Abstract
Docosahexaenoic acid (DHA, 22:6n-3) is a unique polyunsaturated fatty acid particularly abundant in nerve cell membrane phospholipids. DHA is a pleiotropic molecule that, not only modulates the physicochemical properties and architecture of neuronal plasma membrane, but it is also involved in multiple facets of neuronal biology, from regulation of synaptic function to neuroprotection and modulation of gene expression. As a highly unsaturated fatty acid due to the presence of six double bonds, DHA is susceptible for oxidation, especially in the highly pro-oxidant environment of brain parenchyma. We have recently reported the ability of DHA to regulate the transcriptional program controlling neuronal antioxidant defenses in a hippocampal cell line, especially the glutathione/glutaredoxin system. Within this antioxidant system, DHA was particularly efficient in triggering the upregulation of Gpx4 gene, which encodes for the nuclear, cytosolic, and mitochondrial isoforms of phospholipid-hydroperoxide glutathione peroxidase (PH-GPx/GPx4), the main enzyme protecting cell membranes against lipid peroxidation and capable to reduce oxidized phospholipids in situ. We show here that this novel property of DHA is also significant in the hippocampus of wild-type mice and, to a lesser extent in APP/PS1 transgenic mice, a familial model of Alzheimer's disease. By doing this, DHA stimulates a mechanism to self-protect from oxidative damage even in the neuronal scenario of high aerobic metabolism and in the presence of elevated levels of transition metals, which inevitably favor the generation of reactive oxygen species. Noticeably, DHA also upregulated a Gpx4 CIRT (Cytoplasmic Intron-sequence Retaining Transcripts), a novel Gpx4 splicing variant, harboring part of the first intronic region, which according to the “sentinel RNA hypothesis” would expand the ability of Gpx4 (and DHA) to provide neuronal antioxidant defense independently of conventional nuclear splicing in cellular compartments, like dendritic zones, located away from nuclear compartment. We discuss here, the crucial role of this novel transcriptional regulation triggered by DHA in the context of normal and pathological hippocampal cell.
Keywords: docosahexaenoic acid, hippocampal cells, glutathione peroxidase 4, phospholipid-hydroperoxide glutathione peroxidase, transcriptional regulation, intron retention, neuroprotection
Introduction
Docosahexaenoic acid (DHA) is the most abundant n-3 long-chain polyunsaturated fatty acid (LCPUFA) in nerve cells. In fact, brain is the organ containing the largest amount of DHA in the whole organism, and it seems that it was selected soon in the evolution of the cephalization process of vertebrates to provide a special biochemical microenvironment to nerve cell membranes, especially during the massive accretion in the evolution of primate brains (Crawford et al., 1999, 2013; Simopoulos, 2011). DHA is a pleiotropic molecule; it is an essential component of nerve cells membranes where it esterify sn-2 position of glycerophospholipids (mainly phosphatidylethanolamine and phosphatidylserine, the most abundant phospholipids in nerve cells) and is largely determinant of the structural and physicochemical properties of plasma membrane. Indeed, properties like membrane viscosity, lateral mobility, phase separation and microdomain segregation, conformational transitions and stability of membrane proteins, lipid-protein and protein-protein interactions, all have been shown to be modulated by DHA (Uauy et al., 2001; Stillwell and Wassal, 2003; Díaz et al., 2012, 2015). Besides its structural role, DHA participates in the modulation of neurogenesis, synaptogenesis and neurite outgrowth, refinement of synaptic connectivity, neurotransmitter release, and in memory consolidation processes (Alessandri et al., 2004; Calderon and Kim, 2004; Innis, 2007; Cao et al., 2009; Moriguchi et al., 2013), but also in the activation of signaling pathways for neuronal survival against oxidative and inflammatory cascades (Oster and Pillot, 2010; Bazinet and Layé, 2014). The importance of DHA for brain health is highlighted by the extensive epidemiological and experimental evidence linking its depletion with the development of neurodegenerative diseases (Huang, 2010; Díaz and Marín, 2013).
The pro-oxidant environment of brain and lipid peroxidation
Chemically, DHA is a 22 carbon atoms fatty acid containing six double bonds. The presence of a double bond in the fatty acid weakens the C–H bonds on the carbon atom nearby the double bond and thus facilitates H• abstraction from a methylene group, giving rise to an unpaired electron on the carbon (–•CH–) susceptible for oxidation. This circumstance is likely to be favored in the brain parenchyma given its high metabolic rate and elevated oxygen consumption, which inevitably will produce significant amounts of reactive oxygen species as by-products, including the highly reactive superoxide anion O−2 that is converted to H2O2 (Dröge, 2002). In addition, brain is rich in redox transition metals, particularly iron, which by virtue of Fenton reaction with endogenous H2O2 produce iron(III) and generate the highly reactive hydroxyl radical OH• at the expense of endogenous reducing agents, i.e., polyunsaturated fatty acids like DHA or arachidonic acid, generating lipoperoxyl radicals. Lipid peroxidation generates hydroperoxides as well as endoperoxides, which undergo fragmentation to produce a broad range of reactive intermediates called reactive carbonyl species (RCS) such as isoprostanes (IsoPs), neuroprotanes, malondialdehyde, unsaturated aldehydes including 4-hydroxy-2-trans-nonenal (HNE), 4-hydroxy-2-trans-hexenal (HHE), and 2-propenal (acrolein), with different degrees of reactivity (Porter et al., 1995; Niki et al., 2005; Catalá, 2009; Fritz and Petersen, 2013; Naudí et al., 2013).
Clearly, the concurrency of these factors in the presence of high amounts of polyunsaturated fatty acids, mainly DHA, is expected to favor the free radical-induced peroxidation of DHA in the brain parenchyma (Van Kuijk et al., 1990; Dröge, 2002; Valko et al., 2007). An important aspect of lipid peroxidation is its self-propagating nature and fundamentally different from other forms of free radical injury in that it is a self-sustaining process capable to provoke extensive brain tissue damage (Catalá, 2009; Singh et al., 2010). As most lipids in the brain are contained in the membrane phospholipids, the main outcome of lipid peroxidation is the structural damage of membranes, causing structural changes that impact membrane fluidity and permeability, neurotransmission, signaling, ion transport, and impaired electrical conduction.
Nerve cells are endowed with different antioxidant systems that render them protected from oxidative damage caused by lipid peroxides. This protection is mainly accomplished by phase II detoxifying enzymes belonging to two antioxidant systems, namely thioredoxin and glutathione systems, which use hydrophilic thiol-containing molecules (thioredoxin and glutathione, respectively), as electron donors to generate conjugated metabolites (Arnér and Holmgren, 2000; Imai and Nakagawa, 2003). However, although members of the thioredoxin system (mammalian thioredoxin reductases) are capable to reduce some non-disulfide-containing molecules, including lipid hydroperoxides independently of thioredoxin (Björnstedt et al., 1995), it is the only within the glutathione system where enzymes exist that are capable to recover oxidized membrane lipids (Imai and Nakagawa, 2003).
Phospholipid-hydroperoxide glutathione peroxidase and membrane protection
Phospholipid hydroperoxide glutathione peroxidase, glutathione peroxidase 4, or GPx4, is a member of glutathione peroxidases family of selenoproteins, most of which bear a selenocystein as catalytically active amino acid, which confers a more efficient reaction with peroxide substrates (Nauser et al., 2006). GPx4 uses preferentially glutathione as electron donors as long as cellular concentrations of the reduced form (GSH) are not limiting, but may accept other thiol groups in proteins as reducing equivalent (Godeas et al., 1997). Therefore, GPx4 can either act as a GSH peroxidase or a thiol peroxidase depending on the availability of GSH (Brigelius-Flohé and Maiorino, 2013). However, unlike other glutathione peroxidases, GPx4 is capable of reducing complex lipid peroxides, like phospholipid hydroperoxides, even when integrated in highly structured lipid-protein assemblies such as lipoproteins and membranes (Imai and Nakagawa, 2003). X-ray data indicated that, in contrast to other GPx isoforms, the active site of GPx4 lacks a surface-exposed loop domain which appears to limit the accessibility of large oxidized substrates, but instead contains a large hydrophobic surface that allows GPx4 to closely associate with membranes and lipoproteins (Scheerer et al., 2007). Its ability to directly reduce phospholipid hydroperoxides in membranes without prior action of phospholipase A2 membrane makes GPx4 unique amongst antioxidative enzymes (Imai et al., 2003; Savaskan et al., 2007). Further, unlike other glutathione peroxidases (GPx1, GPx2, GPx3, GPx5, and GPx6) which exist as homo(tetra)-oligomeric proteins, GPx4 is a monomeric protein of about 20 kDa and misses the tetramer interfaces for oligomer formation (Scheerer et al., 2007; Brigelius-Flohé and Maiorino, 2013).
GPx4 is also peculiar amongst glutathione peroxidases because of its additional regulatory functions. Thus, GPx4 is considered not only a phase II antioxidant enzyme but is also endowed with functions associated with the regulation of apoptosis, gene expression, eicosanoid biosynthesis, and embryo development (Imai and Nakagawa, 2003; Savaskan et al., 2007; Ufer and Wang, 2011). The essential nature of GPX4 is demonstrated by the fact that genetic disruption of the entire Gpx4 gene in homozygous Gpx4 knockout causes in utero fetal death by midgestation (Imai et al., 2003; Yant et al., 2003). This intrauterine lethality has been related to increased apoptosis and cell death leading to malformation of embryonic structures and major defects in brain development (Imai et al., 2003; Yant et al., 2003). This contrasts profoundly with the knockouts on Gpx1–3 genes, which are viable though more sensitive to stressors (Brigelius-Flohé and Maiorino, 2013), indicating a degree of redundancy of these genes for at least some of their biochemical functions, i.e., oxidant scavengers, in vivo.
In mammalian cells GPx4 is found in the cytoplasm, nucleus, mitochondria, and also in the endoplasmic reticulum (Imai and Nakagawa, 2003). From the molecular point of view, GPX4 is also singular because the three isoenzymes, namely cytosolic GPx4 (c-GPx4), mitochondrial GPx4 (m-GPx4), and nuclear GPx4 (n-GPx4), all derive from a single gene (Figure 1), but that can be distinguished by their N-terminal sequences (Imai and Nakagawa, 2003; Savaskan et al., 2007). The Gpx4 gene comprises 7 exons, of which exons 2–7 encoding for the functional enzyme are shared by the three isoforms. The differential N-terminal sequences are attained by three major start sites for translation (Figure 1) that reside in two alternative exons 1 (E1a and E1b). Thus, exon 1a contains two in-frame translational start sites (5′AUG and 3′AUG) separated by a sequence that encode for a mitochondrial leader peptide. Translation initiation at the 5′AUG results in the generation of m-GPx4 isoenzyme, while translation from the 3′AUG, that lacks this leader signal, yields c-GPx4. Because the mitochondrial leader peptide is cleaved off after import into mitochondria, c-GPx4 and m-GPx4 cannot be differentiated on the basis of their primary structure. Finally, the alternative first exon (E1b) encodes the N-terminal part of the nuclear isoform, n-GPx4, and contains a nuclear targeting sequence which is apparently retained after nuclear import (Pfeifer et al., 2001), and makes this isoform distinguishable from m/c-GPx4 at protein level.
Figure 1.
Structural organization of mouse Gpx4 gene. Boxes indicate coding exons (red) and UTRs (orange). Bold lines represent intronic sequences. Sites targeted by amplification primers are shown as arrowed lines.
Unexpected DHA-induced transcriptional regulation of Gpx4 gene
We have recently reported that DHA upregulates several members of both glutathione/glutaredoxin and thioredoxin/peroxiredoxin antioxidant systems in mouse hippocampal HT22 cells (Casañas-Sánchez et al., 2014). Noticeably, within the glutathione/glutaredoxin system, largest changes in gene expression were observed for the Gpx4 isoforms, around 150%, and affecting all transcripts analyzed, including the mitochondrial, cytosolic, and nuclear isoforms. The change induced by DHA on glutathione peroxidase genes was specific for Gpx4 since no variation was observed for the cytosolic Gpx1 gene (Casañas-Sánchez et al., 2014). Paralleling these changes, DHA treatment significantly increased GPx4 (and total GPx) activities, following a time-course that was compatible with the necessary delay for Gpx4 mRNA translation. Importantly, these effects of DHA were specific for DHA and were not be detected when DHA was replaced by arachidonic acid, another highly abundant polyunsaturated fatty acid in nerve cells, under identical experimental conditions (Casañas-Sánchez et al., 2014).
Absolute quantification of Gpx4 isoform expression in unstimulated HT22 cells revealed that the largest expression values corresponded to the cytoplasmic variant and the lowest for the nuclear mRNA variant, with a difference of about 3 orders of magnitude (Figure 2). Furthermore, in absolute terms, we observed that DHA treatment increases nearly 1.5-fold the expression of m/c-Gpx4, which must be attributable to the enhancement of the mRNA coding for the cytoplasmic isoform, whose abundance is about one order of magnitude higher than for m-Gpx4. The nuclear isoform was also up-regulated by DHA, but its absolute magnitude was negligible compared to m/c-Gpx4 (Figure 2).
Figure 2.
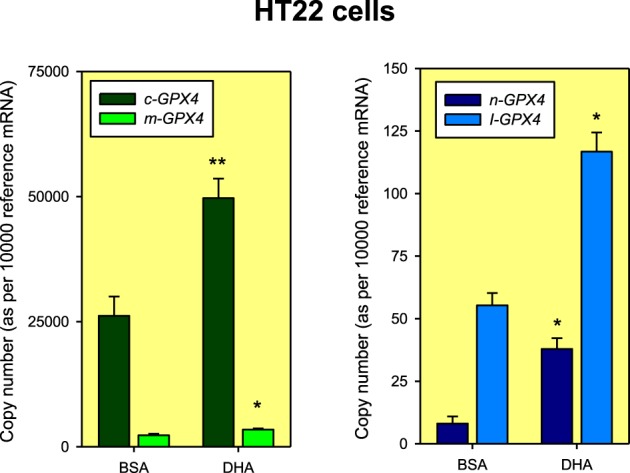
Absolute quantification of Gpx4 mRNA isoforms in HT22 cells exposed to BSA (Vehicle: Bovine serum albumin) or DHA for 48 h. The normalization factor (reference mRNA) for each cDNA sample was calculated as the geometric mean of the expression values of reference genes Hprt1 and Tbp genes using standard curves generated from purified amplicons as described in Expósito-Rodríguez et al. (2011). Data are expressed as mean ± SEM from four different experiments. Statistical comparisons were assessed using Mann-Whitney U-test for independent samples. *p < 0.05 and **p < 0.01 compared to BSA.
Using identical amplicons (Table 1), we further explored for the presence of these different Gpx4 isoforms in the hippocampus of C57BL/6 mice. The results revealed the presence of all different isoforms, and noticeably, following a similar expression pattern, with m/c-Gpx4 being the most abundant transcripts and n-Gpx4 showing lowest expression. Interestingly, animals exposed to High or Low-DHA diets also exhibited differential Gpx4 transcriptional regulation in response to DHA-containing or DHA-impoverished diets and differentially affected between wild-type and APP/PS1, a familial model of Alzheimer's disease (Figure 3). Thus, Absolute quantification of Gpx4 isoforms in WT animals, revealed that Low-DHA diets leads to the stimulation of gene expression of all isoforms (between 1.43 and 1.99 times), being the largest stimulation observed for the cytosolic isoform, which, in turn is the most abundant Gpx4 isoform under any circumstance (Figure 3). These observations strongly suggest a compensatory genetic strategy aimed to ensure protection of membrane DHA from oxidative damage under conditions of limited DHA availability. Overall these data indicates that the lack of sufficient DHA or its deficient supply is accompanied by increased Gpx4 mRNAs expression and GPx4 protein synthesis, which consequently augment cellular resistance to oxidative damage of DHA-containing phospholipids. A different behavior was observed in APP/PS1 animals, where the high-DHA containing diet stimulated the expression of cytoplasmic and mitochondrial Gpx4 mRNAs. The different expression levels observed in APP/PS1 animals, especially for the m-Gpx4 isoform, suggest a genotype-related transcriptional regulation, which fits well with the increased levels of hippocampal oxidative stress demonstrated in transgenic animals (Aso et al., 2012) and also the lower levels of hippocampal DHA compared to WT animals (Fabelo et al., 2012).
Table 1.
Oligonucleotides used as primers for quantification of different Gpx4 mRNA isoforms and for mapping 5′ end of I-Gpx4 mRNA in HT22 cells and mouse hippocampus.
| Targeted gene | Primers | Exon/intron | Amplicon size (bp) | gDNA discrimination |
|---|---|---|---|---|
| m-Gpx4 | F-89: CCgCCgAgATgAgCTgg | E1a | 128 | No |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| m/c-Gpx4 | F-16: TggTCTggCAggCACCAT | E1a | 201 | No |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| n-Gpx4 | F240: gTTCCTgggCTTgTgTgCAT | E1b-E2 | 140 | No |
| R379: AggCCACgTTggTgACgAT | E3 | |||
| n/I-Gpx4 | F205: CTgCAAgAgCCTCCCCAgT | E1b | 157/370 | Yes |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| I-Gpx4 | FI1b176: ggACCTgggTTAggACACTCA | I1b | 147 | Yes |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| Mapping (+1) | F+1: gTgggCTACTggGAACTTgg | I1a | 907 | |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| Mapping (+2) | F+2: gggAAAgCggAgCCTgATAg | I1a | 787 | |
| R112:TgCACACgAAACCCCTgTACT | E2-E3 | |||
| Mapping (+3) | F+3: CTTggCTACCggCTCTTTg | I1a | 630 | |
| R112: TgCACACgAAACCCCTgTACT | E2-E3 | |||
| Hprt1 | F: TCAgACTgAAgAgCTACTgTAATgA | E3-E4 | 136 | Yes |
| R: AAgTTTgCATTgTTTTACCAgTg | E6 | |||
| Tbp | F: gACCCACCAgCAgTTCAgTAg | E6 | 136 | Yes |
| R: CTCTgCTCTAACTTTAgCACCTgT | E7-E8 |
Determination of absolute mRNA levels by real-time quantitative PCR (RT-qPCR) were performed as described in Expósito-Rodríguez et al. (2011). Hprt1, Hypoxanthine guanine phosphoribosyl transferase 1; Tbp, TATA binding protein.
Figure 3.
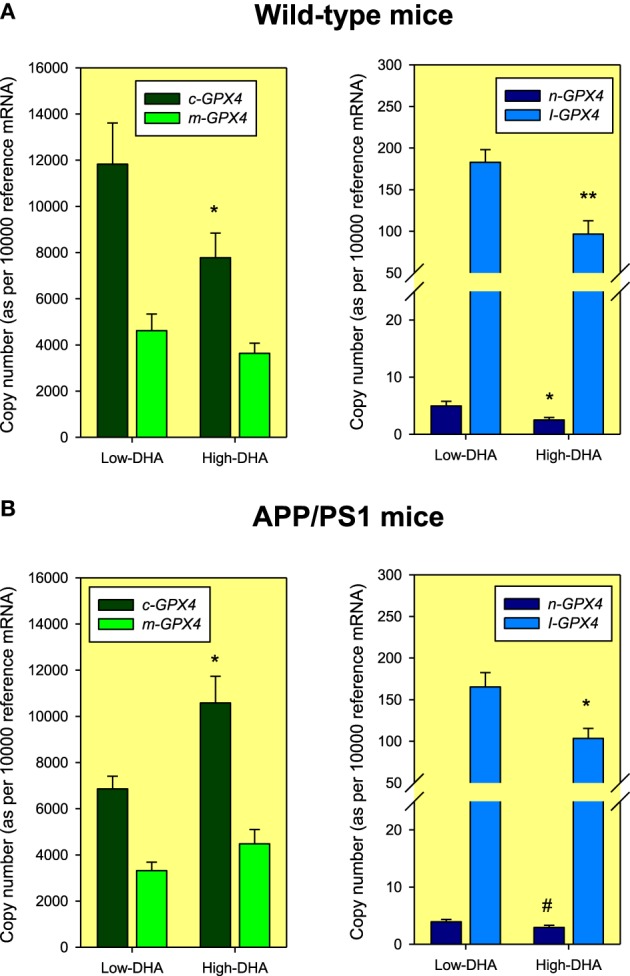
Absolute quantification of Gpx4 mRNA isoforms in 6 months old Wild-type (A) and APP/PS1 (B) mice fed Low-DHA or High-DHA diets for 3 months. Data are expressed as mean ± SEM from four different animals under each dietary condition and genotype. The normalization factor (reference mRNA) for each cDNA sample was calculated as the geometric mean of the expression values of reference genes Hprt1 and Tbp genes using standard curves generated from purified amplicons as described in Expósito-Rodríguez et al. (2011). Statistical comparisons were assessed using Mann-Whitney U-test for independent samples. *p < 0.05, **p < 0.01, and #p < 0.1 compared to Low-DHA.
Finally, using appropriate amplicons (Table 2) we tested for the presence and absolute abundance of the different GPX4 transcripts in the human cell line SHSY-5Y. The results revealed that all GPX4 isoforms are expressed in this cell line and, more interestingly, that their absolute abundance followed the same sequence observed in HT22 cells, though in this case the amount of the most prominent isoform, namely c-GPX4, was notably higher (about four orders of magnitude) than any other isoform.
Table 2.
Oligonucleotides used as primers for quantification of different GPX4 mRNA isoforms and for mapping 5′ end of I-GPX4 mRNA in SHSY-5Y cells.
| Targeted gene | Primers | Exon/intron | Amplicon Size (bp) | gDNA discrimination |
|---|---|---|---|---|
| m-GPX4 | F-E1m: CATTggTCggCTggACgAg | E1a | 242 | Yes |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| m/c-GPX4 | F-E1c: CCTggCCgggACCATg | E1a | 123 | Yes |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| n- GPX4 | F-E2si: CAggCAgCggTgCCAgAg | E1b | 170 | Yes |
| R-E23: gggACgCgCACgggTC | E1b-E2 | |||
| n/I- GPX4 | F-E2N: gATCCACgAATgTCCCAAgTC | E1b | 138/394 | Yes |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| I- GPX4 | F-I1b: gAggAgCgTTCAggTCTTCAg | I1b | 242 | No |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| Mapping (+1) | F+1: gTgAgCTAgCgCCgCg | I1a | 1165 | |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| Mapping (+2) | F+2: CCCTCCAggCCgTTgTAgg | I1a | 978 | |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| Mapping (+3) | F+3: CggAgggCTggAAATCCC | I1a | 730 | |
| R-E34: CACACgAAgCCCCggTACT | E2-E3 | |||
| HPRT1 | F: gACCAgTCAACAggggACAT | 173 | Yes | |
| R:AACACTTCgTggggTCCTTTTC | ||||
| RPL32 | F: CATCTCCTTCTCggCATCA | 153 | Yes | |
| R:AACCCTgTTgTCAATgCCTC |
Determination of absolute mRNA levels by real-time quantitative PCR (RT-qPCR) were performed as described in Expósito-Rodríguez et al. (2011). HPRT1, Hypoxanthine guanine phosphoribosyl transferase 1; RPL32, Ribosomal protein L32.
Defining a unified rationale for DHA-induced regulation of Gpx4 gene expression
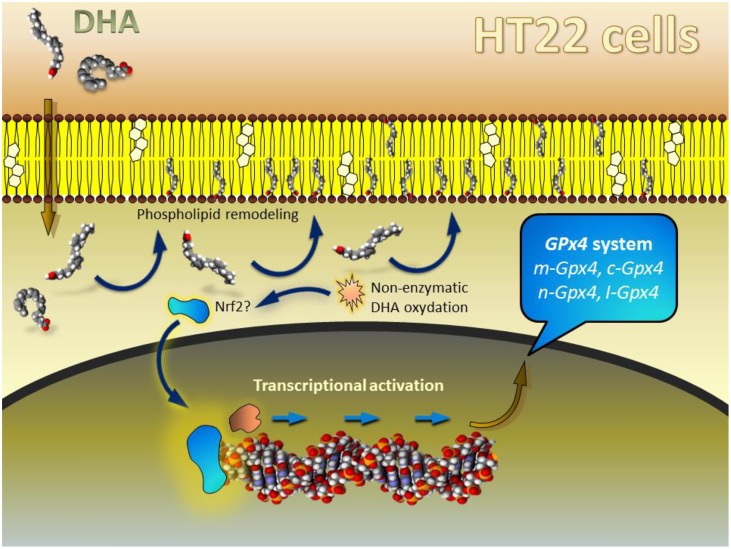
Our initial observations in HT22 indicated that the upregulation of Gpx4 expression occurred with a significant delay form the initial exposure to DHA. Thus, it was necessary to expose cells for 48 h to detect the significant change in the expression levels of all Gpx4 isoforms (Casañas-Sánchez et al., 2014). Our interpretation of these findings was related to the fact that control unstimulated HT22 cells (as most neuronal cell lines studied so far) contain extremely low levels of DHA in their membrane phospholipids (Martín et al., 2006). Supplementation of culture medium with DHA causes a nearly immediate incorporation by deacylation-reacylation mechanisms, whereby some monounsaturated fatty acids (mainly oleic acid) are readily replaced with DHA in membrane phospholipids (Farooqui et al., 2000; Martín et al., 2006). The activation of the transcriptional process occurred only after membrane phospholipids were replenished, because only then enough DHA remained unesterified in the cytoplasm of neuronal cells, being then susceptible for non-enzymatic oxidation in the prooxidant cellular environment (Figure 4). Indeed we could only detect significant levels of the specific DHA-derived lipoperoxide 4-hydroxy-2-hexenal (HHE) after 36 h exposure to DHA, just before upregulation of Gpx4 expression occurred. These findings were interpreted as HHE providing the signal to trigger transcriptional regulation, likely through activation of Nrf2 transcription factor. Indeed, it is known that Nrf2 is a master factor for the regulation of antioxidant response elements (ARE) in several antioxidant and phase II detoxifying proteins in different cell types, including nerve cells (Kobayashi and Yamamoto, 2005; Zhang et al., 2013), and several reports have shown that although DHA itself does not bind Nrf2, it may stimulate transcriptional activity of ARE-containing genes through activation of Nrf2 upon generation of 4-hydroxy-2-hexenal, which indeed activates Nrf2 (Ishikado et al., 2013; Kusunoki et al., 2013; Yang et al., 2013).
Figure 4.
Schematic model depicting the hypothetical mechanism of DHA-mediated regulation of Gpx4 in HT22 cells. For details see “Defining a unified rationale for DHA-induced regulation of Gpx4 gene expression.” Modified from Casañas-Sánchez et al. (2014).
The results obtained in the hippocampus of WT mice were particularly interesting, since transcriptional activation of all Gpx4 isoforms was significantly higher under the Low-DHA dietary condition compared to the High-DHA diet. Though apparently these results might contradict the results obtained in HT22 cells, the outcomes suggest that it is under a low DHA diet with limited availability of n-3 precursors, when it is especially important to activate mechanisms (like upregulation of Gpx4 isoform expression) that may ensure the preservation of DHA in brain membranes. In fact, analyses of hippocampal lipid composition in WT and APP/PS1 animals revealed a significant reduction of DHA contents in Low-DHA diet compared to the High-DHA diet, yet the amount of DHA in the Low-DHA condition was surprisingly higher than expected from the dietary supply (Table 3), which reflects an extremely efficient ability to preserve brain DHA even when subjected to negligible levels in the diet (Díaz et al., accepted). This exceptional capability is mainly attributable to the favorable gradient for fatty acids to cross the blood-brain barrier, either passively or via fatty acid binding proteins (FATPs), since once in the brain, unesterified fatty acids are converted to CoA thioesters by the long-chain-fatty-acid-CoA synthase (ACSL) family of proteins, therefore maintaining a continuous concentration gradient (see (Bazinet and Layé, 2014) for comprehensive review).
Table 3.
Fatty acid composition of total lipids in the hippocampus of 6 months old WT and APP/PS1 mice, and fed for 3 months with either Low-DHA or High-DHA diets.
| Fatty acid | WT | APP/PS1 | ||
|---|---|---|---|---|
| Low-DHA | High-DHA | Low-DHA | High-DHA | |
| 14: 0 | 0.10 ± 0.00*# | 0.12 ± 0.00# | 0.12 ± 0.01 | 0.13 ± 0.00 |
| 16: 0 DMA | 2.16 ± 0.01 | 2.12 ± 0.01 | 2.11 ± 0.04 | 2.13 ± 0.04 |
| 16: 0 | 18.41 ± 0.06# | 18.58 ± 0.16 | 17.80 ± 0.16 | 18.25 ± 0.20 |
| 18: 0 DMA | 4.07 ± 0.04 | 4.09 ± 0.06 | 4.01 ± 0.15 | 4.04 ± 0.05 |
| 18:1 n-9 DMA | 1.51 ± 0.04 | 1.54 ± 0.03 | 1.52 ± 0.08 | 1.52 ± 0.06 |
| 18:1 n-7 DMA | 1.94 ± 0.05 | 2.01 ± 0.04 | 2.09 ± 0.05 | 1.99 ± 0.08 |
| 18: 0 | 19.60 ± 0.10* | 19.05 ± 0.01 | 19.54 ± 0.12 | 19.14 ± 0.30 |
| 18: 1 n-9 | 15.81 ± 0.06*# | 17.04 ± 0.11 | 16.32 ± 0.16 | 17.00 ± 0.25 |
| 18: 1 n-7 | 3.59 ± 0.06* | 3.40 ± 0.01 | 4.18 ± 0.60 | 4.11 ± 0.56 |
| 18: 2 n-6 | 0.49 ± 0.01 | 0.48 ± 0.02 | 0.50 ± 0.02 | 0.48 ± 0.03 |
| 20: 0 | 0.28 ± 0.01 | 0.26 ± 0.01 | 0.29 ± 0.01 | 0.26 ± 0.02 |
| 20: 1 n-9 | 1.76 ± 0.05 | 1.80 ± 0.03 | 1.96 ± 0.07 | 1.74 ± 0.11 |
| 20: 1 n-7 | 0.34 ± 0.01 | 0.36 ± 0.00 | 0.37 ± 0.02 | 0.36 ± 0.02 |
| 20: 3 n-6 | 0.28 ± 0.01* | 0.57 ± 0.00# | 0.28 ± 0.01* | 0.54 ± 0.01 |
| 21: 0 | 0.02 ± 0.01 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| 20: 4 n-6 | 9.87 ± 0.16*# | 7.64 ± 0.02 | 9.34 ± 0.07* | 7.54 ± 0.19 |
| 20:5 n-3 | 0.00 ± 0.00* | 0.19 ± 0.01 | 0.00 ± 0.00* | 0.21 ± 0.01 |
| 22:0 | 0.22 ± 0.01 | 0.21 ± 0.00 | 0.24 ± 0.01 | 0.21 ± 0.02 |
| 22: 4 n-6 | 3.05 ± 0.06* | 1.85 ± 0.02 | 3.14 ± 0.10* | 1.82 ± 0.07 |
| 22: 5 n-6 | 1.19 ± 0.02*# | 0.09 ± 0.00 | 0.87 ± 0.04* | 0.09 ± 0.01 |
| 22: 5 n-3 | 0.03 ± 0.01* | 0.39 ± 0.01 | 0.01 ± 0.01* | 0.39 ± 0.01 |
| 24: 0 | 0.26 ± 0.01 | 0.26 ± 0.01 | 0.29 ± 0.02 | 0.27 ± 0.03 |
| 22: 6 n-3 | 12.69 ± 0.21*# | 16.66 ± 0.10 Δ | 11.19 ± 0.11* | 15.04 ± 0.55 |
| 24: 1 n-9 | 0.85 ± 0.03# | 0.83 ± 0.04# | 1.19 ± 0.09* | 0.70 ± 0.01 |
| TOTALS AND INDEXES | ||||
| Saturates | 45.35 ± 0.05* | 44.84 ± 0.12 | 44.61 ± 0.38 | 44.59 ± 0.49 |
| Unsaturated | 54.54 ± 0.05* | 55.05 ± 0.12 | 55.27 ± 0.39 | 55.28 ± 0.50 |
| DMAs | 9.67 ± 0.08 | 9.76 ± 0.12 | 9.74 ± 0.31 | 9.68 ± 0.15 |
| n-9 | 20.34 ± 0.18*# | 21.62 ± 0.16 | 21.53 ± 0.15 | 21.43 ± 0.56 |
| n-3 | 12.80 ± 0.21* | 16.31 ± 0.09 | 11.27 ± 0.12* | 15.16 ± 0.55 |
| n-6 | 14.99 ± 0.22*# | 10.70 ± 0.04 | 14.23 ± 0.12* | 10.53 ± 0.27 |
| n-3/n-6 | 0.85 ± 0.02* | 1.52 ± 0.01 | 0.86 ± 0.01* | 1.53 ± 0.03 |
| 18:1 n-9/n-3 H | 1.24 ± 0.02*# | 1.06 ± 0.01 | 1.34 ± 0.02* | 1.07 ± 0.06 |
| Unsaturation Index | 163.12 ± 0.99*# | 166.48 ± 0.30 | 158.64 ± 0.53 | 165.50 ± 2.94 |
| saturates/n-3 | 3.55 ± 0.05* | 2.75 ± 0.01 | 3.64 ± 0.03* | 2.77 ± 0.07 |
| saturates/n-9 | 2.23 ± 0.02*# | 2.07 ± 0.02 | 2.07 ± 0.02 | 2.09 ± 0.07 |
Results are expressed as mole % and represent means ± SEM of four different animals under each dietary condition and genotype. DMA: Dimethyl acetals.
p < 0.05 compared to High-DHA.
, Δ : p < 0.05 and p < 0.1 compared to APP/PS1. Fatty acid extraction and determination was performed as described in Fabelo et al. (2012) and Martín et al. (2006). Data were analysed by One-Way ANOVA followed by Tukey's multiple comparison test. Comparisons between Low-DHA and High-DHA treatments were performed using Student's t-test or Mann-Whitney U-test where appropriate. Main monoenoic (oleic acid, 18:1n9), n-6 LCPUFA (arachidonic acid, 20:4n-6), and n-3 LCPUFA (DHA, 22:6n-3) are highlighted on pink background.
As mentioned before, APP/PS1 animals also responded to the different diets with changes in the transcriptional levels of Gpx4 expression. However, we only observed increased expression of c-Gpx4 isoform, and in the High-DHA diet. The apparent discrepancy between WT and APP/PS1 animals is likely related to the involvement of constitutive oxidative stress associated to amyloid generation in the brain of transgenic animals (Aso et al., 2012; Fabelo et al., 2012). Indeed, it is now widely accepted that amyloid peptide induces oxidative stress, and inflammation and oxidative stress generate more Aβ, giving rise to a vicious cycle between Aβ and free radical formation/lipid peroxidation in the presence of transition metals (Behl et al., 1994; Pamplona et al., 2005; Solfrizzi et al., 2006). In line with this matter, increased levels of LCPUFA–derived lipid hydroperoxides like HHE, HNE, and other endoperoxides like isoprostanes have been observed in the brain of transgenic models of Alzheimer's disease, including 3xTgAD and APP/PS1 animals, and more relevantly, in different areas of human brain and cerebrospinal fluid of AD patients (Marcus et al., 1998; Markesbery and Lovell, 1998; Praticò et al., 1998; Valko et al., 2007; Gwon et al., 2012). In this order of ideas, it seems plausible that in the high-DHA diet, lipid-derived hydroperoxides must be elevated in the more prooxidant environment of brain parenchyma in transgenic animals, including DHA-related HHE, which may trigger the stimulation of Nrf2 transcription factor, which eventually would favor the activation of a transcriptional program to increase Gpx4 isoforms expression, especially the cytoplasmic isoform. In agreement with our findings, recent genomic gene expression profiles by microarray analyses in a “gene dose-response” model of Nrf2-null model have revealed that graded activation of Nrf2 increased the transcription of a large number of genes involved in Nrf2-mediated oxidative stress response, glutathione metabolism, and xenobiotic metabolism, including several genes responsible for the reduction of superoxide ions using GSH, i.e., glutathione peroxidases Gpx2 and Gpx4, which augmented by 2260 and 105%, respectively, in Keap1-HKO mice compared to WT mice (Wu et al., 2011).
Alternatively, altered genetic regulation of Gpx4 expression caused by overexpression of mutated APP and PS1 in transgenic animals, may have disturbed the normal gene expression pattern, as it has been recently demonstrated to occur in the frontal cortex and hippocampus of transgenic mice carrying the Δ E9 mutant hPS1 gene (Unger et al., 2005). Likewise, recent comparative studies in sporadic (icv-STZ Mouse) and familial (3xTg-AD mouse) mice models of AD have revealed the transcriptional alteration of more than 80 genes related to synapse function, apoptosis and autophagy, AD-related protein kinases, glucose metabolism, insulin signaling, and mTOR pathway in the hippocampus and the cerebral cortex, not only when compared to WT animals, but also between both AD models (Chen et al., 2012).
In summary, we hypothesize that by upregulating Gpx4 gene expression and increasing GPx4 protein levels and enzyme activity, DHA initiates a self-protective biochemical strategy in the hippocampal neuron to control non-enzymatic peroxidation of DHA-containing phospholipids in the membrane from cellular and subcellular compartments. Given the extremely low capacity of hippocampal cells to biosynthesize DHA (reviewed in Plourde and Cunnane, 2007; Bazinet and Layé, 2014), and the essential nature of this fatty acid in the nervous system, this strategy provides an adaptive mechanism to keep the high DHA levels in hippocampal phospholipids.
Retained intronic sequences in Gpx4 mRNA
In initial experimental strategy used for the detection of the different Gpx4 mRNAs, we designed a couple of primers targeted to exons E1b (forward primer) and the boundary between exons E2/E3 (reverse primer) for the specific detection of nuclear Gpx4 mRNA. Unexpectedly, in these experiments we detected the presence of two amplicons: one of the expected molecular size (157 bp) for a fully processed n-Gpx4 mRNA, and another fragment 370 bp. Once sequenced, we observed that this later amplicon contained an extra 213 bp sequence corresponding to intron I1b located between exons E1b and E2. This unexpected sequence (named here I-Gpx4) was detected in control HT22 cells (treated with vehicle), but more interestingly, that its expression level was subjected to significant upregulation by DHA treatment (near 100% compared to control cells), which was in the range observed for the changes in expression for the set of other Gpx4 mRNA variants (Figure 2). In absolute terms, I-Gpx4 quantification revealed that this isoform was nearly 3 times more abundant than n-Gpx4 under unstimulated conditions, and about 7 times higher after DHA treatment, pointing to a significant biological role of this intron-retaining isoform in hippocampal cells. Nonetheless, I-Gpx4 was expressed a much lower amount than c-Gpx4, even after DHA treatment.
In order to gain a deeper insight on the transcription start site for the intron-retained transcript, we designed several amplification primers directed to intron I1a (Figure 1 and Table 1). While the primers targeted to E1b exon and intron I1b amplified the expected amplicons, all other primers targeting E1b exon upstream sequences were unsuccessful in these PCR assays. These results strongly indicate that a Gpx4 CIRT transcript represents an unprocessed variant of the n-Gpx4 mRNA isoform.
In order to assess for the biological relevance of this intron-retaining Gpx4 isoform, we used these specific amplicons to unveil the presence in the hippocampus of C57Bl/6 mice. We observed that not only the amplicon was detected, but also that its expression was modulated by the contents of DHA in the diet. Thus, levels of I-Gpx4 isoform were significantly higher (around 2 times) in animals receiving Low-DHA diet compared to animals receiving a High-DHA diet. A similar result was obtained in transgenic APP/PS1 animals, though in this case the increase in I-Gpx4 gene expression was around 1.6 times higher in animals maintained in a Low-DHA diet (Figure 3). Absolute comparisons between WT and APP/PS1 animals revealed that I-Gpx4 was higher in WT animals than in the APP/PS1 genotype. Overall these data indicates that the lack of sufficient DHA or its deficient supply is accompanied by an increase in I-Gpx4. Again, these higher expression levels observed in APP/PS1 animals suggests a genotype-related transcriptional regulation, which may be related to increased hippocampal oxidative stress in transgenic animals (Aso et al., 2012).
Whether this regulated Gpx4 mRNA variant constitutes an intermediate processing stage of Gpx4 splicing or represents a different level of transcriptional regulation by intron retention remains unrevealed. However, the notion of intron retention has been gaining consistency in the last decade, as an alternative processing of conventional (nuclear) mRNA, adding a novel level of complexity in gene expression regulation. These sequences, named as CIRTs (Cytoplasmic Intron-sequence Retaining Transcripts), comprise only few introns and are limited to small fractions of introns within the gene (Buckley et al., 2014). Indeed, in recent years, a number of transcripts containing intronic sequences and subjected to cytoplasmic splicing have been unveiled across a number of cell types and species (Buckley et al., 2014), and the list keeps growing steadily. Noticeably, a recent study performed in mouse brains have revealed the broad presence of cytoplasmic intron-sequence retaining transcripts in hippocampal neurons, with 59.4% of total gene transcripts in dendrites containing intronic reads (Khaladkar et al., 2013). Examples of retained introns from individual transcripts of relevance in neuronal cells include the large-conductance calcium-dependent potassium channel (BK) encoded by KCNMA1 gene (Bell et al., 2008, 2010), γ-subunit of GABA-A receptor (encoded by GABRG3) (Buckley et al., 2011), ionotropic Glutamate receptor (encoded by GRIK1 gene) and type 1 NMDA receptor (encoded by GRIN1) (Buckley et al., 2011) or the α-1 subunit of T type, voltage-dependent Calcium channel (encoded by CACNA1H) (Zhong et al., 2006). Though the biological relevance of CIRTs is still largely unknown, their abundance in neuronal dendrites (and also in enucleated platelets) suggest that endogenous cytoplasmic transcripts harboring retained intronic substrates could be subject to extranuclear splicing, thereby obviating the need for shuttling molecules to and from the nucleus for processing (Buckley et al., 2014). In this manner, dendritic splicing could represent a novel method of post-transcriptional gene regulation.
In this order of ideas, it is tempting to speculate that CIRT in Gpx4-I gene may be part of the normal endogenous post-transcriptional regulation pathway. Such regulated extranuclear splicing variants may be part of what Buckley et al. (2014) designed as “sentinel RNA hypothesis,” according to which a master transcript harboring CIRTs would serve to generate transcript variants within the cytoplasm in response to specific stimulation, or under conditions in which a rapid processing is required and adaptive. A plausible scenario for this stimulus-dependent cytoplasmic splicing of I-Gpx4 in hippocampal cells, is that this transcript processing in cellular domains distant from cellular nucleus, as is the case of dendrites, would enhance the antioxidant buffering capacity against the acute generation of lipid peroxides by oxidative insults, which would commit the integrity of membrane unsaturated phospholipids, especially those enriched in DHA.
Conclusions and hypothesis
In summary, current available data indicate that Gpx4 gene is subjected to a complex transcriptional regulation that renders nuclear, cytoplasmic, and mitochondrial isoforms, but also generate an intron-retaining variant, likely corresponding to a CIRT, all derived from the same gene. At least in hippocampal HT22 cells and in the hippocampus of wild type mice, the transcriptional control of most of these Gpx4 isoforms is modulated by DHA following an unprecedented homeostatic strategy aimed to buffer its potential oxidation and to ensure its preservation in the hippocampal neuronal membrane. The observations in transgenic APP/PS1 mice, suggest that such homeostatic mechanism is disturbed by the presence of mutated APP and/or PS1, and may be relevant in Alzheimer's disease, as suggested by the generalized observation that DHA contents are reduced in the hippocampus of AD brains.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Supported by grants SAF2010-22114-C02-01/02 and SAF2014-52582R from Ministerio de Economía y Competitividad (Spain). VC and DQ hold fellowships from Gobierno de Canarias and Fundación Cajacanarias, respectively. We are grateful to Biosearch Life-Puleva Biotech for continuous technical support to the project at the Laboratory of Membrane Physiology and Biophysics.
References
- Alessandri J. M., Guesnet P., Vancassel S., Astorg P., Denis I., Langelier B., et al. (2004). Polyunsaturated fatty acids in the central nervous system: evolution of concepts and nutritional implications throughout life. Reprod. Nutr. Dev. 44, 509–538. 10.1051/rnd:2004063 [DOI] [PubMed] [Google Scholar]
- Arnér E. S. J., Holmgren A. (2000). Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 267, 6102–6109. 10.1046/j.1432-1327.2000.01701.x [DOI] [PubMed] [Google Scholar]
- Aso E., Lomoio S., López-Gonzalez I., Joda L., Carmona M., Fernández-Yagüe N., et al. (2012). Amyloid generation and dysfunctional immunoproteasome activation with disease progression in animal model of familial Alzheimer disease. Brain Pathol. 22, 636–653. 10.1111/j.1750-3639.2011.00560.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet R. P., Layé S. (2014). Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat. Rev. Neurosci. 15, 771–785. 10.1038/nrn3820 [DOI] [PubMed] [Google Scholar]
- Behl C., Davis J. B., Lesley R., Schubert D. (1994). Hydrogen peroxide mediates amyloid beta protein toxicity. Cell 77, 817–827. 10.1016/0092-8674(94)90131-7 [DOI] [PubMed] [Google Scholar]
- Bell T. J., Miyashiro K. Y., Sul J. Y., Buckley P. T., Lee M. T., McCullough R., et al. (2010). Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc. Natl. Acad. Sci. U.S.A. 107, 21152–21157. 10.1073/pnas.1015264107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell T. J., Miyashiro K. Y., Sul J. Y., McCullough R., Buckley P. T., Jochems J., et al. (2008). Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 1901–1906. 10.1073/pnas.0711796105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnstedt M., Hamberg M., Kumar S., Xue J., Holmgren A. (1995). Human thioredoxin reductase directly reduces lipid hydroperoxides by NADPH and selenocystine strongly stimulates the reaction via catalytically generated selenols. J. Biol. Chem. 270, 11761–11764. 10.1074/jbc.270.20.11761 [DOI] [PubMed] [Google Scholar]
- Brigelius-Flohé R., Maiorino M. (2013). Glutathione peroxidases. Biochim. Biophys. Acta. 1830, 3289–3303. 10.1016/j.bbagen.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Buckley P. T., Khaladkar M., Kim J., Eberwine J. (2014). Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip. Rev. RNA 5, 223–230. 10.1002/wrna.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley P. T., Lee M. T., Sul J. Y., Miyashiro K. Y., Bell T. J., Fisher S. A., et al. (2011). Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron 69, 877–884. 10.1016/j.neuron.2011.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderon F., Kim H. Y. (2004). Docosahexaenoic acid promotes neurite growth in hippocampal neurons. J. Neurochem. 90, 979–988. 10.1111/j.1471-4159.2004.02520.x [DOI] [PubMed] [Google Scholar]
- Cao D., Kevala K., Kim J., Moon H. S., Jun S. B., Lovinger D., et al. (2009). Docosahexaenoic acid promotes hippocampal neuronal development and synaptic function. J. Neurochem. 111, 510–521. 10.1111/j.1471-4159.2009.06335.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casañas-Sánchez V., Pérez J. A., Fabelo N., Herrera-Herrera A. V., Fernández C., Marín R., et al. (2014). Addition of docosahexaenoic acid, but not arachidonic acid, activates glutathione and thioredoxin antioxidant systems in murine hippocampal HT22 cells: potential implications in neuroprotection. J. Neurochem. 131, 470–483. 10.1111/jnc.12833 [DOI] [PubMed] [Google Scholar]
- Catalá A. (2009). Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem. Phys. Lipids 157, 1–11. 10.1016/j.chemphyslip.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Chen Y., Tian Z., Liang Z., Sun S., Dai C., ling, Lee M. H., et al. (2012). Brain gene expression of a sporadic (icv-STZ Mouse) and a familial mouse model (3xTg-AD mouse) of Alzheimer's disease. PLoS ONE 7:e51432. 10.1371/journal.pone.0051432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford M. A., Bloom M., Broadhurst C. L., Schmidt W. F., Cunnane S. C., Galli C., et al. (1999). Evidence for the unique function of docosahexaenoic acid during the evolution of the modern hominid brain. Lipids 34, S39–S47. 10.1007/BF02562227 [DOI] [PubMed] [Google Scholar]
- Crawford M. A., Broadhurst C. L., Guest M., Nagar A., Wang Y., Ghebremeskel K., et al. (2013). A quantum theory for the irreplaceable role of docosahexaenoic acid in neural cell signalling throughout evolution. Prostagl. Leukot. Essent. Fatty Acids 88, 5–13. 10.1016/j.plefa.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Díaz M., Fabelo N., Marin R. (2012). Genotype-induced changes in biophysical properties of frontal cortex lipid raft from APP/PS1 transgenic mice. Front. Physiol. 3:454. 10.3389/fphys.2012.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz M., Fabelo N., Martín V., Ferrer I., Gómez T., Marín R. (2015). Biophysical alterations in lipid rafts from human cerebral cortex associate with increased BACE1/Aβ PP interaction in early stages of Alzheimer's disease. J. Alzheimers Dis. 43, 1185–1198. 10.3233/JAD-141146 [DOI] [PubMed] [Google Scholar]
- Díaz M., Marín R. (2013). Brain polyunsaturated lipids and neurodegenerative diseases, in Nutraceuticals and Functional Foods: Natural Remedy, eds Brar S. K., Kaur S., Dhillon G. S. (Hauppage, NY: Nova Science Publishers; ), 387–412. [Google Scholar]
- Dröge W. (2002). Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95. 10.1152/physrev.00018.2001 [DOI] [PubMed] [Google Scholar]
- Expósito-Rodríguez M., Borges A. A., Borges-Pérez A., Pérez J. A. (2011). Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol. Biochem. 49, 782–791. 10.1016/j.plaphy.2011.02.022 [DOI] [PubMed] [Google Scholar]
- Fabelo N., Martín V., Marín R., Santpere G., Aso E., Ferrer I., et al. (2012). Evidence for premature lipid raft aging in APP/PS1 double-transgenic mice, a model of familial Alzheimer disease. J. Neuropathol. Exp. Neurol. 71, 868–881. 10.1097/NEN.0b013e31826be03c [DOI] [PubMed] [Google Scholar]
- Farooqui A. A., Horrocks L. A., Farooqui T. (2000). Deacylation and reacylation of neural membrane glycerophospholipids. J. Mol. Neurosci. 14, 123–135. 10.1385/JMN:14:3:123 [DOI] [PubMed] [Google Scholar]
- Fritz K. S., Petersen D. R. (2013). An overview of the chemistry and biology of reactive aldehydes. Free Radicals Biol. Med. 59, 85–91. 10.1016/j.freeradbiomed.2012.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godeas C., Tramer F., Micali F., Soranzo M., Sandri G., Panfili E. (1997). Distribution and possible novel role of phospholipid hydroperoxide glutathione peroxidase in rat epididymal spermatozoa. Biol. Reprod. 57, 1502–1508. 10.1095/biolreprod57.6.1502 [DOI] [PubMed] [Google Scholar]
- Gwon A. R., Park J. S., Arumugam T. V., Kwon Y. K., Chan S. L., Kim S. H., et al. (2012). Oxidative lipid modification of nicastrin enhances amyloidogenic γ-secretase activity in Alzheimer's disease. Aging Cell 11, 559–568. 10.1111/j.1474-9726.2012.00817.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. L. (2010). Omega-3 fatty acids, cognitive decline, and Alzheimer's disease: a critical review and evaluation of the literature. J. Alzheimers Dis. 21, 673–690. 10.3233/JAD-2010-090934 [DOI] [PubMed] [Google Scholar]
- Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., et al. (2003). Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Biophys. Res. Commun. 305, 278–286. 10.1016/S0006-291X(03)00734-4 [DOI] [PubMed] [Google Scholar]
- Imai H., Nakagawa Y. (2003). Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 34, 145–169. 10.1016/S0891-5849(02)01197-8 [DOI] [PubMed] [Google Scholar]
- Innis S. M. (2007). Dietary (n-3) fatty acids and brain development. J. Nutr. 137, 855–859. 10.1016/j.brainres.2008.08.078 [DOI] [PubMed] [Google Scholar]
- Ishikado A., Morino K., Nishio Y., Nakagawa F., Mukose A., Sono Y., et al. (2013). 4-Hydroxy hexenal derived from docosahexaenoic acid protects endothelial cells via Nrf2 activation. PLoS ONE 8:e69415. 10.1371/journal.pone.0069415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaladkar M., Buckley P. T., Lee M. T., Francis C., Eghbal M. M., Chuong T., et al. (2013). Subcellular RNA sequencing reveals broad presence of cytoplasmic intron-sequence retaining transcripts in mouse and rat neurons. PLoS ONE 8:e76194. 10.1371/journal.pone.0076194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Yamamoto M. (2005). Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox. Signal. 7, 385–394. 10.1089/ars.2005.7.385 [DOI] [PubMed] [Google Scholar]
- Kusunoki C., Yang L., Yoshizaki T., Nakagawa F., Ishikado A., Kondo M., et al. (2013). Omega-3 polyunsaturated fatty acid has an anti-oxidant effect via the Nrf-2/HO-1 pathway in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 430, 225–230. 10.1016/j.bbrc.2012.10.115 [DOI] [PubMed] [Google Scholar]
- Marcus D. L., Thomas C., Rodriguez C., Simberkoff K., Tsai J. S., Strafaci J. A., et al. (1998). Increased peroxidation and reduced antioxidant enzyme activity in Alzheimer's disease. Exp. Neurol. 150, 40–44. 10.1006/exnr.1997.6750 [DOI] [PubMed] [Google Scholar]
- Markesbery W. R., Lovell M. A. (1998). Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer's disease. Neurobiol. Aging 19, 33–36. 10.1016/S0197-4580(98)00009-8 [DOI] [PubMed] [Google Scholar]
- Martín V., Almansa E., Fabelo N., Díaz M. (2006). Selective enrichment in polyunsaturated fatty acids in phospholipids from neuronal-derived cell lines. J. Neurosci. Meth. 153, 230–238. 10.1016/j.jneumeth.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Moriguchi T., Harauma A., Salem N. (2013). Plasticity of mouse brain docosahexaenoic acid: modulation by diet and age. Lipids 48, 343–355. 10.1007/s11745-013-3775-5 [DOI] [PubMed] [Google Scholar]
- Naudí A., Jové M., Ayala V., Portero-Otín M., Barja G., Pamplona R. (2013). Membrane lipid unsaturation as physiological adaptation to animal longevity. Front. Physiol. 4:372. 10.3389/fphys.2013.00372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauser T., Dockheer S., Kissner R., Koppenol W. H. (2006). Catalysis of electron transfer by selenocysteine. Biochemistry 16, 6038–6043. 10.1021/bi0602260 [DOI] [PubMed] [Google Scholar]
- Niki E., Yoshida Y., Saito Y., Noguchi N. (2005). Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem. Biophys. Res. Commun. 338, 668–676. 10.1016/j.bbrc.2005.08.072 [DOI] [PubMed] [Google Scholar]
- Oster T., Pillot T. (2010). Docosahexaenoic acid and synaptic protection in Alzheimer's disease mice. Biochim. Biophys. Acta 1801, 791–798. 10.1016/j.bbalip.2010.02.011 [DOI] [PubMed] [Google Scholar]
- Pamplona R., Dalfó E., Ayala V., Bellmunt J., Prat J., Ferrer I., et al. (2005). Proteins in human brain cortex are modified by oxidation, glycoxidation, and lipoxidation: effects of Alzheimer disease and identification of lipoxidation targets. J. Biol. Chem. 280, 21522–21530. 10.1074/jbc.M502255200 [DOI] [PubMed] [Google Scholar]
- Pfeifer H., Conrad M., Roethlein D., Kyriakopoulos A., Brielmeier M., Bornkamm G. W., et al. (2001). Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 15, 1236–1238. 10.1096/fj.00-0655fje [DOI] [PubMed] [Google Scholar]
- Plourde M., Cunnane S. C. (2007). Extremely limited synthesis of long chain polyunsaturates in adults: implications for their dietary essentiality and use as supplements. Appl. Physiol. Nutr. Metab. 32, 619–634. 10.1139/H07-034 [DOI] [PubMed] [Google Scholar]
- Porter N. A., Caldwell S. E., Mills K. A. (1995). Mechanisms of free radical oxidation of unsaturated lipids. Lipids 30, 277–290. 10.1007/BF02536034 [DOI] [PubMed] [Google Scholar]
- Praticò D., M. Y., Lee V., Trojanowski J. Q., Rokach J., Fitzgerald G. A. (1998). Increased F2-isoprostanes in Alzheimer's disease: evidence for enhanced lipid peroxidation in vivo. FASEB J. 12, 1777–1783. [DOI] [PubMed] [Google Scholar]
- Savaskan N. E., Ufer C., Kühn H., Borchert A. (2007). Molecular biology of glutathione peroxidase 4: from genomic structure to developmental expression and neural function. Biol. Chem. 388, 1007–1017. 10.1515/BC.2007.126 [DOI] [PubMed] [Google Scholar]
- Scheerer P., Borchert A., Krauss N., Wessner H., Gerth C., Hohne W., et al. (2007). Structural basis for catalytic activity and enzyme polymerization of phospholipid hydroperoxide glutathione peroxidase-4 (GPx4). Biochemistry 46, 9041–9049. 10.1021/bi700840d [DOI] [PubMed] [Google Scholar]
- Simopoulos A. P. (2011). Evolutionary aspects of diet: the Omega-6/Omega-3 Ratio and the brain. Mol. Neurobiol. 44, 203–215. 10.1007/s12035-010-8162-0 [DOI] [PubMed] [Google Scholar]
- Singh M., Dang T. N., Arseneault M., Ramassamy C. (2010). Role of by-products of lipid oxidation in Alzheimer's disease brain: a focus on acrolein. J. Alzheimers. Dis. 21, 741–756. 10.3233/JAD-2010-100405 [DOI] [PubMed] [Google Scholar]
- Solfrizzi V., D'Introno A., Colacicco A. M., Capurso C., Todarello O., Pellicani V., et al. (2006). Circulating biomarkers of cognitive decline and dementia. Clin. Chim. Acta 364, 91–112. 10.1016/j.cca.2005.06.015 [DOI] [PubMed] [Google Scholar]
- Stillwell W., Wassal S. R. (2003). Docosahexaenoic acid: membrane properties of a unique fatty acid. Chem. Phys. Lipids 126, 1–27. 10.1016/S0009-3084(03)00101-4 [DOI] [PubMed] [Google Scholar]
- Uauy R., Hoffman D. R., Peirano P., Birch D. G., Birch E. E. (2001). Essential fatty acids in visual and brain development. Lipids 36, 885–895. 10.1007/s11745-001-0798-1 [DOI] [PubMed] [Google Scholar]
- Ufer C., Wang C. C. (2011). The roles of glutathione peroxidases during embryo development. Front. Mol. Neurosci. 4:12. 10.3389/fnmol.2011.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger T., Korade Z., Lazarov O., Terrano D., Schor N. F., Sisodia S. S., et al. (2005). Transcriptome differences between the frontal cortex and hippocampus of wild-type and humanized presenilin-1 transgenic mice. Am. J. Geriatr. Psychiatry 13, 1041–1051. 10.1097/00019442-200512000-00003 [DOI] [PubMed] [Google Scholar]
- Valko M., Leibfritz D., Moncol J., Cronin M. T., Mazur M., Telser J. (2007). Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 39, 44–84. 10.1016/j.biocel.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Van Kuijk F. J., Holte L. L., Dratz E. A. (1990). 4-Hydroxyhexenal: a lipid peroxidation product derived from oxidized docosahexaenoic acid. Biochim. Biophys. Acta 1043, 116–118. 10.1016/0005-2760(90)90118-H [DOI] [PubMed] [Google Scholar]
- Wu K. C., Cui J. Y., Klaassen C. D. (2011). Beneficial role of Nrf2 in regulating NADPH generation and consumption. Toxicol. Sci. 123, 590–600. 10.1093/toxsci/kfr183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. C., Lii C. K., Wei Y. L., Li C. C., Lu C. Y., Liu K. L., et al. (2013). Docosahexaenoic acid inhibition of inflammation is partially via cross-talk between Nrf2/heme oxygenase 1 and IKK/NF-κ B pathways. J. Nutr. Biochem. 24, 204–212. 10.1016/j.jnutbio.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Yant L. J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J. G., et al. (2003). The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 34, 496–502. 10.1016/S0891-5849(02)01360-6 [DOI] [PubMed] [Google Scholar]
- Zhang M., An C., Gao Y., Leak R. K., Chen J., Zhang F. (2013). Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog. Neurobiol. 100, 30–47. 10.1016/j.pneurobio.2012.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X., Liu J. R., Kyle J. W., Hanck D. A., Agnew W. S. (2006). A profile of alternative RNA splicing and transcript variation of CACNA1H, a human T-channel gene candidate for idiopathic generalized epilepsies. Hum. Mol. Genet. 15, 1497–1512. 10.1093/hmg/ddl068 [DOI] [PubMed] [Google Scholar]