Abstract
Bacteria predominantly reside in microbial communities known as biofilms, where cells are encapsulated and protected by the extracellular matrix (ECM). While all biofilm cells benefit from the ECM, only a subgroup of cells carries the burden of producing this public good. This dilemma provokes the question of how these cells balance the cost of ECM production. Here we show that ECM producing cells have a higher gene expression response to quorum sensing (QS) signals, which can lead to a private benefit. Specifically, we constructed a synthetic quorum-sensing system with designated “Sender” and “Receiver” cells in Bacillus subtilis. This synthetic QS system allowed us to uncouple and independently investigate ECM production and QS in both biofilms and single cells. Results revealed that ECM production directly enhances the response to QS signals, which may offset the cost of ECM production.
Introduction
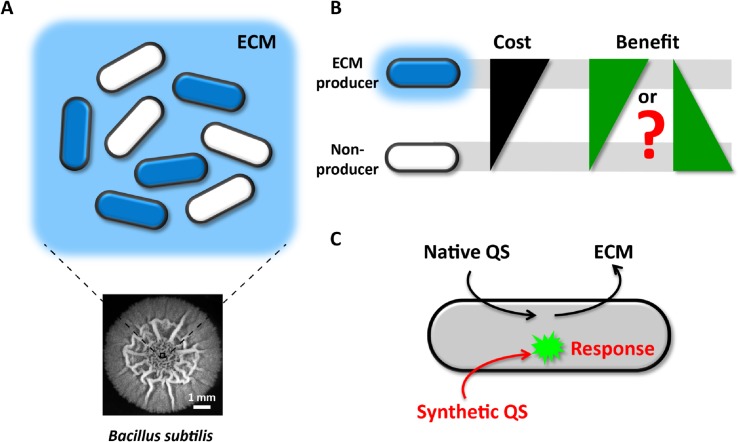
Even though bacteria are unicellular organisms, they predominantly reside in structured communities known as biofilms [1,2]. One of the defining characteristics of biofilms is the presence of an extracellular matrix that encapsulates all cells within the community and provides the biofilm with structural integrity [3,4]. The major ECM constituents of most biofilms, for example those formed by Bacillus subtilis, are polysaccharides and amyloid-like protein filaments [5–8]. It has been shown that only a subpopulation of cells synthesizes and secretes these ECM components in B. subtilis biofilms [9,10] (Fig 1A). Those ECM producer cells are clearly burdened with a cost at a time of environmental stress and high cell density. This cost is evident by the observation that an ECM-deficient strain can outcompete the ECM producing strain in a mixed culture [11]. However, all cells within the biofilm, even those that do not contribute to ECM production, are believed to benefit from the ECM protection [12–14]. This provokes the question of how the subpopulation of cells that are burdened with ECM production can be sustained within the biofilm community [11,15–19]. It has been argued that expression of ECM by a subpopulation of cells may constitute a primitive form of altruism in bacteria [10] (Fig 1B). Here we investigated whether ECM producers could also enjoy a private benefit, countering the cost of this public good production (Fig 1B).
Fig 1. Cost-benefit dilemma for ECM producers in biofilms.
(A) In biofilms, only a subgroup of cells produces and secretes ECM. However, once secreted, ECM benefits all cells, including non-producers. The bottom picture is the top view of a three-day-old biofilm. (B) ECM producers carry a higher cost than non-producers due to production of ECM. Will these producers accordingly get a higher benefit from ECM or not? (C) An orthogonal (inert) synthetic QS system was constructed in B. subtilis. This synthetic QS system does not genetically regulate ECM production, thus enabling us to independently study how the response to QS signals may be affected by ECM production.
How could ECM-producing cells obtain a private benefit? Here we focused on whether ECM expression may affect the response to quorum sensing signals. The response to QS signals can provide a private benefit by inducing intracellular metabolic enzyme expression to better cope with increasing cell density and environmental changes [20–23]. Interestingly, a recent in vitro study indicated that quorum sensing molecules could transiently associate with the ECM [24]. Therefore, QS signals may be locally concentrated by the ECM, increasing their availability to cells. However, it is unclear if the transient association of QS signals and the ECM affects the response of ECM-producing cells in vivo. Here we investigated whether ECM producing cells may locally concentrate QS signals and thereby obtain a private benefit through an enhanced QS response, by quantitatively measuring a synthetic QS signaling system in biofilm communities and single cells.
ECM expression and QS are tightly coupled and cannot be perturbed independently, since native QS regulates ECM expression [25–31] (Fig 1C). Therefore, we constructed an orthogonal (inert) synthetic QS system in B. subtilis to study how the response to QS signals may be affected by ECM production (Fig 1C). This system consists of dedicated “Sender” and “Receiver” cells that can secrete or respond to QS signals, respectively. This synthetic QS system provides the unique opportunity to study QS without interfering with native B. subtilis QS or ECM production (Fig 1C). Investigating synthetic QS in the background of ECM gene deletion strains revealed that ECM expressing cells are more responsive to QS signals. Additionally, we find that the ECM produced by cells retains QS peptides in vivo, enhancing the QS response. Finally, we confirm that results from our synthetic QS system also apply to the native QS response of B. subtilis. Our findings thus support a model where the cellular cost of ECM production can be compensated by an enhanced QS response.
Materials and Methods
Strains and growth conditions
All Bacillus subtilis strains used in this study were derived from NCIB3610 (a gift from the laboratory of Wade Winkler, University of Maryland, College Park, MD [32]), an undomesticated wild type strain. Strains are listed in S1 Table. Cells were grown in Luria-Bertani (LB) broth (EMD) or on LB agar plates at 37°C. When appropriate, antibiotics were added at the following concentration: 9 μg/mL neomycin (Fisher BioReagents), 5 μg/mL chloramphenicol (Sigma-Aldrich), 300 μg/mL spectinomycin (Sigma-Aldrich), and 100 μg/mL ampicillin (Sigma-Aldrich).
MSgg medium was used when testing cells’ response to AIP. MSgg medium containing 5 mM potassium phosphate (pH 7.0, Fisher BioReagents), 100 mM MOPS (pH 7.0, Sigma-Aldrich), 2mM MgCl2 (Fisher BioReagents), 700 μM CaCl2 (Fisher BioReagents),50 μM MnCl2 (Sigma-Aldrich), 100 μM FeCl3 (Sigma-Aldrich), 1 μM ZnCl2 (Sigma-Aldrich),2 μM thiamine (Fisher BioReagents), 0.5% glycerol (Sigma-Aldrich) and 0.5% glutamate (Sigma-Aldrich)[8].
Strain constructions
The agr system or fluorescent proteins fused to PrpsD were integrated into B. subtilis using a standard chromosomal integration method. First, target genes were generated through fusion PCR and cloned into chromosomal integration vectors. Then the resulted constructs were confirmed by direct sequencing. The agr system was PCR amplified using Phusion high-fidelity DNA polymerase (NEB) and appropriate primers from Staphylococcus epidermidis ATTC 14990. Vectors designed for integration into the gltA, amyE and sacA loci were pGlt-Kan (ECE173, constructed by R Middleton and obtained from the Bacillus Genetic Stock Center), pDL30 (a kind gift from Jonathan Dworkin, Columbia University), pSac-Cm [33] (ECE174, constructed by R Middleton and obtained from the Bacillus Genetic Stock Center) respectively. Then, vectors containing target genes were transformed into B. subtilis using a standard one-step transformation procedure [34]. Strains with targeted integration were confirmed through colony PCR with appropriate primers.
The ΔepsH strain is a kind gift from the Roberto Kolter laboratory (Harvard Medical School, Cambridge, MA) [8]. All other strains harboring epsH deletion were constructed based on this ΔepsH strain. To construct the ΔtapA operon strain, upstream and downstream regions of the tapA operon were cloned into per449, a generic integration vector constructed for integration into the gene of interest (kind gift from Wade Winkler, University of Maryland, College Park, MD). The resulting vector was then transformed into B. subtilis.
Synthesis of S. epidermidis AIP
S. epidermidis AIP was synthesized by Protein Chemistry Technology Core in University of Texas Southwestern Medical Center. The synthesis method was previously described by Otto M, et al. [35].
Biofilm formation
The biofilm formation assay is adapted from previous protocols [8]. B. subtilis strains were grown on LB agar plates with appropriate antibiotics at 37°C for overnight. For each strain, a single colony was picked and grown in 1mL LB liquid culture at 37°C for 6h until saturation. When needed, cultures from different strains were mixed together at a certain ratio and diluted 5000-fold with MSgg medium. Then 1μL culture was spotted on MSgg solid plate (1.5% agar, 3 mm thickness, dried overnight). During the imaging process, cells were incubated at 30°C.
Microscopy
Movies and snapshots were acquired with an IX71 inverted fluorescence microscope (Olympus), IX83 inverted fluorescence microscope (Olympus) or DeltaVision microscopy imaging system (GE Healthcare). Biofilm movies were taken from the bottom with a 2.5x objective lens (MPLFLN 2.5x/0.08, Olympus) to film the whole biofilm or a 10x objective lens (UPLFLN 10x/0.3, Olympus) to zoom in on certain regions. Images were taken every 40 min. Single cell movies were taken with a 100x objective lens (UPLFLN 100x/1.3, Olympus) while growing 10-fold diluted cell cultures on an MSgg pad at 30°C. Top-view of biofilms was acquired by a Retiga 2000R digital camera (QImaging) via an SZX10 fluorescent stereomicroscope (Olympus).
Image analysis
ImageJ (National Institutes of Health, http://imagej.nih.gov/ij/) and MATLAB (MathWorks) were used for image processing and analysis. The detailed image analysis methods for biofilms and single cells are described below respectively.
Biofilm image segmentation and quantification
In a mixed biofilm, regions of each strain were segmented out based on fluorescence images of constitutive markers. PrpsD fused with a fluorescent protein (CFP or mCherry) were integrated into each strain except one. For strains containing PrpsD driven fluorescent proteins, a fluorescent image was taken accordingly. Then this fluorescent image was processed using ImageJ to determine the region of the strain in two steps. First, the edge was detected using the Otsu method in auto local threshold function. Second, the fluorescence positive region was determined by the analyze particle function. After the region of each fluorescently marked strain was determined, the rest of the area was assigned to the one strain without a fluorescent marker.
After segmentation, the distance of each pixel within Receiver microcolonies to the edge of Sender microcolonies was calculated. In S4 Fig, the average P3-yfp fluorescent intensity of all pixels within every 12.9μm (10 pixels) distance was calculated and plotted against its distance from the edge of the Sender microcolony for WT Receiver and ΔepsH Receiver, respectively. The average P3-yfp fluorescence intensity of WT Receiver and ΔepsH Receiver within 12.9μm from the edge of Sender microcolony (grey region in S4 Fig) was calculated and normalized according to the P3-yfp fluorescent intensity of WT Receiver. This relative P3-yfp intensity of WT Receiver and ΔepsH Receiver was measured and the mean was calculated across different movies.
Single cell segmentation
Single cells were segmented based on phase contrast images using the analyze particle function in ImageJ or image processing and statistics toolboxes in MATLAB, as previously described [36]. When mixing WT Receiver and ECM-deficient Receiver cells together, WT Receiver cells were marked by PrpsD-cfp, which enabled us to distinguish between these two cell types.
Density measurement
Cell density was determined using bright field images as previously described and validated by A. Seminara et al. [37]. Relative cell density was defined as–log(I/I0), where I is the average image intensity in the region of a certain strain and I0 is the average image intensity in regions outside of the biofilm.
Results
Construction of a synthetic quorum sensing (QS) system in undomesticated B. subtilis
To investigate how ECM expression may affect the response to QS signals, we constructed a synthetic QS system. In particular, we transplanted the Auto-Inducing-Peptide (AIP) based QS system (agr system) from Staphylococcus epidermidis into the biofilm forming B. subtilis NCIB 3610 strain (Fig 2A). We chose the S. epidermidis QS system because of the following four main reasons: (1) S. epidermidis is a gram-positive bacteria, which increases the likelihood that this QS system could be functional in gram-positive B. subtilis cells. (2) The S. epidermidis QS system is well characterized and simple [38]. It involves only four components (AgrA-D) that are expressed from a single operon. The agrD gene encodes the AIP peptide and AgrB is responsible for posttranslational modification and export of AIP [39]. AgrC is the AIP receptor that activates AgrA, the transcription factor that promotes expression of the agr operon driven by the P2 promoter, as well as gene expression of downstream targets driven by P3 promoters [40,41]. (3) The S. epidermidis AIP is a short peptide containing 8 amino acids [35] (S1 Fig) and is similar in size to the major phr encoded QS peptides in B. subtilis that range in size from 5–6 amino acids [42,43]. (4) There is no known agr system in B. subtilis, suggesting that the transplanted QS system would not interfere with native B. subtilis QS. Therefore, transplantation of the S. epidermidis QS system into B. subtilis provides a unique opportunity to directly investigate how ECM expression may affect the QS response without interfering with native ECM production or QS.
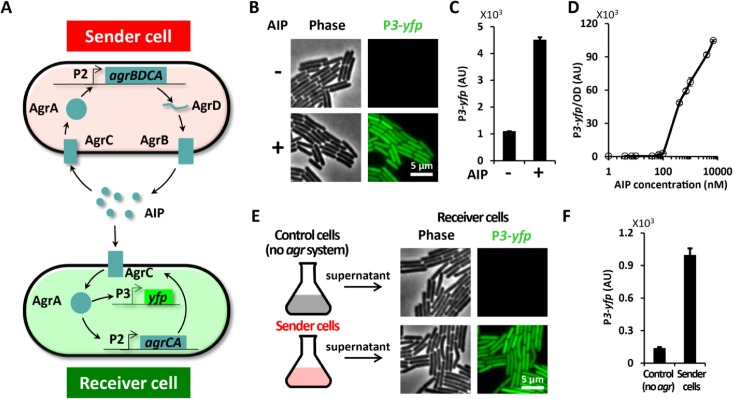
Fig 2. A synthetic quorum sensing (QS) system was built and its functionality and modularity was confirmed in B. subtilis.
(A) Diagram of the synthetic agr systems in Sender cells (top) and Receiver cells (bottom). The Sender cell contains the entire agr operon (P2-agrBDCA) and can thus both secrete and sense AIP, the autoinducing peptide. The Receiver cell, on the other hand, carries P2-agrCA and P3-yfp and thus can respond to AIP by expressing YFP. (B) Snapshots of Receiver cells grown on MSgg pads with 0 (-) or 100nM (+) chemically synthesized AIP. yfp expression from P3 is shown in green. (C) Mean P3-yfp fluorescence intensity from individual Receiver cells with 0 (-) or 100 nM (+) AIP (mean ± SEM, n = 78 cells, p<0.0001). (D) Dose response curve of Receiver cells to AIP (mean ± SEM, n = 2). (E) Images of Receiver cells grown on MSgg pad supplemented with 50% conditioned media from control cells (no agr system) or Sender cells. (F) Mean P3-yfp fluorescence intensity in Receiver cells grown with conditioned media from control cells or Sender (mean ± SEM, n = 97 cells, p<0.0001).
We began by testing the modularity and functionality of the agr system after it was transformed into B. subtilis. Specifically, we constructed two types of synthetic QS systems in B. subtilis, namely “Receiver” and “Sender” cells (Fig 2A). The Sender cells contain the entire agr operon (agrA,B,C and D) and are capable of sensing and secreting AIP. In contrast, Receiver cells can only respond to AIP, but not synthesize it as they lack the necessary genes. Receiver cells only contain the AgrC receptor and downstream AgrA transcription factor that regulates expression of the P3 promoter. Therefore, we can use Receiver cells to measure the response to AIP by monitoring expression of a fluorescent protein reporter YFP from the P3 promoter (P3-yfp).
Receiver cells can respond to chemically synthesized AIP
Quantitative fluorescence microscopy measurements confirmed that B. subtilis Receiver cells containing the synthetic QS system could indeed respond to extracellular AIP (Fig 2B). In particular, we find that addition of 100 nM chemically synthesized AIP induced a nearly 5-fold increase in P3 expression (Fig 2C). We also obtained the AIP dose response curve for P3 expression in Receiver cells (Fig 2D). Receiver cells begin to elicit a detectable response to approximately 70 nM extracellular AIP. The response increased with increasing AIP concentrations and surprisingly did not saturate even with addition of 10 μM AIP, a concentration 3 orders higher than the minimal activating concentration. We also performed two control experiments to determine adverse effects of S. epidermidis gene integrations or AIP peptide addition to B. subtilis biofilm morphology. Results show that chromosomal integration of agr genes into B. subtilis does not affect biofilm formation (S2 Fig). Furthermore, extracellular addition of 10 μM AIP does not alter B. subtilis biofilm development or morphology, suggesting that native ECM production is not affected by the presence of S. epidermidis AIP (S3 Fig). Together, these data show that the S. epidermidis agr system in Receiver cells is functional and does not interfere with native B. subtilis biofilm development.
Sender cells secrete functional AIP
Next we tested the ability of Sender cells to synthesize and secrete functional AIP. Specifically, Sender cells were grown in LB for 3 hours after which the supernatant was collected and added to Receiver cells (Fig 2E). Exposure of Receiver cells to supernatant collected from Sender cells triggered P3 expression in Receiver cells ( Fig 2E and 2F ). Therefore, Sender cells secrete sufficient functional AIP to trigger a QS response in Receiver cells. In contrast, supernatant collected from genetically unmodified B. subtilis cells did not elicit a response in Receiver cells ( Fig 2E and 2F ). This data also shows that B. subtilis cells do not express any peptide that can stimulate the agr system, further indicating the lack of interference between the synthetic and natural QS systems. Together, these data indicate that the synthetic QS is functional in both B. subtilis Sender and Receiver cells and performs as intended.
ECM enhances QS response in biofilms
Next, we investigated directly in biofilm communities whether QS signals would be affected by ECM expression. Specifically, we grew mixed biofilms that contained Sender cells together with two types of Receiver cells, where one Receiver strain was deficient in ECM production (Fig 3A). The ECM deficient Receiver strain was obtained by deleting the B. subtilis epsH gene (S1 Table). The epsH gene encodes a critical enzyme necessary for extracellular polysaccharide synthesis and it has been shown that ΔepsH cells are deficient in ECM production and biofilm development [8]. We grew mixed biofilms that were comprised of adjacent clusters of Sender and Receiver strains (Fig 3B). We then measured the QS response of Receiver cells with and without the epsH gene to AIP secreted by an adjacent cluster of Sender cells (Fig 3C). Results show that the ΔepsH strain has approximately four-fold lower QS response (P3-yfp) compared to the Receiver strain with an intact epsH gene (Fig 3D). Control experiments show that despite the reduced response to AIP, the ΔepsH Receiver strain was still functional and capable of exhibiting an AIP dependent dose response curve (S5 Fig). Furthermore, we confirmed that the difference in the response between Receiver strains was not due to differences in cell density (S6 Fig). These findings suggest that ECM deficient cells have a reduced response to AIP-mediated QS and are therefore less effective in sensing signals from distant cells.
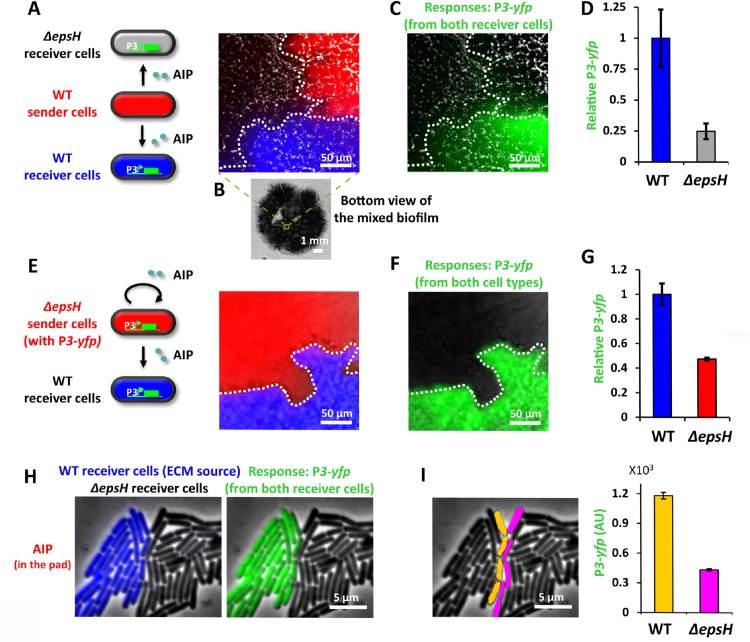
Fig 3. ECM enhances the response of producer cells to QS signal in biofilms.
(A) A local region in a mixed biofilm where clusters of WT Sender cells (red), WT Receiver cells (blue) and ΔepsH Receiver cells (grey) merge. Sender cells are marked by PrpsD-mCherry and WT Receiver cells are marked by PrpsD-cfp, where PrpsD is a constitutive promoter. In this experiment, WT Sender cells (red) secrete AIP that diffuses to nearby WT (blue) and ΔepsH (grey) Receiver cells. Both Receiver cells respond to AIP by expressing YFP. (B) Bottom view of the mixed biofilm. (C) P3-yfp overlaid on bright field image showing the same biofilm region as in A. (D) Relative mean P3-yfp intensity of WT Receiver cells and ΔepsH Receiver cells (mean ± SEM, n = 3, p<0.09). (E) A local region in a mixed biofilm where clusters of ΔepsH Sender cells (with P3-yfp, red) and WT Receiver cells (false colored as blue) merge. ΔepsH Sender cells (with P3-yfp) are marked by PrpsD-cfp. In this experiment, ΔepsH Sender cells (red) secrete AIP which induces a response in the ΔepsH Sender cells (red) themselves. Meanwhile some AIP will diffuse to nearby WT Receiver cells and trigger a response. (F) P3-yfp overlaid on bright field image showing the same biofilm region as in E. (G) Relative mean P3-yfp intensity of WT Receiver cells and ΔepsH Sender cells (with P3-yfp) (mean ± SEM, n = 3, p<0.05). (H) A snapshot of WT (blue) and ΔepsH (grey) Receiver cells growing near each other on an MSgg agar pad containing 100nM AIP. WT Receiver cells are marked by PrpsD-cfp. The response, P3-yfp, is shown in green on the right panel. (I) Response of WT (false colored as orange) and ΔepsH Receiver cells (false colored as magenta) on the interface where these two strains merge (mean ± SEM, n = 39 cells, p<0.0001).
We asked whether the above-described deficiency in QS response to signals from distant cells would be alleviated if Receiver cells were also directly the source of the AIP signal. Interestingly, we found that even if ΔepsH cells are the ones to produce the AIP, the more distant WT Receiver cells exhibited a higher QS response. Specifically, we grew mixed biofilms that contained ΔepsH Sender cells and wild type Receiver cells (Fig 3E). We find that the ΔepsH strain had approximately two-fold lower QS response compared to the wild type Receiver strain ( Fig 3F and 3G ). Even though ΔepsH Sender cells would experience a higher local AIP concentration, distant wild type Receiver cells generated a higher QS response. Therefore, even when the QS signal is not required to travel among cells, ECM deficient cells have a reduced response to locally made QS signals. These data suggest that the higher QS response of ECM producing cells constitutes a private property that is independent of whether cells respond to their own AIP signal or those expressed by distant cells.
QS related ECM benefit does not extend to directly adjacent cells
To directly test the finding that enhanced QS appears to be limited exclusively to ECM producing cells, we performed experiments at the single cell level. In particular, we wanted to determine if ECM expression could enhance the QS response of directly adjacent cells. We thus turned to experiments in mixed microcolonies that enabled quantitative measurement of the QS response in single cells (Fig 3H). Results show that the QS response of WT Receiver cells, capable of ECM production, is clearly higher compared to the response measured in the nearest physically adjacent ΔepsH Receiver cells (Fig 3I). Furthermore, the low QS response of ΔepsH Receiver cells did not decrease further as a function of increasing distance from the WT Receiver cells (S7 Fig). Together, these results indicate that enhanced QS response in ECM producing cells is indeed a private property at the single cell level that does not extend to adjacent cells.
ECM production dependent QS response also affects single cells in liquid cultures
Finally we investigated whether the reduced QS response observed in ECM deficient strains was limited to structured communities such as biofilms, or could even be observed in liquid cultures. Accordingly, we grew an equal mixture of wild type and ΔepsH Receiver strains for one hour in liquid culture that was supplemented with 1μM of chemically synthesized AIP (Fig 4A). We then imaged single cells sampled from this liquid culture. Consistent with our previous results, quantitative fluorescence microscopy showed that ECM deficient ΔepsH Receiver cells have a lower response to AIP than wild type Receiver cells, even when grown together in shaking liquid cultures (Fig 4B).
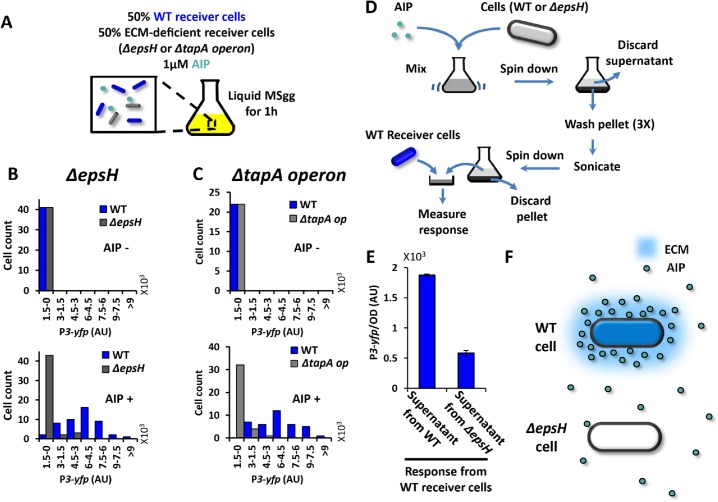
Fig 4. ECM retains AIP around ECM producer cells and enhances their response to AIP even in liquid culture.
(A) Illustration of experimental setup for B-C. ECM-deficient Receiver cells (ΔepsH or ΔtapA operon) and WT Receiver cells (PrpsD-cfp, blue) are growing together in liquid culture containing 0 or 1 μM AIP for 1h before the response (P3-yfp) is quantified. (B) Histograms of P3-yfp fluorescence intensity from ΔepsH Receiver cells and WT Receiver cells (n = 41 cells for the upper panel and n = 48 cells for the bottom panel). The expression of P3-yfp was measured for individual cells from microscopic snapshots. (C) Histograms of P3-yfp fluorescence intensity from ΔtapA operon Receiver cells and WT Receiver cells (n = 22 cells for the upper panel and n = 40 cells for the bottom panel). The expression of P3-yfp was measured for individual cells from microscopic snapshots. (D) Scheme of the experimental setup in E. (E) Response of WT Receiver cells to AIP extracted from WT cells or ΔepsH cells (mean ± SEM, n = 4, p<0.0001). (F) A diagram illustrates that WT cells concentrate AIP in their vicinity, while ΔepsH cells do not.
Could the reduced QS response be specifically linked to the deletion of the epsH gene, or perhaps be a general consequence of ECM deficiency? To address this question, we repeated the same experiment using an equal mixture of wild type and a tapA operon deletion strain (S1 Table). The tapA operon encodes another critical component of the ECM, namely proteins that can form amyloid-like protein filaments [7]. We find that similar to ΔepsH Receiver cells, the tapA operon deleted Receiver cells also exhibit a reduced QS response compared to wild type Receiver cells (Fig 4C). Therefore, regardless of whether the ECM deficiency is caused by lack of exopolysaccharides or amyloid-like protein fibers, the QS response to AIP is reduced in both types of ECM deficient cells. We also ruled out the possibility that the reduced QS response observed in ECM deficient cells was due to a general problem with protein expression by measuring the activity of a ribosomal gene promoter (PrpsD) in wild type and ECM deficient cells (S8 Fig). Collectively, these results show that the reduced QS response is not tied to the deletion of any particular gene, but appears to be caused by an overall deficiency in ECM production. Furthermore, the reduced QS response in ECM deficient strains not only arises in the context of the biofilm community or microcolonies, but appears to be a single cell level property that even applies in liquid cultures.
ECM concentrates extracellular AIP in the vicinity of ECM producing cells
Motivated by the recent in vitro study that indicated transient physical binding of QS molecules to the ECM [24], we wanted to test whether the elevated QS response we observed was due to local concentration of AIP by the ECM. To determine whether AIP accumulates near ECM producing cells, we added chemically synthesized AIP to wild type cells capable of ECM production. This culture was mixed, spun down and washed three times to eliminate unbound AIP. We then subjected the culture to mild sonication in order to release AIP that may have been retained by the ECM back into the media. In parallel, we subjected ΔepsH cells to the same protocol to serve as a control. The supernatant collected after sonication from both WT and ΔepsH cells was added to WT Receiver cells and their QS response was measured (Fig 4D). We found that WT Receiver cells elicited a higher QS response when exposed to the supernatant from WT cells compared to that obtained from ΔepsH cells (Fig 4E). This result provides a mechanistic explanation for the enhanced QS response of ECM producing cells, by indicating that the ECM concentrates AIP in the vicinity of cells and therefore increases the likelihood of binding to receptors on the membrane (Fig 4F).
The QS response positively correlates with natural cell-to-cell variation in ECM production
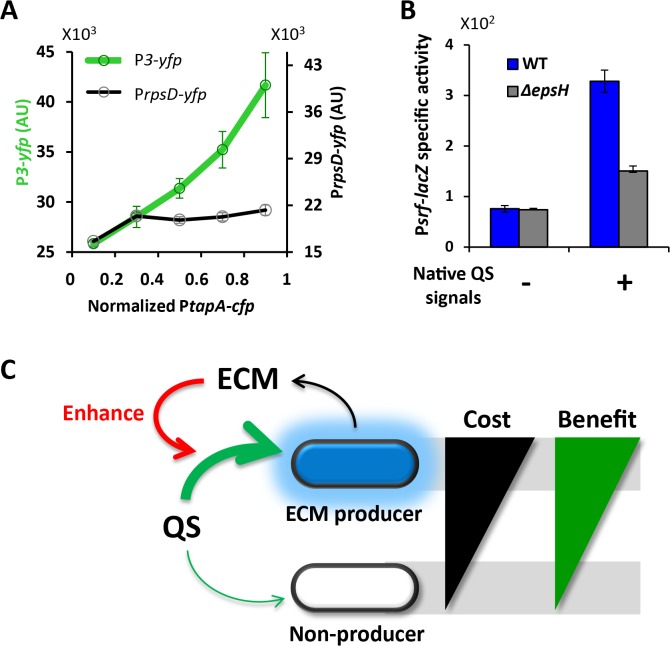
The amount of ECM produced by individual cells can vary [9,44]. Therefore, we tested whether the natural cell-to-cell variation that is inherent to ECM production would affect the synthetic QS response. Specifically, we introduced into the Receiver strain a second fluorescent protein reporter to measure the natural cell-to-cell variation in tapA expression (PtapA-cfp). Single cell analysis of the two-color reporter strain for P3 and PtapA expression shows that the QS response in Receiver cells positively correlates with the natural variation in B. subtilis ECM expression (Fig 5A). In contrast, the control experiment showed that variation in PtapA-cfp expression did not correlate with expression of the PrpsD promoter for a ribosomal gene (Fig 5A). Therefore, in addition to the reduced QS response observed in ECM gene deletion strains, response to the QS signal is also affected by the natural variation in ECM expression that is inherent to B. subtilis.
Fig 5. The observation that ECM elevates response applies to native systems.
(A) Average P3-yfp and PrpsD-yfp as a function of normalized PtapA activity (mean ± SEM, total n = 646 cells). WT Receiver cells containing PtapA-cfp were grown in liquid MSgg culture with 1μM AIP. The expression from the reporters PtapA-cfp and P3-yfp were measured for individual cells from microscopic snapshots. Relationship between PtapA-cfp and PrpsD-yfp were measured accordingly using the strain containing PtapA-cfp and PrpsD-yfp. (B) Response of WT and ΔepsH to conditioned media, which contains native quorum sensing signals that activate Psrf. Response are measured by Psrf-lacZ (mean ± SEM, n = 6, p<0.01 for native QS signals + group). (C) Model: ECM production can exclusively benefit producer cells by enhancing their response to QS, thereby compensating for the cost of ECM production.
ECM elevates response to native B. subtilis quorum sensing signals as well
We asked whether our results obtained using a synthetic QS system would also be relevant to the native QS response of B. subtilis. Accordingly, we used what is known as conditioned, or spent media that naturally contains QS signals expressed by B. subtilis cells, such as ComX, and then monitored expression of Psrf, a B. subtilis native QS responsive gene promoter [45]. We find that upon addition of conditioned media containing QS signals, WT cells display a higher expression of Psrf-lacZ compared to ΔepsH cells (Fig 5B). This result shows that the amplitude of the native QS response also depends on ECM production, indicating that results obtained with our synthetic QS system may be generally applicable to native QS systems in B. subtilis.
Discussion
The results presented in this study indicate that the cost of ECM expression may be balanced by an enhanced private response to QS signaling (Fig 5C). As the name implies, QS refers to a collective process by which individual cells can sense global population density. While QS contains information about the population, the response to a QS signal is an intracellular process that can be considered “private” to the cell. In fact, recent studies have shown that response to QS can give rise to an advantageous metabolic response in individual cells [20–23]. Therefore, our findings together with results published by other groups suggest a possible private benefit for ECM production. In particular, our data revealed that ECM expressing cells might perceive changes in global cell density earlier and respond accordingly. Earlier detection of an environmental change such as crowding can be beneficial by speeding up stress responses that, for example, induce changes in metabolic activity. Furthermore, ECM deficient cells exhibit a lower response not only to signals produced by distant cells (paracrine), but also self-made signals (autocrine). Therefore, ECM expression appears to promote both paracrine and autocrine mediated QS signaling. Balancing the cost of public good production with a private benefit would allow ECM producing cells to be more easily sustained within the biofilm and also reduce the ability of cheater cells to dominate the community.
The enhanced QS response, however, may not provide a private benefit in all conditions. For example, cells can respond to some QS signals by producing and secreting public goods, such as virulence factors and degradative enzymes [46,47]. If the ECM elevates the response to such QS signals, the response could place an additional burden on ECM producing cells. Furthermore, we note that the relationship between the ECM and QS described here may vary depending on the chemical characteristics and composition of the ECM and QS molecules. As suggested by a previous study [24], hydrophobicity can for example determine the affinity between the ECM and QS signals. If a QS molecule is hydrophilic, rather than attracting it, ECM may even prevent the QS from approaching cells. Therefore, the chemical nature of QS molecules and/or the composition of the ECM may provide bacterial cells with an additional means to tune their QS response.
In addition to serving as a tool to interrogate the relationship between ECM expression and QS, the synthetic QS system we developed here has the advantage of functioning in an undomesticated biofilm community. Therefore, this synthetic QS system can in the future be utilized to investigate various other questions regarding biofilm development. Specifically, biofilm communities are comprised of distinct cell types, and this cellular differentiation appears to be spatially organized. It has been suggested that QS plays a critical role in the spatial organization of cellular differentiation. Therefore, the synthetic QS system presented here may aid in uncovering the principles that govern the spatial organization of cell types within biofilms.
Supporting Information
It has a thiolester linkage between carboxyl-terminus and the middle cysteine.
(TIF)
WT cells were grown on MSgg plates with 0 or 10 μM AIP for 3 days. Then the bright field image was taken from the top and compared to ΔepsH cells grown on MSgg plates for 3 days.
(TIF)
Four-day-old biofilm structure of control strain (no agr system integrated) and Sender strain (with P3-yfp).
(TIF)
(TIF)
Dose-response curve of ΔepsH Receiver cells to AIP (mean ± SEM, n = 2)
(TIF)
(A) A local region in a mixed biofilm where clusters of Sender cells, ΔepsH Receiver cells and WT Receiver cells merge. Sender cells are marked by PrpsD-mCherry (red) and WT Receiver cells are marked by PrpsD-cfp (blue), where PrpsD is a constitutive promoter. This is the same figure as Fig 3B. (B) Bright field image of the same biofilm region as in A. (C) Relative cell density (–log(I/I0)) of WT Receiver cell clusters and ΔepsH Receiver cell clusters (mean ± SEM, n = 3).
(TIF)
(A) ΔepsH Receiver cells grown nearby WT cells (PrpsD-cfp, blue) on the MSgg agar pad containing 100nM AIP. ΔepsH Receiver cells with different distance from the border of WT cells are false colored with magenta (layer 1), cyan (layer 2) and yellow (layer 3). (B) Response (P3-yfp) of these three layers of ΔepsH Receiver cells (mean ± SD, n = 49 cells).
(TIF)
(A) Snapshot of WT and ΔepsH cells grow in MSgg liquid culture. PrpsD-cfp is shown in green. (B) P3-yfp fluorescence intensity in WT and ΔepsH cells (mean ± SEM, n = 20 cells).
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank J Garcia-Ojalvo, M. Asally, T. Cagatay and members of the G.M.S. laboratory for comments on the manuscript; W. Winkler, R. Kolter and J. Dworkin for kindly providing bacterial strains and plasmids; members of the K. Pogliano laboratory for help with β-galactosidase assay, use of microplate reader and sonicator.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the National Institutes of Health, National Institute of General Medical Sciences Grant R01 GM088428 (http://www.nigms.nih.gov) and the National Science Foundation Grant MCB-1450867 (http://www.nsf.gov), both to GMS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. O’Toole G, Kaplan HB, Kolter R. Biofilm formation as microbial development. Annu Rev Microbiol. 2000;54: 49–79. 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- 2. Bryers JD. Medical biofilms. Biotechnol Bioeng. 2008;100: 1–18. 10.1002/bit.21838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005/01/11 ed. 2005;13: 20–26. 10.1016/j.tim.2004.11.006 [DOI] [PubMed] [Google Scholar]
- 4. Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. Nature Publishing Group; 2010;8: 623–33. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 5. Romero D, Vlamakis H, Losick R, Kolter R. Functional analysis of the accessory protein TapA in Bacillus subtilis amyloid fiber assembly. J Bacteriol. 2014;196: 1505–13. 10.1128/JB.01363-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107: 2230–4. 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006/01/25 ed. 2006;59: 1229–1238. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- 8. Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R, González-Pastor JE. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001/09/27 ed. 2001;98: 11621–6. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008/04/03 ed. 2008;22: 945–53. 10.1101/gad.1645008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2008/12/05 ed. 2009;33: 152–163. 10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- 11. Van Gestel J, Weissing FJ, Kuipers OP, Kovács AT. Density of founder cells affects spatial pattern formation and cooperation in Bacillus subtilis biofilms. ISME J. 2014;8: 2069–79. 10.1038/ismej.2014.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rainey PB, Rainey K. Evolution of cooperation and conflict in experimental bacterial populations. 2003;425: 72–74. 10.1038/nature01942.1 [DOI] [PubMed] [Google Scholar]
- 13. Borenstein DB, Meir Y, Shaevitz JW, Wingreen NS. Non-local interaction via diffusible resource prevents coexistence of cooperators and cheaters in a lattice model. PLoS One. 2013;8: e63304 10.1371/journal.pone.0063304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8: 15–25. 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nadell CD, Bassler BL. A fitness trade-off between local competition and dispersal in Vibrio cholerae biofilms. Proc Natl Acad Sci U S A. 2011;108: 14181–5. 10.1073/pnas.1111147108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brockhurst M a, Buckling A, Gardner A. Cooperation peaks at intermediate disturbance. Curr Biol. 2007;17: 761–5. 10.1016/j.cub.2007.02.057 [DOI] [PubMed] [Google Scholar]
- 17. Xavier JB, Foster KR. Cooperation and conflict in microbial biofilms. Proc Natl Acad Sci USA. 2007;104: 876–81. 10.1073/pnas.0607651104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Drescher K, Nadell CD, Stone H a, Wingreen NS, Bassler BL. Solutions to the public goods dilemma in bacterial biofilms. Curr Biol. Elsevier Ltd; 2014;24: 50–5. 10.1016/j.cub.2013.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schluter J, Nadell CD, Bassler BL, Foster KR. Adhesion as a weapon in microbial competition. ISME J. Nature Publishing Group; 2015;9: 139–49. 10.1038/ismej.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dandekar A a, Chugani S, Greenberg EP. Bacterial quorum sensing and metabolic incentives to cooperate. Science. 2012;338: 264–6. 10.1126/science.1227289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vivant A-L, Garmyn D, Gal L, Piveteau P. The Agr communication system provides a benefit to the populations of Listeria monocytogenes in soil. Front Cell Infect Microbiol. 2014;4: 1–7. 10.3389/fcimb.2014.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schuster M, Lostroh CP, Ogi T, Greenberg EP. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185: 2066–79. 10.1128/JB.185.7.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao K, Li Y, Yue B, Wu M. Genes as early responders regulate quorum-sensing and control bacterial cooperation in Pseudomonas aeruginosa. PLoS One. 2014;9: e101887 10.1371/journal.pone.0101887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Seviour T, Hansen SH, Yang L, Yau YH, Wang VB, Stenvang MR, et al. Functional Amyloids Keep Quorum Sensing Molecules in Check. J Biol Chem. 2015; 10.1074/jbc.M114.613810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50: 101–104. 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- 26. Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190: 2527–36. 10.1128/JB.01756-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hammer BK, Bassler BL. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate cyclic dimeric GMP levels to control biofilm formation. J Bacteriol. 2009;191: 169–77. 10.1128/JB.01307-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shank EA, Kolter R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr Opin Microbiol. 2011/10/26 ed. Elsevier Ltd; 2011;14: 741–747. 10.1016/j.mib.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lopez D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2008/12/31 ed. 2009;106: 280–285. 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. López D, Vlamakis H, Losick R, Kolter R, Lopez D. Paracrine signaling in a bacterium. Genes Dev. 2009/07/17 ed. 2009;23: 1631–1638. 10.1101/gad.1813709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol. 2007;189: 5383–6. 10.1128/JB.00137-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Irnov I, Winkler WC. A regulatory RNA required for antitermination of biofilm and capsular polysaccharide operons in Bacillales. Mol Microbiol. 2010;76: 559–75. 10.1111/j.1365-2958.2010.07131.x [DOI] [PubMed] [Google Scholar]
- 33. Middleton R, Hofmeister A. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid. 2004;51: 238–45. 10.1016/j.plasmid.2004.01.006 [DOI] [PubMed] [Google Scholar]
- 34. Jarmer H, Berka R, Knudsen S, Saxild HH. Transcriptome analysis documents induced competence of Bacillus subtilis during nitrogen limiting conditions. FEMS Microbiol Lett. 2002;206: 197–200. 10.1111/j.1574-6968.2002.tb11009.x [DOI] [PubMed] [Google Scholar]
- 35. Otto M, Süßmuth R, Jung G, Götz F, Süssmuth R. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 1998;424: 89–94. 10.1016/S0014-5793(98)00145-8 [DOI] [PubMed] [Google Scholar]
- 36. Süel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440: 545–50. 10.1038/nature04588 [DOI] [PubMed] [Google Scholar]
- 37. Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, et al. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A. 2012/01/11 ed. 2012;109: 1116–1121. 10.1073/pnas.1109261108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42: 541–64. 10.1146/annurev.genet.42.110807.091640 [DOI] [PubMed] [Google Scholar]
- 39. Zhang L, Gray L, Novick RP, Ji G. Transmembrane topology of AgrB, the protein involved in the post-translational modification of AgrD in Staphylococcus aureus. J Biol Chem. 2002;277: 34736–42. 10.1074/jbc.M205367200 [DOI] [PubMed] [Google Scholar]
- 40. Novick RP, Projan SJ, Kornblum J, Ross HF, Ji G, Kreiswirth B, et al. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248: 446–58. Available: http://www.ncbi.nlm.nih.gov/pubmed/7565609 [DOI] [PubMed] [Google Scholar]
- 41. Novick RP, Ross HF, Projan SJ, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12: 3967–75. Available: http://www.ncbi.nlm.nih.gov/pubmed/413679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pottathil M, Lazazzera BA. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front Biosci. 2003;8: d32–45. 10.2741/913 [DOI] [PubMed] [Google Scholar]
- 43. Solomon JM, Lazazzera B a, Grossman a D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10: 2014–2024. 10.1101/gad.10.16.2014 [DOI] [PubMed] [Google Scholar]
- 44. Lopez D, Kolter R. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol Rev. 2009/12/25 ed. 2010;34: 134–149. 10.1111/j.1574-6976.2009.00199.x [DOI] [PubMed] [Google Scholar]
- 45. Magnuson R, Solomon J, Grossman a D. Biochemical and genetic characterization of a competence pheromone from B. subtilis. Cell. 1994;77: 207–16. Available: http://www.ncbi.nlm.nih.gov/pubmed/8168130 [DOI] [PubMed] [Google Scholar]
- 46. Rutherford ST, Bassler BL. Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2: 1–26. 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol Microbiol. 2005;57: 1159–74. 10.1111/j.1365-2958.2005.04749.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
It has a thiolester linkage between carboxyl-terminus and the middle cysteine.
(TIF)
WT cells were grown on MSgg plates with 0 or 10 μM AIP for 3 days. Then the bright field image was taken from the top and compared to ΔepsH cells grown on MSgg plates for 3 days.
(TIF)
Four-day-old biofilm structure of control strain (no agr system integrated) and Sender strain (with P3-yfp).
(TIF)
(TIF)
Dose-response curve of ΔepsH Receiver cells to AIP (mean ± SEM, n = 2)
(TIF)
(A) A local region in a mixed biofilm where clusters of Sender cells, ΔepsH Receiver cells and WT Receiver cells merge. Sender cells are marked by PrpsD-mCherry (red) and WT Receiver cells are marked by PrpsD-cfp (blue), where PrpsD is a constitutive promoter. This is the same figure as Fig 3B. (B) Bright field image of the same biofilm region as in A. (C) Relative cell density (–log(I/I0)) of WT Receiver cell clusters and ΔepsH Receiver cell clusters (mean ± SEM, n = 3).
(TIF)
(A) ΔepsH Receiver cells grown nearby WT cells (PrpsD-cfp, blue) on the MSgg agar pad containing 100nM AIP. ΔepsH Receiver cells with different distance from the border of WT cells are false colored with magenta (layer 1), cyan (layer 2) and yellow (layer 3). (B) Response (P3-yfp) of these three layers of ΔepsH Receiver cells (mean ± SD, n = 49 cells).
(TIF)
(A) Snapshot of WT and ΔepsH cells grow in MSgg liquid culture. PrpsD-cfp is shown in green. (B) P3-yfp fluorescence intensity in WT and ΔepsH cells (mean ± SEM, n = 20 cells).
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.