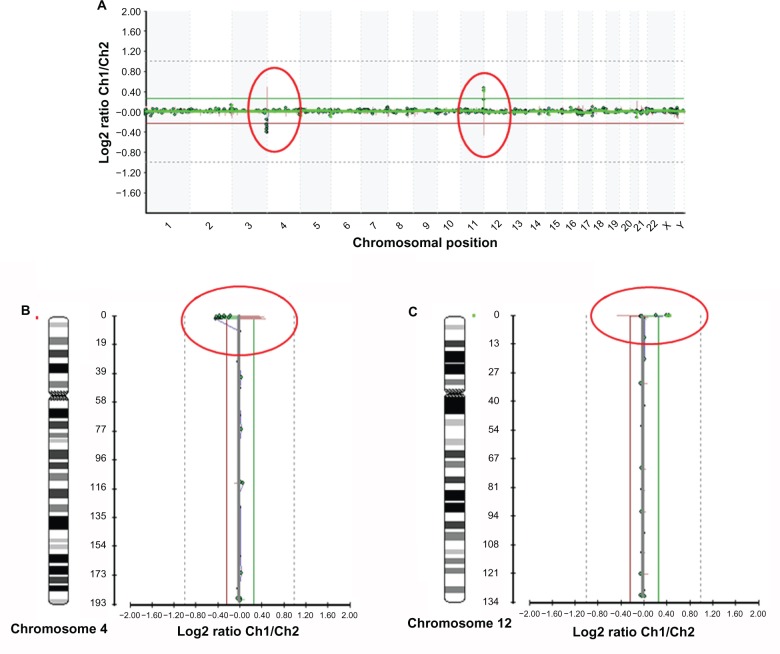
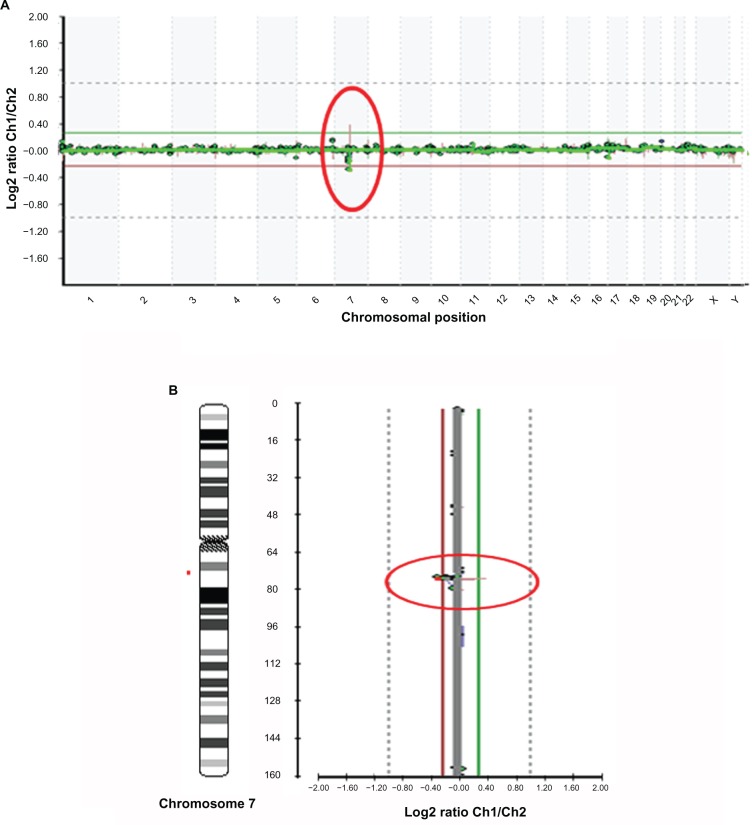
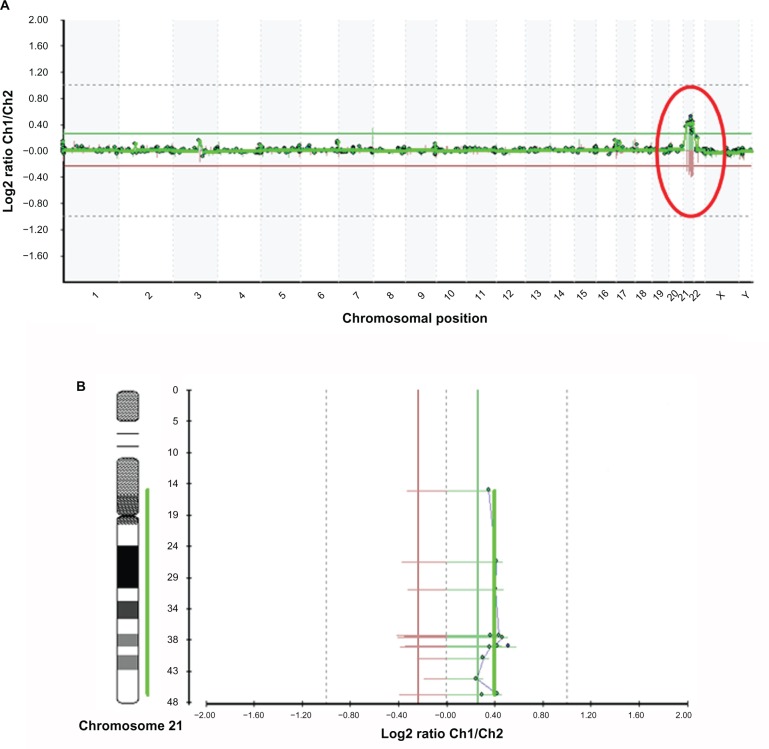
Chromosomal abnormalities included microdeletions [del4p16.3, del7q11.2] and aneuploidies (trisomy 21) respectively associated to Wolf-Hirschhorn, Williams and Down syndrome are shown in Figures 2, 3 and 4.
Figure 2.
A. Microarray profile of Wolf-Hirschhorn syndrome (del 4p16.3) in a patient also carrying dup12p13.33, identified by microarray-based comparative genomic hybridization (array CGH) using GOLD Chip. Each clone represented on the array is arranged along the x-axis according to its location on the chromosome with the most distal/telomeric p-arm clones on the left and the most distal/telomeric q-arm clones on the right. The green lines represent the log2 ratios from the first experiment (patient Cy3/control Cy5), whereas the red plots represent the log2 ratios obtained from the second experiment in which the dyes have been reversed (patient Cy5/control Cy3). B and C. Microarray profiles of chromosomes 4 (B) and 12 (C). The green line represents the patient-to-control fluorescence intensity ratios (gain of DNA); the red line represents dye-reversed control-to-patient fluorescence ratios (loss of DNA). Deletion in 4p16.3 is represented from BAC clones C4P-023, C4P-022, C4P-021, C4P-020, C4P-018, C4P-016, C4P-014, C4P-013, C4P-011, C4P-010, C4P-008; duplication in 12p13.33 is represented from BAC clones C12P-010 and C12P-009.
Figure 3.
A. Microarray profile for Williams syndrome (7q11.2) identified by microarray-based comparative genomic hybridization (array CGH) using GOLD Chip. Each clone represented on the array is arranged along the x-axis according to its location on the chromosome with the most distal/telomeric p-arm clones on the left and the most distal/telomeric q-arm clones on the right. The green lines represent the log2 ratios from the first experiment (patient Cy3/control Cy5), whereas the red plots represent the log2 ratios obtained from the second experiment in which the dyes have been reversed (patient Cy5/control Cy3). B. Microarray profiles of chromosome 7. The green line represents the patient-to-control fluorescence intensity ratios (gain of DNA); the red line represents dye-reversed control-to-patient fluorescence ratios (loss of DNA). Deletion in 7q11.2 is represented from BAC clones C7Q-015 and C7Q-012.
Figure 4.
A. Microarray profile for Down syndrome (47,XX+21) identified by microarray-based comparative genomic hybridization (array CGH) using GOLD Chip. Each clone represented on the array is arranged along the x-axis according to its location on the chromosome with the most distal/telomeric p-arm clones on the left and the most distal/telomeric q-arm clones on the right. The green lines represent the log2 ratios from the first experiment (patient Cy3/control Cy5), whereas the red plots represent the log2 ratios obtained from the second experiment in which the dyes have been reversed (patient Cy5/control Cy3). B. Microarray profiles of chromosome 21. The green line represents the patient-to-control fluorescence intensity ratios (gain of DNA); the red line represents dye-reversed control-to-patient fluorescence ratios (loss of DNA). Some clones appear in the same horizontal line because they are overlapping. In the image are shown only the 13 clones that, after data analysis and normalization, were selected by Bluefuse considering the parameters of the experiments. Some regions of chromosome 21 (p-arm) are not covered by BAC clones.
In the Wolf-Hirschhorn sample (Fig. 2), deletion in 4p16.3 was identified by the loss of copy number of 11 BAC clones on chromosome 4 (C4P-023, C4P-022, C4P-021, C4P-020, C4P-018, C4P-016, C4P-014, C4P-013, C4P-011, C4P-010, C4P-008). Moreover, duplication in 12p13.33 was identified by BAC clones C12P-010 and C12P-009.
Deletion in Williams syndrome (del7q11.2) (Fig. 3) is represented from BAC clones C7Q-015 and C7Q-012, and a-CHG analysis of the sample with trisomy 21 (Fig. 4) had a gain in copy number of clones corresponding to chromosome 21.
Discussion
Microarrays have been successfully constructed for all or parts of the human genome. Snijders et al21 constructed one of the first ‘‘whole-genome’’ arrays using 2,400 BAC clones to scan for genome-wide copy-number alterations. More recently different arrays have been developed, consisting of overlapping clones spanning the entire genome,22 covering the subtelomeric regions23 or focusing on specific chromosomes and chromosome regions.24–29
These and other arrays constructed for research purposes are designed to screening chromosomal segments or the whole-genome for DNA gains or losses at unprecedented resolution.
Thus, whole genome arrays are likely to generate data that are difficult to be interpreted and are subjected to multiple FISH verifications per patient. Furthermore, alterations in regions of the genome that do not have established clinical relevance may be difficult to interpret in a clinical setting. Moreover, with a whole-genome approach, polymorphisms are expected to be abundant. This assumption is based on the data from subtelomeric FISH analysis revealing many telomeric alterations with no apparent clinical significance.30 Supporting this, two recent studies have reported the prevalence of large-scale copy-number variations (LCVs) throughout the human genome.31,32
The adoption of such arrays into clinical diagnostics is unwise and may lead to many false positive diagnoses that necessitate expensive follow-up confirmatory tests by FISH or other methods, additional blood draws from unaffected relatives to determine the segregation of these deletions, duplications, or polymorphisms and unnecessary anxiety for the families.
Array-CGH hybridization results for single clones that show dosage difference would need to be examined and each clinical case may result in a mini-research project. Thus, the genome-wide dense arrays that are currently available for research use are not appropriate to use in a clinical setting.
A diagnostically useful microarray must accurately detect the chromosome abnormalities assayed, and must provide interpretable results. Additionally, clinical confidence must be established using microarrays that interrogate regions of known clinical relevance. Respecting the mentioned rules for microarray with diagnostic use, we designed a targeted array called GOLD (Gain or Loss Detection) Chip consisting of 900 FISH mapped non-overlapping BAC clones spanning the whole genome, to enhance the coverage of about 66 unique human genomic regions involved in microdeletion/microduplication syndromes (Table 1).
Targeted-arrays were designed for specific regions of the genome to study specific chromosome or chromosomal segment or to identify and evaluate specific DNA dosage abnormalities in individuals with suspected microdeletion syndromes or unbalanced subtelomeric rearrangements.24,33–35
Our targeted microarray has a crucial goal in medical practice, to provide clinically useful results for diagnosis, genetic counselling, prognosis, and clinical management of unbalanced cytogenetic abnormalities. However, it is well know that BAC-based CGH microarray has some drawbacks. Since its resolving power depends on the number of clones printed and the genomic distance between the clones, a microdeletion or microduplication may be overlooked if the clones printed are less dense. Furthermore, CGH microarray cannot detect balanced rearrangements, polyploidies and low mosaics.36 An alternative is offered by synthetic oligonucleotides microarrays, for which the exact sequence and length for each element on the arrays is known. For PCR amplified BACs, this is not the case since the amplification procedure is not linear and is variable for each amplification round.
The odds are that the array-CGH field is evolving towards high resolution oligonucleotide array-CGH for the measurement of chromosomal copy number changes in human genetics and cancer, analogous to the way cDNA arrays for expression profiling have been replaced by oligonucleotide arrays. For specific applications there will still be a place for BAC arrays, like in the case of methylation studies.37
At this time our results provide evidence that our BAC array can be used for the identification of the most commons microdeletion syndromes and common aneuploidies. Probably, it has the potential to replace karyotyping for prenatal cytogenetic analysis, but at the same time a deep clinical trial is strongly required to confirm sensitivity and specificity in clinical operating conditions, to establish guide lines to array-CGH uses in prenatal diagnosis.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors report no conflicts of interest.
References
- 1.Caspersson T, Farber S, Foley GE, et al. Chemical differentiation along metaphase chromosomes. Exp Cell Res. 1968;49:219–22. doi: 10.1016/0014-4827(68)90538-7. [DOI] [PubMed] [Google Scholar]
- 2.Shaffer LG, Bejjani BA. A cytogeneticist’s perspective on genomic microarrays. Hum Reprod Update. 2004;10:221–6. doi: 10.1093/humupd/dmh022. [DOI] [PubMed] [Google Scholar]
- 3.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 4.Cirigliano V, Voglino G, Canadas MP, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR. Assessment on 18,000 consecutive clinical samples. Mol Hum Reprod. 2004;10:839–46. doi: 10.1093/molehr/gah108. [DOI] [PubMed] [Google Scholar]
- 5.Mann K, Fox SP, Abbs SJ, et al. Development and implementation of a new rapid aneuploidy diagnostic service within the UK National Health Service and implications for the future of prenatal diagnosis. Lancet. 2001;358:1057–61. doi: 10.1016/S0140-6736(01)06183-9. [DOI] [PubMed] [Google Scholar]
- 6.Mansfield ES. Diagnosis of down syndrome and other aneuploidies using quantitative polymerase chain reaction and small tandem repeat polymorphisms. Hum Mol Genet. 1993;2:43–50. doi: 10.1093/hmg/2.1.43. [DOI] [PubMed] [Google Scholar]
- 7.Klinger K, Landes G, Shook D, et al. Rapid detection of chromosome aneuploidies in uncultured amniocytes by using fluorescence in situ hybridization (FISH) Am J Hum Genet. 1992;51:55–65. [PMC free article] [PubMed] [Google Scholar]
- 8.Kallioniemi A, Kallioniemi OP, Sudar D, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992 Oct 30;258(5083):818–21. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 9.Pinkel D, Segraves R, Sudar D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 10.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 11.Shaw Smith C, Redon R, Rickman L, et al. Microarray based comparative genomic hybridisation (arrayCGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–8. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vissers LE, de Vries BB, Osoegawa K, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73:1261–70. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bejjani BA, Saleki R, Ballif BC, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: is less more? Am J Med Genet A. 2005;134:259–67. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- 14.Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 15.Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 16.Toujani S, Dessen P, Ithzar N, et al. High resolution genome-wide analysis of chromosomal alterations in Burkitt’s lymphoma. PLoS One. 2009 Sep 17;4(9):e7089. doi: 10.1371/journal.pone.0007089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaffer LG, Bejjani BA, Torchia B, Kirkpatrick S, Coppinger J, Ballif BC. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am J Med Genet C Semin Med Genet. 2007 Nov 15;145C(4):335–45. doi: 10.1002/ajmg.c.30152. Review. [DOI] [PubMed] [Google Scholar]
- 18.Rickman L, Fiegler H, Shaw-Smith C, et al. Prenatal detection of unbalanced chromosomal rearrangements by array CGH. Med Genet. 2006;43(4):353–61. doi: 10.1136/jmg.2005.037648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaeffer AJ, Chung J, Heretis K, Wong A, Ledbetter DH, Lese Martin C. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet. 2004;74:1168–74. doi: 10.1086/421250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiegler H, Carr P, Douglas EJ, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–74. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 21.Snijders AM, Nowak N, Segraves R, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet. 2001;29:263–4. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 22.Ishkanian AS, Malloff CA, Watson SK, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nature Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 23.Veltman JA, Schoenmakers EF, Eussen BH, et al. High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet. 2002;70:1269–76. doi: 10.1086/340426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu W, Ballif BC, Kashork CD, Heilstedt HA, et al. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-characterized 1p36 deletions. Hum Mol Genet. 2003;12:2145–52. doi: 10.1093/hmg/ddg230. [DOI] [PubMed] [Google Scholar]
- 25.Locke DP, Segraves R, Nicholls RD, et al. BAC microarray analysis of 15q11–q13 rearrangements and the impact of segmental duplications. J Med Genet. 2004;41:175–82. doi: 10.1136/jmg.2003.013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Veltman JA, Yntema HG, Lugtenberg D, et al. High resolution profiling of X chromosomal aberrations by array comparative genomic hybridisation. J Med Genet. 2004;41:425–32. doi: 10.1136/jmg.2004.018531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinkel D, Segraves R, Sudar D, Clark S, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–11. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 28.Bruder CE, Hirvela C, Tapia-Paez I, et al. High resolution deletion analysis of constitutional DNA from neurofibromatosis type 2 (NF2) patients using microarray-CGH. Hum Mol Genet. 2001;10:271–82. doi: 10.1093/hmg/10.3.271. [DOI] [PubMed] [Google Scholar]
- 29.Buckley PG, Mantripragada KK, Benetkiewicz M, et al. A full-coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum Mol Genet. 2002;11:3221–9. doi: 10.1093/hmg/11.25.3221. [DOI] [PubMed] [Google Scholar]
- 30.Ballif BC, Sulpizio SG, Lloyd RM, et al. The clinical utility of enhanced subtelomeric coverage in array CGH. Am J Med Genet Part A. 2007;143A:1850–7. doi: 10.1002/ajmg.a.31842. [DOI] [PubMed] [Google Scholar]
- 31.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 32.Sebat J, Lakshmi B, Troge J, Alexander J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 33.Van Buggenhout G, Melotte C, Dutta B, et al. Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet. 2004;41:691–8. doi: 10.1136/jmg.2003.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw CJ, Shaw CA, Yu W, et al. Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet. 2004;41:113–9. doi: 10.1136/jmg.2003.012831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bejjani BA, Saleki R, Ballif BC, et al. Use of targeted array-based CGH for the clinical diagnosis of chromosomal imbalance: Is less more? Am J Med Genet A. 2005;134:259–67. doi: 10.1002/ajmg.a.30621. [DOI] [PubMed] [Google Scholar]
- 36.Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet. 2004 Dec;66(6):488–95. doi: 10.1111/j.1399-0004.2004.00322.x. Review. [DOI] [PubMed] [Google Scholar]
- 37.Ching TT, Maunakea AK, Junn P, et al. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nature Genet. 2005;37:645–51. doi: 10.1038/ng1563. [DOI] [PubMed] [Google Scholar]