Abstract
Regeneration involves the integration of new and old tissues in the context of an adult life history. It is clear that the core conserved signalling pathways that orchestrate development also play central roles in regeneration, and further study of conserved signalling pathways is required. Here we have studied the role of the conserved JNK signalling cascade during planarian regeneration. Abrogation of JNK signalling by RNAi or pharmacological inhibition blocks posterior regeneration and animals fail to express posterior markers. While the early injury-induced expression of polarity markers is unaffected, the later stem cell-dependent phase of posterior Wnt expression is not established. This defect can be rescued by overactivation of the Hh or Wnt signalling pathway to promote posterior Wnt activity. Together, our data suggest that JNK signalling is required to establish stem cell-dependent Wnt expression after posterior injury. Given that Jun is known to be required in vertebrates for the expression of Wnt and Wnt target genes, we propose that this interaction may be conserved and is an instructive part of planarian posterior regeneration.
KEY WORDS: Regeneration, Stem cells, JNK signalling, Jun, Wnt, Planarian, Schmidtea mediterranea
Summary: The conserved JNK signalling pathway promotes Wnt signalling in stem cells to allow the regeneration of posterior structures in injured flatworms.
INTRODUCTION
Regenerative phenomena are widespread across the animal kingdom, although our understanding of how they work, whether they are conserved and how they evolve is still sparse. A number of model systems have been used to expand our understanding of the cellular and molecular mechanisms underpinning regeneration (reviewed by Poss, 2010; Tanaka and Reddien, 2011). This work has established that conserved signalling cascades with central roles in the embryogenesis of all animals are also active during regeneration, with roles that are also anatomically and physiologically reminiscent of their functions during development.
This is particularly the case in the planarian flatworm model system. For example, planarians use Wnt and Hedgehog (Hh) signalling for anterior-posterior patterning and BMP signalling for dorsal-ventral patterning in line with the widely conserved roles of these pathways in specifying fates during the embryogenesis of many animal phyla (reviewed by Aboobaker, 2011; Almuedo-Castillo et al., 2012; Reddien, 2011; Rink, 2013). In addition to Wnt and Hh, a number of other conserved signalling molecules and transcription factors have recently been shown to be involved in different aspects of regeneration, in some cases providing entirely novel insights into the developmental and physiological roles of conserved genes (Almuedo-Castillo et al., 2014; Cebrià et al., 2002a; Cowles et al., 2013; Fraguas et al., 2014; González-Estévez et al., 2012; Molina et al., 2011; Solana et al., 2013; Vogg et al., 2014).
Planarian regeneration and the planarian life history allow the roles of genes and pathways essential to embryogenesis in other animals to be studied in an adult context, easily circumventing the problem of embryonic lethality masking later life history functions. Many genes are likely to have pleiotropic roles in adult animals but the ability to study specific regenerative scenarios coupled with RNAi-mediated studies of gene function can allow these roles to be picked apart. We have exploited this particular feature to study the role of the broadly conserved c-Jun N-terminal protein kinase (JNK) pathway and uncover an undescribed role in controlling posterior regeneration.
JNK, also referred to as stress-activated protein kinase, is a member of the large mitogen-activated protein kinase family. The JNK pathway is implicated in multiple physiological processes, including cell proliferation, programmed cell death, nervous system development and T-cell-mediated immunity (reviewed by Davis, 2000; Weston and Davis, 2002, 2007). It has recently been suggested that JNK-dependent compensatory proliferation is itself induced by JNK-mediated apoptosis. This finding might explain how JNK appears to have both protective and inductive roles in different types of cancer (Chen, 2012). Many of the functions of JNK signalling are obviously of high relevance to the processes of regeneration, such as organismal and local responses to wounding and the control of growth for tissue remodelling. In planarians, potential downstream components of the JNK signalling cascade have been implicated in controlling the clonal expansion of stem cells and are upregulated upon wounding (Wagner et al., 2012; Wenemoser et al., 2012). Two previous studies describe that the signalling pathway and the planarian orthologue of JNK itself are required for wound healing, for driving stem cell mitosis and for correctly activating cell death during regeneration (Almuedo-Castillo et al., 2014; Tasaki et al., 2011).
Here, we have elucidated another role for the JNK signalling cascade and, in particular, for the transcriptional effector Jun in driving axial polarity. Pharmacological or RNAi-mediated abrogation of JNK signalling leads to a failure in posterior regeneration. JNK signalling is required for extension of posterior tissues and establishment of the posterior pole of Wnt-expressing cells, possibly through the involvement of Jun in activating the expression of Wnts in stem cell progeny. In junl-1(RNAi) animals, we also uncover an anterior midline regeneration defect that is caused by expansion of slit expression as correct wnt5 expression fails to re-establish during regeneration.
These data suggest a model in which JNK signalling is required downstream of initial wound-induced Wnt activity to drive the formation of a posterior Wnt-expressing pole from differentiating stem cells at the posterior regeneration blastema. Similar interactions between the JNK and Wnt signalling pathways have been described previously in mammals and other vertebrates, suggesting that this might be a conserved signalling pathway interaction within the Bilateria that is important for posterior identity (Gan et al., 2008; Nateri et al., 2005; Saadeddin et al., 2009).
RESULTS
JNK signalling components are required for tail regeneration
We used informatics searches of the planarian genome and consolidated transcriptome data sets to identify orthologues of Hemipterous/Map kinase kinase 7 (hem), JNK (jnk) and two potential orthologues of the Jun transcription factor. One of these, previously called junl-1 (Wagner et al., 2012), appears to be closely related to other platyhelminth and protostome Jun genes. The other, jun-1 (Wenemoser et al., 2012), does not have a clear orthologue in extant parasitic platyhelminth data or other protostomes and appears to have undergone relatively rapid sequence evolution (supplementary material Fig. S1).
In order to investigate the role of the JNK signalling pathway during regeneration we used RNA interference (RNAi) to knock down the expression of the core JNK signalling components (see supplementary material Fig. S2A for RNAi protocol). After two rounds of injections we amputated animals in front of and behind the pharynx and followed regeneration (supplementary material Fig. S2B). Whereas anterior regeneration proceeded normally, in nearly all animals we observed a clear impairment in tail regeneration, with all head fragments and most trunk fragments failing to regenerate a tail (Fig. 1A; supplementary material Fig. S3). A small proportion of tail fragments failed to regenerate their eyes appropriately, displaying smaller eyes than controls (see below).
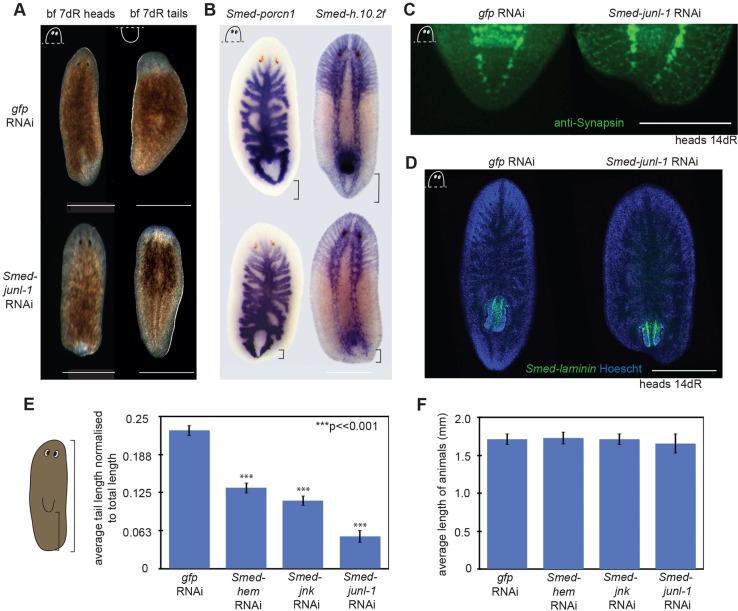
Fig. 1.
RNAi of JNK signalling pathway members disrupts tail formation. (A) Posterior blastemas of control animals [gfp(RNAi)] regenerated tails, whereas the tails did not regenerate after Smed-junl-1(RNAi). Anterior blastemas appeared normal after Smed-junl-1(RNAi). (B) WISH with the gut marker Smed-porcn1 (gfp n=4/4, junl-1 a=7/7) and the nervous system marker Smed-h.10.2f (gfp n=6/6, junl-1 a=9/9) showed that the tails do not extend after Smed-junl-1(RNAi). All stains were performed on heads at 7 days of regeneration (dR). (C) Immunohistochemistry of the nervous system with anti-Synapsin revealed that the ventral nerve chords (VNCs) at 14 dR do not fuse at the posterior after Smed-junl-1(RNAi) (gfp n=7/7, junl-1 a=6/7; in a separate experiment, animals that were fixed at 7 dR: gfp n=26/26, junl-1 a=24/29). (D) FISH with the pharynx marker Smed-laminin demonstrates a failure of regeneration posterior to the pharynx (green) in the regenerating head at 14 dR stained with Hoechst 33342 (blue) (gfp n=10/10, junl-1 a=10/10). (E) The tail lengths of Smed-hem(RNAi), Smed-jnk(RNAi) and Smed-junl-1(RNAi) in regenerating head pieces were significantly shorter than in gfp controls. Tail length was measured from the tip of the pharynx to the end of the animal and normalised to total length. A minimum of 60 animals was measured with the Measure tool in Fiji, using bright-field pictures. The bar chart shows mean±s.e.m.; ***P<0.001 (two-tailed t-test) compared with gfp(RNAi) animals. (F) The average total length (mean±s.e.m.) of the animals from E is not significantly different to controls, suggesting a specific failure in tail regeneration and potential compensatory changes elsewhere. Counts are for normal (n) or abnormal (a) animals as described per condition. Scale bars: 500 μm.
To further characterise the tailless phenotype, WISH was performed using a gut marker (Smed-porcn1) and a nervous system marker (Smed-h.10.2f) at 7 days of regeneration (dR). Although the gut and the nervous system clearly regenerate, the ventral nerve chords (VNCs) do not join at the posterior, and the posterior gut branches appeared closer to the posterior tip of all knockdown animals (Fig. 1B). At 14 dR, gfp(RNAi) controls fully regenerated the VNCs, which joined at the posterior tip, whereas junl-1(RNAi) tails had truncated VNCs and the posterior tip failed to regenerate correctly (Fig. 1C). FISH with the pharynx marker Smed-laminin revealed that this organ does regenerate in junl-1(RNAi) knockdowns, albeit in a relatively more posterior position than in controls (Fig. 1D).
jnk(RNAi) animals shared the same tailless phenotype and hem(RNAi) animals displayed a milder defect with respect to VNC regeneration (supplementary material Figs S3 and S4). Trunk and tail pieces regenerate the anterior normally without any effect on eye regeneration after both jnk(RNAi) and hem(RNAi) (supplementary material Fig. S3). However, by performing double-RNAi experiments with pathway components, more severe tailless phenotypes could be generated in hem/jnk(RNAi) animals and more severe effects on regeneration were observed in jnk/junl-1(RNAi) animals (supplementary material Fig. S3). Measurement of Smed-jnk transcript levels (supplementary material Fig. S5) remaining after jnk(RNAi) revealed that levels were similar to those reported in an earlier study (Almuedo-Castillo et al., 2014). Given the likely pleiotropic roles of JNK signalling, the posterior regeneration defect caused by our RNAi knockdown schedule of all three JNK pathway components presented a focused opportunity to study a particular role of JNK signalling during planarian regeneration.
In order to quantify and confirm our phenotypic observations, the distance between the posterior tip of the pharynx and the end of each animal was measured and normalised to the total length of each animal to provide a measure of tail length. Tail length was analysed at 14 dR on head fragments, with more than 60 animals for each RNAi condition. JNK pathway RNAi animals had significantly shorter tails than the controls, junl-1(RNAi) having the largest effect (Fig. 1E). Conversely, the length from the anterior to the tip of the pharynx increased in all three RNAi phenotypes (supplementary material Fig. S2C). In line with these two findings, the overall total length of animals was not affected (Fig. 1F). This confirms that the observed phenotype is a failure in posterior regeneration and also results in the relative position of the pharynx being more to the posterior. Taken together, these data suggest that our RNAi experiments reveal a role of JNK signalling that is specific to posterior regeneration and does not reflect a general growth defect.
The tailless phenotype can be phenocopied by chemical inhibition of JNK signalling
To further investigate the role of JNK signalling the JNK inhibitor SP600125 (Bennett et al., 2001) was used to treat animals during regeneration. We found that the high dose of this inhibitor (25 µM) previously used for planarians (Tasaki et al., 2011) was lethal to S. mediterranea within 30 min to 1 h of exposure. At this high dose it is also likely to have off-target effects on other kinases (Bain et al., 2007; Tanemura et al., 2009). A dose curve was established to find a concentration at which S. mediterranea animals could be cultured without any general toxic effects (supplementary material Fig. S6A). Treatment with doses up to 5 µM did not have any apparent immediate adverse effects but these animals could not perform wound healing and died between 2 and 3 days after amputation. A 5 µM dose applied after wound healing allowed 70% survival until 5 dR, and these survivors had no posterior blastema, a reduced anterior and all died over the following 5 days.
At 1 µM SP600125 over 95% of animals survived for the whole regenerative timecourse. At this concentration, tailless regeneration phenotypes were observed that phenocopied JNK signalling RNAi (Fig. 2A; supplementary material Fig. S6B), including a failure of the VNCs to join (Fig. 2A). At 1 µM no effect on anterior regeneration was observed (Fig. 2B). These data provide a second line of evidence that JNK signalling is required for posterior regeneration.
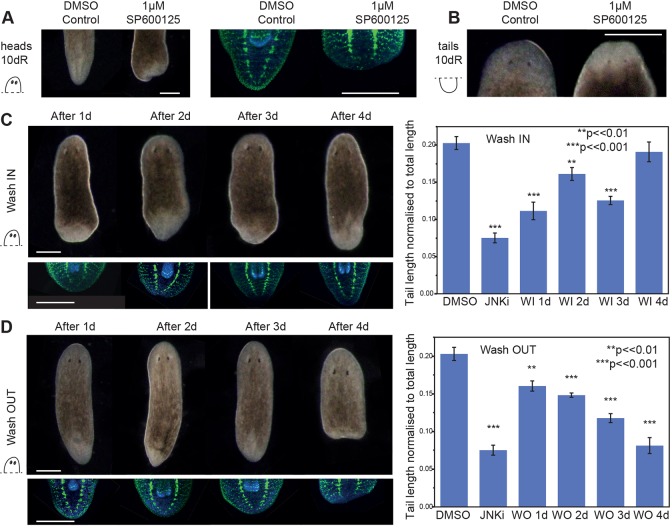
Fig. 2.
Pharmacological inhibition of JNK signalling phenocopies the tailless phenotype. (A) Treatment with 1 μM SP600125 JNK inhibitor (JNKi) results in tailless animals after 10 dR, as compared with controls (0.05% DMSO control n=40/40, JNKi a=37/40, across three experiments). (B) Anterior regeneration was normal in both DMSO controls (n=40/40) and JNKi (n=40/40) across three experiments. Immunohistochemistry with anti-Synapsin shows that the VNCs do not extend or join at the tip (a=10/10). (C) Wash-in experiment. Animals were amputated pre-pharyngeally and placed in planarian water supplemented with 0.05% DMSO. The water was replaced with 1 μM JNKi after 1, 2, 3 or 4 dR (wash-in day, WI d), and the animals were left to regenerate until day 10, when they were scored, measured, fixed and stained with anti-Synapsin. Adding JNKi during the first 3 dR results in reduced tail length, whereas adding JNKi after 4 dR has no effect (WI 1 d a=8/10, WI 2 d a=8/10 and WI 3 d a=11/11 show reduced tail length and VNCs that join at the tip; WI 4 d n=11/11 have normal tails and VNCs). (D) Wash-out experiment. Animals were cut pre-pharyngeally and immediately placed in JNKi solution. The solution was replaced with planarian water containing 0.05% DMSO after 1, 2, 3 or 4 dR (wash-out day, WO d), after which the animals were left to regenerate. Imaging, fixing and staining was performed at 10 dR. Addition of the inhibitor for just 1 day resulted in reduced tail length, with the most pronounced effect occurring after exposure for 4 days, after which the phenotype was not reversible (WO 1 d a=8/9, WO 2 d a=8/10 and WO 3 d a=11/11 have reduced tail lengths compared with controls and VNCs that joined at the tip; WO 4 d a=9/11 were indistinguishable from JNKi controls, had very small or no posterior blastema and the VNCs did not join at the posterior). Bar charts show mean±s.e.m.; ***P<0.001, **P<0.01 (one-tailed t-test). Both wash-in and wash-out experiments had matched controls for 0.05% DMSO and 1 μM JNKi for 10 dR (A). Counts are for normal (n) or abnormal (a) animals as described per condition. Scale bars: 500 μm.
The efficacy of SP600125 presented the opportunity to study the time at which the posterior phenotype takes effect and to ascertain whether the effect on tail formation is reversible. To examine the time window at which JNK signalling is required for tail specification, we performed timed exposure experiments during posterior regeneration. First, we performed ‘wash-in’ assays, amputating animals and adding 1 µM SP600125 after 1, 2, 3 or 4 dR, and observing tail regeneration at 10 dR. We found that if the inhibitor was added within the first 3 dR, although tails could regenerate to a greater extent than in animals under maintained exposure, they were significantly shorter than control animals exposed to vehicle alone. The greatest effects on tail length were observed after adding the inhibitor after 1 or 3 dR (Fig. 2C). Adding the inhibitor at 4 dR had no effect on tail length at 10 dR, even though the majority of elongation occurs in this time window. In wash-in experiments between 2 and 4 days after amputation, VNC regeneration was more complete than in animals subjected to constant SP600125 exposure or in RNAi animals. When 1 µM SP600125 was added after just 1 dR, VNC regeneration defects were similar to those observed upon constant exposure to the inhibitor or in RNAi animals (Fig. 2B).
The opposite ‘wash-out’ experiment, in which 1 µM SP600125 was removed after 1, 2, 3 or 4 dR, revealed that the longer the animals were exposed to the inhibitor the shorter the tails became, until they were indistinguishable from controls exposed to the inhibitor for the whole 10 dR (Fig. 2D). Removing the inhibitor after 4 dR did not result in a restoration of tail length.
Taken together, these data suggest that knocking down or inhibiting members of the JNK signalling cascade leads to defects in planarian tail regeneration and that JNK signalling is necessary to allow posterior regeneration during the first 4 dR, but not thereafter.
Smed-junl1 is required for correct anterior midline specification
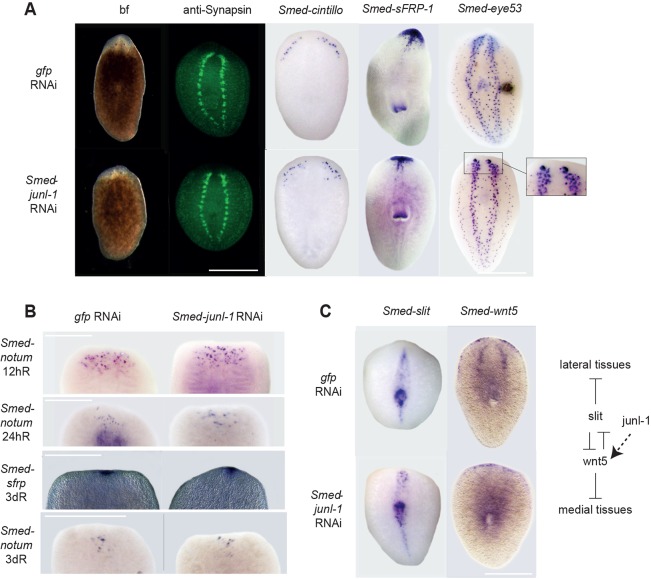
Close observation of anterior regeneration of Smed-junl-1(RNAi) animals revealed a low penetrance effect on eye formation (smaller or misplaced) in 12/81 regenerating tails and 7/85 regenerating trunks (Fig. 3A). Further structural characterisation revealed that there were some underlying defects in anterior regeneration that were likely to be responsible for causing the mild effects on eye size and placement. Immunostaining for the neuronal marker Synapsin revealed that, although the brain and nervous system regenerated, they were patterned incorrectly (Fig. 3A). This was also apparent from staining for other neuronal markers, such as Smed-eye-53 and Smed-cintillo (Fig. 3A). The brain lobes were laterally spread compared with controls and failed to connect. Ectopic cintillo+ cells were observed away from their normal position at the lateral anterior margins (Fig. 3A). The anterior polarity marker Smed-sFRP had reduced lateral expression compared with controls at 14 dR. Early anterior polarity marker expression (Smed-notum and Smed-sFRP) was indistinguishable from that of controls (Fig. 3B), confirming that there was not a defect in setting anterior polarity per se, but with later patterning events.
Fig. 3.
Smed-junl1 is required for correct anterior midline specification. (A) After 10 dR, tail pieces regenerated an anterior blastema with eyes as scored by bright-field (bf) (gfp n=80/80, junl-1 n=78/85). However, cephalic ganglia (CG) did not connect and were spread laterally in Smed-junl-1(RNAi) animals as revealed by staining of the brain with anti-Synapsin (gfp n=29/29, junl-1 a=25/27, in three separate experiments at 10 dR) and staining with Smed-eye-53 at 7 dR (gfp n=25/25, junl-1 a=18/25). The lateral expression of anterior marker Smed-sFRP was reduced at 14 dR (gfp n=6/6, junl-1 a=10/12), and there were ectopic cintillo+ cells in Smed-junl-1(RNAi) animals by 10 dR (gfp n=4/4, junl-1 a=4/4). (B) Early anterior polarity appeared unaffected in junl-1(RNAi) animals. The markers used were Smed-notum at 12 hours of regeneration (hR) (gfp n=27/27, junl-1 n=24/24), 24 hR (gfp n=10/10, junl-1 n=11/11) and 3 dR (gfp n=5/5, junl-1 n=5/5) and Smed-sFRP (gfp n=6/6, junl-1 n=7/7) at 3 dR. (C) The expression of midline marker Smed-slit appeared expanded at the anterior blastema in junl-1(RNAi) regenerating tails at 7 dR, consistent with a midline phenotype (gfp n=12/12, junl-1 a=6/8). Additionally, the expression of medial tissue patterning gene Smed-wnt5 is reduced after junl-1(RNAi) (gfp n=6/7, junl-1 a=10/11). Counts are for normal (n) or abnormal (a) animals as described per condition. Scale bars: 500 μm.
The defects observed in anterior regeneration suggested a potential midline expansion defect. This possibility was investigated with the midline marker Smed-slit. We found this marker to have an expanded domain of expression at the anterior regeneration tip of junl-1(RNAi) animals (Fig. 3C). This provides a plausible explanation for the phenotype that we observe and the position of the brain lobes, which in wild-type animals regenerate around the midline. We did not observe expansion of Smed-slit expression in animals treated with 1 µM SP600125 (supplementary material Fig. S7A).
The expansion in slit expression we observe is reminiscent of that seen after Smed-wnt5(RNAi) (Gurley et al., 2010). The mediolateral expression of Smed-wnt5 in junl-1(RNAi) animals was indeed reduced compared with controls. These data indicate that the effects of junl-1(RNAi) on anterior regeneration might be the result of a reduction in Smed-wnt5 expression leading to an expansion in Smed-slit expression and thus midline specification (Fig. 3C). We did not see a loss of wnt5 expression in 1 µM SP600125-treated animals (supplementary material Fig. S7B) or in Smed-hem(RNAi) or Smed-jnk(RNAi) animals (supplementary material Fig. S7C), suggesting that a specific effect of junl-1(RNAi) leads to this subtle anterior phenotype.
JNK signalling is required for the stem cell-dependent phase of Wnt gene expression
Given the high penetrance of the junl-1(RNAi) posterior phenotype, it seemed pertinent to focus on this transcription factor as a terminal effector of JNK signalling activity. Previous knowledge of JNK signalling and its described roles in planarians (Almuedo-Castillo et al., 2014; Tasaki et al., 2011; Wagner et al., 2012) led to the consideration of three initial plausible roles for junl-1 in posterior regeneration: (1) a requirement to drive regeneration by facilitating new tissue growth; (2) a requirement specifically for new tissue growth at posterior-facing wounds; (3) a role in establishing posterior identity, which then leads to correct tail growth.
To test whether growth was affected in a way that could plausibly cause a defect in posterior regeneration the proliferative response of stem cells after junl-1(RNAi) was measured by counting mitotic cells. Although all animals were tailless, no negative effects on stem cell proliferation during the first 72 h of regeneration were observed, arguing against an effect on growth, as reflected in stem cell proliferation, during the time window that JNK signalling is required for posterior regeneration (supplementary material Fig. S8A). We also measured the absolute number of stem cells at posterior-facing wounds after 4 dR to see whether a decrease in neoblast numbers could explain the posterior defects, but we observed no difference between junl-1(RNAi) and control animals (supplementary material Fig. S8B). Given that, in the vast majority of animals, the anterior regeneration of trunks and tail fragments appears normal, the finding that global proliferation is not affected argues against a general effect on growth in our experimental paradigm. A previous report highlighted effects of jnk(RNAi) in reducing apoptosis levels during tissue remodelling (Almuedo-Castillo et al., 2014). We used whole-mount terminal uridine nick-end labelling (TUNEL) to look at apoptosis at 4 h and 72 h post amputation, but observed no difference between junl-1(RNAi) and control animals. We also investigated the effect of 1 µM SP600125 treatment on apoptosis (supplementary material Fig. S9A,B) at 4 h and 72 h post amputation, and observed no difference between SP600125 treatment and control animals.
In order to determine whether the effect of junl-1(RNAi) was specific to regenerating any new tissue in posterior regions, reflecting a specific effect on growth at posterior-facing amputations, double beta-catenin1/junl-1(RNAi) was performed. It has previously been demonstrated that beta-catenin1(RNAi) leads to anterior structures regenerating at all wounds and, when used in double-RNAi experiments, has proven to be a useful test of epistastic relationships and pleiotropic roles during planarian regeneration (Blassberg et al., 2013; Felix and Aboobaker, 2010; Gurley et al., 2008; Iglesias et al., 2008; Petersen and Reddien, 2008). In contrast to junl-1/gfp(RNAi) animals, which were all tailless, beta-catenin1/junl-1(RNAi) animals were able to regenerate anterior structures at posterior-facing wounds to a similar extent as beta-catenin1/gfp(RNAi) worms (supplementary material Fig. S8C), with similarly sized blastemas. The neural marker Smed-h.10.2f, the anterior polarity marker Smed-sFRP and the brain-specific marker Smed-GPAS were all expressed at posterior-facing wounds, confirming ectopic anterior regeneration in both beta-catenin1/gfp(RNAi) and beta-catenin1/junl-1(RNAi) animals (supplementary material Fig. S8D). Together, these data suggest that the posterior phenotype observed in junl-1(RNAi) animals is unlikely to be due to a failure to regenerate new tissue by stem cells at posterior wounds, but is instead a specific failure to regenerate posterior tissues.
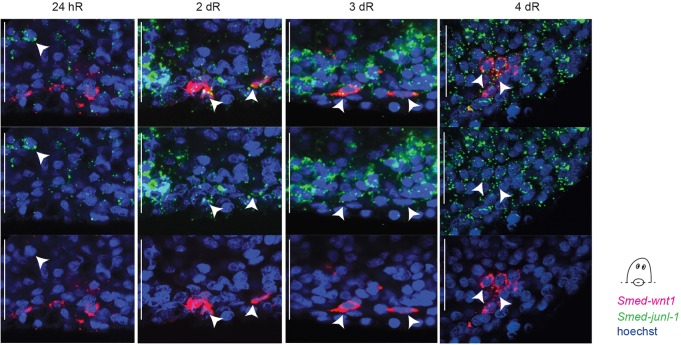
Previous work has shown that posterior regeneration requires a defined programme of Wnt signalling activity in response to wounding. Wounding results in stem cell-independent Smed-wnt1 expression in muscle cells at all wound sites (Petersen and Reddien, 2009b; Witchley et al., 2013). This early wnt1 expression is required for stem cells at the posterior wound site to differentiate to form a Wnt gene-expressing pole in the regenerating blastema (Petersen and Reddien, 2009b). Active Hh signalling is required for early wound-induced Wnt gene expression and a number of other genes have now been shown to be required for the formation of the stem cell-dependent phase of later Wnt gene expression (Blassberg et al., 2013; Chen et al., 2013; Currie and Pearson, 2013; Hayashi et al., 2011; Marz et al., 2013; Rink et al., 2009; Yazawa et al., 2009). In junl-1(RNAi) worms, the early phase of wound-induced wnt1 expression was not affected and was equivalent to that observed for control animals at both anterior and posterior wounds (Fig. 4A; supplementary material Fig. S10A). Additionally, early wnt1 expression was unaffected by 1 µM SP600125 treatment (supplementary material Fig. S10B). At 4 dR the stem cell-dependent Smed-wnt1 expression that is normally established at the posterior pole failed to appear in junl-1(RNAi) worms (Fig. 4A). This loss of later Smed-wnt1 was also observed for SP600125-treated animals (Fig. 4B) and for jnk(RNAi) animals (supplementary material Fig. S10C). Other posterior determinants and markers, including Smed-wnt11-2, Smed-wnt11-5 and Smed-fzl4, were all absent in junl-1(RNAi) animals (Fig. 4C).
Fig. 4.
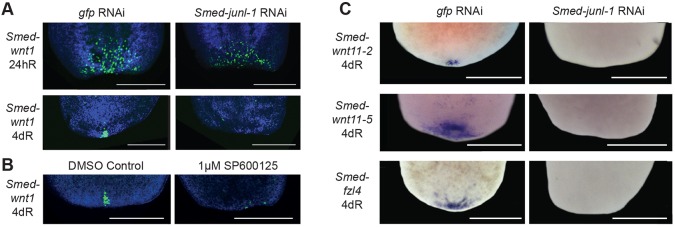
JNK-signalling is required for stem cell-dependent wnt1 expression. (A) The wound-induced expression of wnt1 (24 hR) was not affected after Smed-junl-1(RNAi) (gfp n=40/40, junl-1 n=40/43, over three experiments). However, stem cell-dependent wnt1 is not expressed at 4 dR (gfp n=14/14, junl-1 a=13/16, over two separate experiments). (B) Stem cell-dependent wnt1 expression is also reduced after incubation with JNKi as compared with the controls (control n=13/14, JNKi a=7/13). (C) Posterior markers Smed-wnt11-2 (gfp n=5/5, junl-1 a=5/5), Smed-wnt11-5 (gfp n=6/6, junl-1 a=5/5) and Smed-fzl4 (gfp n=7/7, junl-1 a=14/15) were also absent at 4 dR following Smed-junl-1(RNAi). Counts are for normal (n) or abnormal (a) animals as described per condition. Scale bars: 500 μm.
Taken together, these data suggest that JNK signalling is not required for early wound-induced expression of wnt1 but is required for later establishment of the stem cell-dependent posterior domain of expression of multiple Wnt genes and for posterior regeneration. These data explain the failure in posterior regeneration caused by abrogation of JNK signalling.
Constitutive Hh and Wnt signalling rescue posterior regeneration defects and junl-1 is required for islet expression at the posterior pole
These results led to the possibility that junl-1 could play a role in the regulation of posterior cell fate by regulating the ability of differentiating cells entering the blastema to react to the wound-induced Wnt signal and to subsequently express appropriate posterior effectors and markers. One prediction of this hypothesis is that overactivating Wnt signalling should rescue the junl-1(RNAi) phenotype.
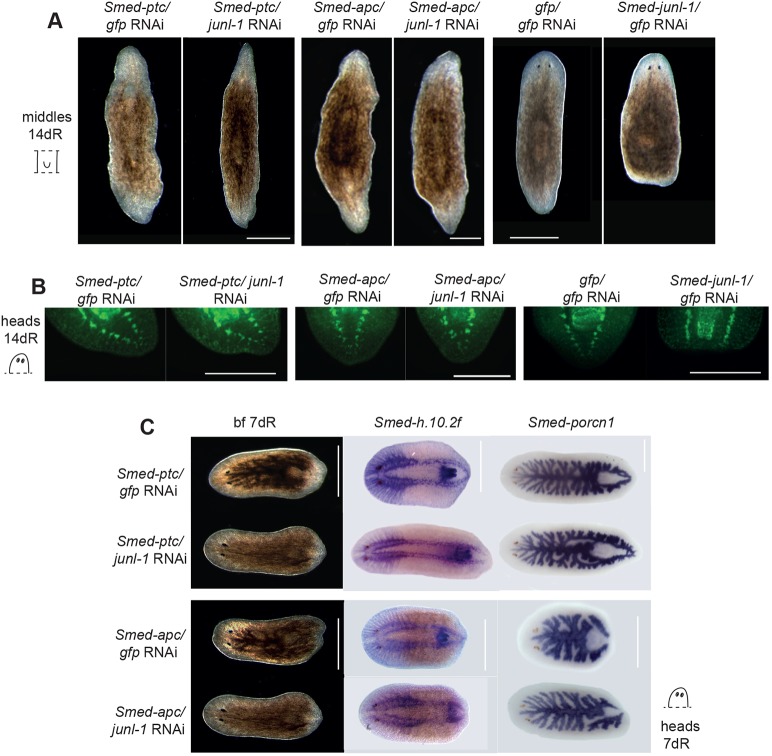
To overactivate Wnt signalling we performed double RNAi with Smed-patched (ptc) and Smed-apc (apc). The phenotypes of ptc and apc have been extensively characterised previously (Evans et al., 2011; Gurley et al., 2008; Rink et al., 2009; Yazawa et al., 2009). Loss of ptc leads to constitutive Hh signalling, which in turn overactivates Wnt signalling and thus posterior fate and tail formation at every wound. Similarly, loss of apc leads to constitutive Wnt signalling and thus posterior fate and tail formation at every wound. Overactivation of Hh and Wnt signalling by double ptc/junl-1(RNAi) or apc/junl-1(RNAi) was sufficient to rescue the junl-1 RNAi tailless phenotype. This was confirmed by bright-field microscopy (Fig. 5A), by staining with anti-Synapsin at 14 dR (Fig. 5B), and by staining for the neural marker Smed-h.10.2f and gut marker Smed-porcn1 at 7 dR (Fig. 5C). We measured junl-1 transcript levels in the different double-RNAi conditions and found no indication that rescue was due to reduced efficacy of junl-1 knockdown (supplementary material Fig. S5).
Fig. 5.
Overactivation of Wnt signalling rescues the tailless phenotype. (A) Double Smed-ptc/junl-1(RNAi) and Smed-apc/junl-1(RNAi) resulted in tails forming at every wound site (xt), compared with the tailless animals (a) that result after Smed-junl-1/gfp(RNAi) (ptc/gfp xt=53/66, ptc/junl-1 xt=67/69, apc/gfp xt=40/40, apc/junl-1 xt=40/40, gfp/gfp n=86/86, junl-1/gfp a=95/101, as scored for tails at both ends for the ptc knockdowns and tails at the posterior for gfp and junl-1). (B) The restoration of the tail was confirmed with the pan-neuronal marker anti-Synapsin on heads at 14 dR (ptc/gfp xt=25/25, ptc/junl-1 xt=30/33, apc/gfp xt=6/6, apc/junl-1 xt=9/9, gfp/gfp n=26/27, junl-1/gfp a=20/23, over three experiments). (C) The rescue of the tail is also apparent in head pieces by 7 dR (ptc/gfp xt=40/40, ptc/junl-1 xt=40/40, apc/gfp xt=40/40, apc/junl-1 xt=40/40). Nervous system and gut markers Smed-h.10.2f (ptc/gfp xt=16/16, ptc/junl-1 xt=15/17, apc/gfp xt=14/14, apc/junl-1 xt=5/9) and Smed-porcn1 (ptc/gfp xt=11/11, ptc/junl-1 xt=7/7, apc/gfp xt=10/10, apc/junl-1 xt=4/4) further confirmed the rescue of the tail. Counts indicate the number of animals showing normal (n), extra tails (xt) or absence of tail (a) per condition. Scale bars: 500 μm.
Taken together, these results demonstrate that stem cell-dependent wnt1 expression, but not wound-induced wnt1 expression, is affected by junl-1(RNAi) and that rescue of this phenotype by ectopic activation of Wnt/Hh signalling supports the finding that the effect of junl-1 is mediated through an effect on Wnt signalling.
Although Wnt and Hh signalling are clearly central players in controlling anterior-posterior polarity in planarians, a number of other genes have also been shown to be required for this process by potentially interacting with these pathways. In particular, Smed-islet (Hayashi et al., 2011), Smed-pitx (Currie and Pearson, 2013; Marz et al., 2013) and Smed-pbx (Blassberg et al., 2013; Chen et al., 2013) have all been shown to be required for correct posterior regeneration. In all cases these genes are necessary for the stem cell-dependent phase of wnt1 expression and the subsequent activation of Smed-wnt11-2 and Smed-wnt11-5. Furthermore, it is known that Smed-islet and Smed-pitx are co-expressed in wnt1-expressing (wnt1+) cells during regeneration, suggesting the potential for transcriptional regulation by these genes. Following junl-1(RNAi), the expression of Smed-islet and Smed-pitx at the posterior pole was absent (supplementary material Fig. S11A-D).
As an independent test of these results we combined both apc(RNAi) and ptc(RNAi) with 1 µM SP600125 treatment. Treatment with 1 µM SP600125 alone leads to a failure in tail regeneration, but this was clearly rescued by combining the treatment with apc(RNAi) and ptc(RNAi) (supplementary material Fig. S12A-E), including the posterior (and anterior) pole expression of wnt1 (supplementary material Fig. S12C,E).
junl-1 is expressed in both stem cells and Smed-wnt1+ cells during regeneration
A possible mechanism by which JNK signalling might regulate the formation of the posterior Wnt signalling centre is through activated junl-1 driving the transcription of stem cell-dependent wnt1 expression. A prerequisite for this to occur is the expression of junl-1 in wnt1+ cells in the stem cell-dependent phase of Wnt expression.
FISH experiments confirmed the previously described stem cell-enriched expression of junl-1 (Wagner et al., 2012) by showing that Smed-H2B+ cells are junl-1+ (Fig. 6A). In addition, 24 h after amputation junl-1 was upregulated at wound sites (Fig. 6B), and at these wound sites there were some junl-1+ cells that were no longer H2B+ in the region of the newly developing blastema (Fig. 6C). This represents a potential population of cells in which stem cell-dependent wnt1 expression might be induced.
Fig. 6.
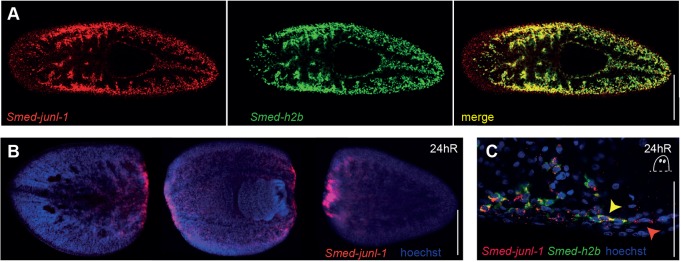
Smed-junl-1 is expressed in the stem cells and regenerating blastemas. (A) Smed-junl-1 colocalised with stem cell marker Smed-H2B in intact animals. (B) Smed-junl-1 is upregulated at the regenerating wounds after amputation (10/10 animals). (C) At the posterior blastema, Smed-junl-1 and Smed-H2B colocalise. 135/135 Smed-H2B+ cells are also junl-1+ (yellow arrowhead), but 22/157 junl-1+cells closest to the wound are negative for Smed-H2B expression (red arrowhead) (counts across six animals). One confocal plane is shown. Scale bars: 1 mm in A; 500 μm in B; 50 μm in C.
At 24 h of regeneration no wnt1/junl-1 double-positive cells were observed. wnt1+ and junl-1+ cells were in different dorsal-ventral planes, consistent with expression in muscle cells (Witchley et al., 2013) and newly forming differentiating cells/stem cells, respectively. This suggests that early wound-induced wnt1 expression is indeed independent of any influence from junl-1. However, at later stages of stem cell-dependent wnt1 expression (2-4 days), wnt1+ cells in the regenerative blastema were also junl-1+ (Fig. 7). Taken together, these data support the contention that JNK signalling and junl-1 act to promote wnt1 expression and posterior fate directly at the posterior regeneration blastema.
Fig. 7.
The stem cell-dependent Smed-wnt1-expressing cells also express Smed-junl-1. Smed-junl-1 does not colocalise with Smed-wnt1 in the wound-induced phase of wnt1 expression (24 hR) (nc=43/43 wnt1+ cells in four animals), but becomes colocalised from 2 dR onwards (2 dR co=17/20 wnt1+ cells in four animals; 3 dR co=87/87 in 16 animals over four experiments; 4 dR co=21/21 in four animals). Arrowheads indicate cells with co-expression of wnt1 and junl-1, except at 24 hR where the arrowhead highlights a single junl-1-expressing cell. Shown are posterior blastemas from regenerating head pieces. One plane, as obtained by confocal microscopy, is shown. Counts indicate the number of cells showing colocalised junl-1/wnt1 expression (co) or no colocalisation (nc). Scale bars: 50 μm.
DISCUSSION
In this work a major new role for JNK signalling in planarian regeneration is presented. Employing both RNAi-mediated and pharmacological abrogation of JNK signalling leads to a block of posterior regeneration as the stem cell-dependent phase of Wnt expression fails to establish at the posterior pole. In addition, we describe a less penetrant anterior phenotype in junl-1(RNAi) animals caused by an expansion of the midline, which we suggest results from a reduction of Smed-wnt-5 expression.
A specific requirement for JNK signalling in posterior regeneration
Although Wnt and Hh signalling are clearly central players in controlling anterior-posterior polarity in planarians, a number of other genes have also been shown to be required for this process, by potentially interacting with these pathways (Blassberg et al., 2013; Chen et al., 2013; Currie and Pearson, 2013; Hayashi et al., 2011; Marz et al., 2013), and to be required for the stem cell-dependent phase of wnt1 expression and subsequent activation of Smed-wnt11-2 and Smed-wnt11-5. In this report it has been shown that JNK signalling is also required for posterior regeneration and that junl-1 is expressed in stem cell-dependent wnt1+ cells within 48 h of regeneration. At 24 h of regeneration, junl-1 is expressed at wound sites in stem cells but also in some cells that do not express the stem cell marker H2B. It is in a subset of medially located junl-1+/H2B– blastema cells that wnt1 also begins to be expressed by 2 dR, and the posterior Wnt-expressing pole can be detected. RNAi of junl-1 or abrogation of JNK activity by SP600125 blocks the formation of these Wnt gene-expressing cells. Our data support previous studies reporting that normal wnt1 expression is required for posterior regeneration (Petersen and Reddien, 2009b; Blassberg et al., 2013; Chen et al., 2013; Currie and Pearson, 2013; Hayashi et al., 2011; Marz et al., 2013; Rink et al., 2009; Yazawa et al., 2009). The fact that we observe pharynx regeneration and that the overall size of animals is unaffected suggests that overall patterning of regions other than the tail is unaffected. Furthermore, we show that Smed-islet and Smed-pitx, which are also normally co-expressed in wnt1+ cells during regeneration, are not correctly expressed after junl-1(RNAi), suggesting that JNK signalling might also be required. The formation of the posterior Wnt-expressing pole is very similar at the cellular level to the emerging description of mechanisms that control the formation of the anterior pole, although the molecular players are distinct (Vásquez-Doorman and Petersen, 2014; Vogg et al., 2014; Scimone et al., 2014).
Additional support for the proposal that the JNK signalling phenotype is a result of loss of the stem cell-dependent phase of wnt1 expression comes from rescue of the tailless phenotype by overactivating the Hh and Wnt pathways using combinatorial RNAi of ptc. Inducing constitutively active Hh, and thereby activating Wnt, by performing double junl-1/ptc(RNAi) rescued the tailless phenotype caused by abrogation of JNK signalling. These data suggest that one role of JNK signalling in posterior regeneration is to promote Wnt expression in stem cell progeny entering the posterior regenerative blastema.
These findings differ from those previously described for other genes required for the stem cell-dependent phase of posterior Wnt expression. First, loss of JNK signalling has no other polarity or position effects on other body regions, such as the pharynx, as reported for Smed-pbx (Blassberg et al., 2013; Chen et al., 2013). Second, both Smed-islet and Smed-pitx appear to be expressed specifically in wnt1+ cells or in a localised medial region in regenerating blastemas (Currie and Pearson, 2013; Hayashi et al., 2011; Marz et al., 2013), whereas junl-1 is expressed more broadly in the regenerative blastema. Although these data suggest that junl-1 might be directly involved in switching on Wnt gene expression at the posterior, other potential direct or indirect cross-regulatory interactions between JNK and Wnt signalling have not been ruled out.
Potentially conserved cross-regulatory interactions between JNK and Wnt mediate posterior identity
Studies of JNK signalling in a number of other systems suggest that cross-regulatory interactions between JNK and both canonical and non-canonical Wnt signalling are involved in a variety of processes, including both the inhibition and activation of Wnt activity. As well as the well-characterised interactions between JNK signalling and the PCP pathway in a non-canonical fashion (Boutros et al., 1998), direct interactions between beta-catenin and Jun have been reported, which in turn mediate the transcription of both Wnt and JNK signalling pathway members (Saadeddin et al., 2009).
Many interactions between the JNK and the canonical Wnt signalling pathways have been reported. Previously described roles of JNK signalling promoting canonical Wnt signalling include recent studies in mammalian cell lines and in zebrafish embryos, which have highlighted extensive crosstalk between JNK and Wnt signalling, including the direct interaction of Jun with beta-catenin, TCF4 and Dishevelled (Gan et al., 2008; Nateri et al., 2005; Toualbi et al., 2007). c-Jun has been suggested to mediate the association of this complex by stabilising its formation at Wnt target gene promoters, and phosphorylated c-Jun protein has been shown to colocalise with TCF4 and with beta-catenin in the nuclei of human adenocarcinomas (Takeda et al., 2008). Given that in planarians wound-induced wnt1 activates later stem cell-dependent Wnt expression in a beta-catenin-dependent manner (Petersen and Reddien, 2009b), it is tempting to speculate that the role of Jun in stabilising this complex might be conserved during posterior planarian regeneration. There is a previous report that suggests a role for JNK in specifying posterior tissues in animals. The majority of zebrafish embryos injected with a c-jun morpholino (MO) exhibited an enlarged anterior and a reduced or absent posterior; this phenotype is very similar to that observed after wnt8 MO injection (Gan et al., 2008). These data are consistent both with our phenotypic observations of tailless animals after knockdown of Jnk pathway members, as well as our finding that junl-1 and wnt1 are expressed in the same cells during posterior specification. Clearly, the role of Wnt in promoting posterior regeneration mirrors its role in controlling posterior fate during the embryogenesis of many bilaterians (Niehrs, 2010; Petersen and Reddien, 2009a).
From an evolutionary perspective, our findings highlight the possibility that regulatory interactions between JNK and Wnt in specifying posterior regions may be conserved across bilaterians. More broadly, there is growing evidence that JNK signalling can regulate the outputs of Wnt signalling and Wnt target genes through direct interaction of Jun with TCF/LEF, beta-catenin and Dishevelled and target promoters (Gan et al., 2008; Nateri et al., 2005; Saadeddin et al., 2009). Our data suggest the possibility that this particular crosstalk between these two pathways described in vertebrates might be more widely conserved and may have been present in the ancestral bilaterian.
Uncovering pleiotropic roles for JNK signalling during planarian regeneration
JNK signalling will be deployed multiple times during an animal's life history in a number of different roles. Previous genetic studies of the JNK pathway in S. mediterranea have focused on the study of the two Jun family members jun1 and junl-1, and the planarian orthologue of jnk. jun-1 has been previously described as a wound-induced gene whose early expression is dependent on active translation (Wenemoser et al., 2012), while junl-1 has been shown to be required for the clonal expansion of stem cells in a sublethal irradiation paradigm along with genes such as vasa and Setd8-like protein (Wagner et al., 2012). The breadth of genes implicated as having roles in clonal expansion suggests it is unlikely that there is a link between this role of junl-1 and the role we describe here. In addition, jnk has recently been shown to be required for limiting stem cell entry into mitosis after wounding and for activating apoptosis, such that knockdown leads to early overproliferation of stem cells after wounding and reduced apoptosis causing defects in regeneration and remodelling (Almuedo-Castillo et al., 2014).
We did not investigate jun-1 but instead focused on junl-1, along with hem and jnk, as our phylogenetic analysis indicated this gene was closer to other animal Jun proteins. Our RNAi knockdown results for hem, jnk and junl-1 reveal a clear role for JNK signalling in promoting posterior regeneration, without global effects on regeneration or effects on stem cell proliferation or stem cell numbers. The previous report of junl-1 being required for the clonal expansion of stem cells after non-lethal regeneration (Wagner et al., 2012) is clearly a very different regenerative context to that tested here. A recently published study of jnk (Almuedo-Castillo et al., 2014) combined three rounds of RNAi injection and two rounds of full regeneration compared with just a single round of regeneration in our study. It is possible that the different experimental paradigms led to different levels of jnk transcript knockdown by RNAi, but we observed broadly similar transcript levels after RNAi as in the previous study. Thus, other differences in experimental set-up between our work and previous work are likely to explain the less severe effects on regeneration that we observe, allowing us to unmask and study the effect of JNK signalling on posterior regeneration outside of its more fundamental cellular roles. We did observe more severe phenotypes in our experimental paradigm when performing double junl-1/jnk(RNAi) (supplementary material Fig. S3), which might reflect the broader effects reported upon use of multiple rounds of regeneration (Almuedo-Castillo et al., 2014).
Our data from using the JNK inhibitor SP600125 is in agreement with our RNAi-based approaches. A previous study used SP600125 in the planarian Dugesia japonica (Tasaki et al., 2011) and found that treatment with a 25 µM dose blocked wound healing, regeneration and stem cell division. Since this concentration of the inhibitor rapidly killed S. mediterranea, this suggests the response to SP600125 differs significantly between the two species. However, we observed a failure of wound healing at a dose of 5 µM, which prevented regeneration and led to death within 3 dR. A lower concentration of 1 µM, shown to minimise potential effects on other kinase families (Bennett et al., 2001; Cui et al., 2007; Valesio et al., 2013), phenocopied the posterior regeneration defect caused by RNAi of JNK signalling pathway members. This provides support for our model whereby JNK signalling is required for posterior regeneration and correlates with our RNAi data, with stronger effects at higher doses of inhibitor. Finally, junl-1 is also required to re-establish correct wnt5 expression during anterior regeneration, which normally acts to limit midline slit expression, to ensure correct mediolateral patterning of the nervous system.
Taken together, extant data not surprisingly delineate multiple roles for JNK signalling during regeneration that can be separated out using different experimental paradigms and by focusing on different aspects of phenotypes revealed by RNAi experiments. The many pleiotropic functions of conserved signalling pathways can be revealed in an adult whole organism context using different experimental paradigms and regenerative challenges in planarians. This represents a significant strength of the planarian model system that can be exploited to understand the complex molecular control mechanisms of regeneration.
MATERIALS AND METHODS
Planarian culture
Asexual Schmidtea mediterranea were cultured as previously described (Felix and Aboobaker, 2010). Animals were starved for 7 days before and during the experiments.
Identification and cloning of Smed-junl-1, Smed-jnk and Smed-hem
We used reciprocal BLAST searches of available S. mediterranea genome and transcriptome data (Blythe et al., 2010; Kao et al., 2013) and genome data from Washington University, St Louis, USA (Robb et al., 2008) and subsequently confirmed full open reading frames for each gene by RT-PCR experiments and sequencing. During this research the sequence of Smed-junl-1 (junl-1) was described in another study and submitted to GenBank with accession number AFD29623. The Smed-hemipterous (hem) and Smed-jnk (jnk) sequences have been submitted to GenBank with accession numbers KM095500 and KM095499.
RNAi experiments
dsRNA was synthesized by in vitro transcription using T7 and SP6 polymerases (Roche and NEB, respectively) from a plasmid PCR template. In single RNAi experiments, 96 nl 2 µg/µl dsRNA was injected on 3 consecutive days 1 week and on 3 consecutive days the following week, prior to amputation on the next day (supplementary material Fig. S1A). Transverse amputations were performed pre- and post-pharyngeally (supplementary material Fig. S1B). Double-RNAi experiments were performed with the same injection schedule, but the dsRNA concentrations were adjusted so that the final concentration for each gene was 2 µg/µl. An exception to the injection schedule was the amputation of animals for the study of wound-induced expression patterns, where the samples were cut 2 days after the last injection. The primers used to amplify and clone gene products for dsRNA synthesis were as follows (5′-3′, forward and reverse): Smed-junl-1, ACCATCTCCAATAACACCGAAT and GCACCGAAACTACTCTCATTCC; Smed-jnk, GCACAAGGATACGTTGTAGCTG and TTCGTTGAAGTGGATCAATGAC; Smed-hem, TGGTCACTTGGGATGAGTTTAGT and CGACGAATTGAGGAGTCTTAGAA; Smed-ptc, ACTGTGATACTAGTAACGCTTCT and TTTGCTTGGAGTGAAATTATCAA; Smed-bcat, TCAGGGATTGCAGATTCTCATTCG and GGCTAATGATCAATTCCAGTCC; Smed-apc, TATCTACGGGATCTGCTGCTAG and TATCATAGTCATCAGGATACG.
Measurement of animal lengths
For all length measurements, bright-field images were taken with a Zeiss Axiovision stereomicroscope and an Axiocam MRc or an EOS600D camera. The distance from the tip of the pharynx to the tail end was measured and normalised to total length, as were measurements of the head to the tip of the pharynx. Measurements were made using Fiji software (Schindelin et al., 2012).
Immunohistochemistry
Animals were fixed and treated as described previously (Cebrià and Newmark, 2005). Primary antibodies: anti-Synapsin (3C11; Developmental Studies Hybridoma Bank), 1:100; anti-phosphorylated (serine 10) Histone H3 (Millipore, 06-570), 1:1000. Secondary antibodies: Alexa Fluor 488 goat anti-mouse IgG (Life Technologies, A-11070), 1:1000; Alexa Fluor 568 goat anti-rabbit IgG (Life Technologies, A-11036), 1:400. Fluorescent images were taken with a Leica MZ16F fluorescence stereomicroscope and DFC300Fx camera (Leica) or with an Olympus Fluoview FV1000 confocal microscope.
In situ hybridisation
Wholemount in situ hybridisation (WISH) was performed as previously described (Gonzalez-Estevez et al., 2009), except that for fluorescent in situ hybridisation (FISH) the development steps and the peroxidase quenching were performed as described elsewhere (King and Newmark, 2013). The efficacy of quenching reactions was confirmed by performing tyramide reactions in another colour for single probes and imaging to ensure that no signal was present. The following probes were used as previously described: Smed-laminin (Cebria and Newmark, 2007), Smed-sFRP (Gurley et al., 2008), Smed-fzl4 (Gurley et al., 2008), Smed-cintillo (Oviedo et al., 2003), Smed-GPAS (Cebrià et al., 2002b; Iglesias et al., 2011), Smed-porcn1 (Gurley et al., 2008), Smed-wnt1 (Adell et al., 2009; Petersen and Reddien, 2009b), Smed-wnt11-2 (Adell et al., 2009; Gurley et al., 2010; Petersen and Reddien, 2009b), Smed-wnt11-5 (Petersen and Reddien, 2009b), Smed-notum (Petersen and Reddien, 2011), Smed-h10.2f (Cebria and Newmark, 2007; Sánchez Alvarado et al., 2002), Smed-wnt5 (Adell et al., 2009; Gurley et al., 2010), Smed-slit (Cebrià et al., 2007), Smed-H2B (Solana et al., 2012) and Smed-eye53 (Cebrià et al., 2002b; Inoue et al., 2004; Zayas et al., 2005). For Smed-pitx the following primers were used to generate a probe for FISH: PitxF, GTCATTCTCCATCGGCTCA; PitxR, TGACAACATTGGCTGTCGAT. For Smed-islet the following primers were used to generate a probe for WISH: IsletF, ACAAGTTTGTGCTGCCTGC; IsletR, TTAGCCCTACTGATATTTCCGC. For FISH of Smed-junl-1, the following primer pairs were used: Smed-junl-1F, ACCATCTCCAATAACACCGAAT; Smed-junl-1R, GCACCGAAACTACTCTCATTCC; and Smed-junl-1F1, AATCGGAATTCGGTATTTTGG; Smed-junl-1 R1, CCAAATTCGCTTGTTTTTCAA [from Wagner et al. (2012), with equivalent results].
Quantitative (q) RT-qPCR
Total RNA from samples of five regenerating trunk pieces at 72 h of regeneration were extracted with Trizol reagent (Invitrogen) according to the manufacturer's instructions for each biological replicate. RNA was treated with TURBO DNase (Ambion). First-strand cDNAs were synthesized with SuperScript III reverse transcriptase (Invitrogen) and qRT-PCR experiments used the Absolute qPCR SYBR Green Master Mix (Thermo Scientific). Experiments were performed on three biological replicates per RNAi condition. Each of the three biological replicates was technically replicated three times, with each technical replicate consisting of three replicate reactions. Smed-ef-2 was used for normalisation using primers described previously (Solana et al., 2013). The primers for Smed-jnk were the same as those used previously (Almuedo-Castillo et al., 2014). The primers for Smed-junl-1 were: junl-1 Fq, TGTTGATCAACAGCGTAGAT; junl-1 Rq, TATTAGTTCTTTTTCCACAG. Statistical significance was measured using Student's t-test, comparing values from each sample to gfp(RNAi) control samples.
Whole-mount TUNEL
Animals were fixed and stained for TUNEL as previously described (Pellettieri et al., 2010), with the modifications described in another recent study (Almuedo-Castillo., 2014).
Use of pharmacological inhibitors
The inhibitor of JNK SP600125 (Sigma-Aldrich) was dissolved in DMSO and applied to the animals at concentrations between 0.1 µM and 25 µM in 0.05% DMSO. Controls were incubated in 0.05% DMSO alone. Animals were cut and immediately transferred to planarian water containing the inhibitor or a control amount of DMSO. Solutions were changed every 24 h for the duration of the experiment. In wash-out or wash-in experiments animals were gently rinsed in 0.05% DMSO or 1 µM SP600125 in 0.05% DMSO several times over the first hour of incubation.
Supplementary Material
Acknowledgements
We thank past and current members of the Aboobaker laboratory and members of the planarian community for discussions and suggestions. We thank two anonymous reviewers for their comments that significantly improved this manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
B.T.-R. was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) Studentship and Y.M. was funded by a UK Medical Research Council (MRC) Studentship, both originally from the University of Nottingham, UK. This work was funded by a BBSRC Research Grant [BB/K007564/1] and MRC Research Grant [MR/M000133/1] awarded to A.A.A. Deposited in PMC for release after 6 months.
Author contributions
B.T.-R., J.-M.C., Y.M., B.N. and A.A.A. performed experiments; B.T.-R. and A.A.A. designed the study; B.T.-R., J.-M.C. and A.A.A. prepared the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.115139/-/DC1
References
- Aboobaker A. A. (2011). Planarian stem cells: a simple paradigm for regeneration. Trends Cell Biol. 21, 304-311. 10.1016/j.tcb.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Adell T., Salo E., Boutros M. and Bartscherer K. (2009). Smed-Evi/Wntless is required for beta-catenin-dependent and -independent processes during planarian regeneration. Development 136, 905-910. 10.1242/dev.033761 [DOI] [PubMed] [Google Scholar]
- Almuedo-Castillo M., Sureda-Gómez M. and Adell T. (2012). Wnt signaling in planarians: new answers to old questions. Int. J. Dev. Biol. 56, 53-65. 10.1387/ijdb.113451ma [DOI] [PubMed] [Google Scholar]
- Almuedo-Castillo M., Crespo X., Seebeck F., Bartscherer K., Salò E. and Adell T. (2014). JNK controls the onset of mitosis in planarian stem cells and triggers apoptotic cell death required for regeneration and remodeling. PLoS Genet. 10, e1004400 10.1371/journal.pgen.1004400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J., Plater L., Elliott M., Shpiro N., Hastie C. J., McLauchlan H., Klevernic I., Arthur J. S. C., Alessi D. R. and Cohen P. (2007). The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297-315. 10.1042/BJ20070797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett B. L., Sasaki D. T., Murray B. W., O'Leary E. C., Sakata S. T., Xu W., Leisten J. C., Motiwala A., Pierce S., Satoh Y. et al. (2001). SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98, 13681-13686. 10.1073/pnas.251194298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg R. A., Felix D. A., Tejada-Romero B. and Aboobaker A. A. (2013). PBX/extradenticle is required to re-establish axial structures and polarity during planarian regeneration. Development 140, 730-739. 10.1242/dev.082982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe M. J., Kao D., Malla S., Rowsell J., Wilson R., Evans D., Jowett J., Hall A., Lemay V., Lam S. et al. (2010). A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS ONE 5, e15617 10.1371/journal.pone.0015617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M., Paricio N., Strutt D. I. and Mlodzik M. (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94, 109-118. 10.1016/S0092-8674(00)81226-X [DOI] [PubMed] [Google Scholar]
- Cebrià F. and Newmark P. A. (2005). Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132, 3691-3703. 10.1242/dev.01941 [DOI] [PubMed] [Google Scholar]
- Cebria F. and Newmark P. A. (2007). Morphogenesis defects are associated with abnormal nervous system regeneration following roboA RNAi in planarians. Development 134, 833-837. 10.1242/dev.02794 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Kobayashi C., Umesono Y., Nakazawa M., Mineta K., Ikeo K., Gojobori T., Itoh M., Taira M., Sanchez Alvarado A. et al. (2002a). FGFR-related gene nou-darake restricts brain tissues to the head region of planarians. Nature 419, 620-624. 10.1038/nature01042 [DOI] [PubMed] [Google Scholar]
- Cebrià F., Kudome T., Nakazawa M., Mineta K., Ikeo K., Gojobori T. and Agata K. (2002b). The expression of neural-specific genes reveals the structural and molecular complexity of the planarian central nervous system. Mech. Dev. 116, 199-204. 10.1016/S0925-4773(02)00134-X [DOI] [PubMed] [Google Scholar]
- Cebrià F., Guo T., Jopek J. and Newmark P. A. (2007). Regeneration and maintenance of the planarian midline is regulated by a slit orthologue. Dev. Biol. 307, 394-406. 10.1016/j.ydbio.2007.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. (2012). JNK-induced apoptosis, compensatory growth, and cancer stem cells. Cancer Res. 72, 379-386. 10.1158/0008-5472.CAN-11-1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-C. G., Wang I. E. and Reddien P. W. (2013). pbx is required for pole and eye regeneration in planarians. Development 140, 719-729. 10.1242/dev.083741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles M. W., Brown D. D. R., Nisperos S. V., Stanley B. N., Pearson B. J. and Zayas R. M. (2013). Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 140, 4691-4702. 10.1242/dev.098616 [DOI] [PubMed] [Google Scholar]
- Cui J., Zhang M., Zhang Y.-q. and Xu Z.-h. (2007). JNK pathway: diseases and therapeutic potential. Acta Pharmacol. Sin. 28, 601-608. 10.1111/j.1745-7254.2007.00579.x [DOI] [PubMed] [Google Scholar]
- Currie K. W. and Pearson B. J. (2013). Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 140, 3577-3588. 10.1242/dev.098590 [DOI] [PubMed] [Google Scholar]
- Davis R. J. (2000). Signal transduction by the JNK group of MAP kinases. Cell 103, 239-252. 10.1016/S0092-8674(00)00116-1 [DOI] [PubMed] [Google Scholar]
- Evans D. J., Owlarn S., Tejada Romero B., Chen C. and Aboobaker A. A. (2011). Combining classical and molecular approaches elaborates on the complexity of mechanisms underpinning anterior regeneration. PLoS ONE 6, e27927 10.1371/journal.pone.0027927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix D. A. and Aboobaker A. A. (2010). The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 6, e1000915 10.1371/journal.pgen.1000915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraguas S., Barberan S., Iglesias M., Rodriguez-Esteban G. and Cebria F. (2014). egr-4, a target of EGFR signaling, is required for the formation of the brain primordia and head regeneration in planarians. Development 141, 1835-1847. 10.1242/dev.101345 [DOI] [PubMed] [Google Scholar]
- Gan X.-q., Wang J.-y., Xi Y., Wu Z.-l., Li Y.-p. and Li L. (2008). Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J. Cell Biol. 180, 1087-1100. 10.1083/jcb.200710050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Estevez C., Arseni V., Thambyrajah R. S., Felix D. A. and Aboobaker A. A. (2009). Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. Int. J. Dev. Biol. 53, 493-505. 10.1387/ijdb.082825cg [DOI] [PubMed] [Google Scholar]
- González-Esteévez C., Felix D. A., Smith M. D., Paps J., Morley S. J., James V., Sharp T. V. and Aboobaker A. A. (2012). SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet. 8, e1002619 10.1371/journal.pgen.1002619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Rink J. C. and Sanchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323-327. 10.1126/science.1150029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley K. A., Elliott S. A., Simakov O., Schmidt H. A., Holstein T. W. and Sanchez Alvarado A. (2010). Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Dev. Biol. 347, 24-39. 10.1016/j.ydbio.2010.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Motoishi M., Yazawa S., Itomi K., Tanegashima C., Nishimura O., Agata K. and Tarui H. (2011). A LIM-homeobox gene is required for differentiation of Wnt-expressing cells at the posterior end of the planarian body. Development 138, 3679-3688. 10.1242/dev.060194 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Gomez-Skarmeta J. L., Salo E. and Adell T. (2008). Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development 135, 1215-1221. 10.1242/dev.020289 [DOI] [PubMed] [Google Scholar]
- Iglesias M., Almuedo-Castillo M., Aboobaker A. A. and Saló E. (2011). Early planarian brain regeneration is independent of blastema polarity mediated by the Wnt/beta-catenin pathway. Dev. Biol. 358, 68-78. 10.1016/j.ydbio.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Inoue T., Kumamoto H., Okamoto K., Umesono Y., Sakai M., Sanchez Alvarado A. and Agata K. (2004). Morphological and functional recovery of the planarian photosensing system during head regeneration. Zoolog. Sci. 21, 275-283. 10.2108/zsj.21.275 [DOI] [PubMed] [Google Scholar]
- Kao D., Felix D. and Aboobaker A. (2013). The planarian regeneration transcriptome reveals a shared but temporally shifted regulatory program between opposing head and tail scenarios. BMC Genomics 14, 797 10.1186/1471-2164-14-797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. S. and Newmark P. A. (2013). In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev. Biol. 13, 8 10.1186/1471-213X-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M., Seebeck F. and Bartscherer K. (2013). A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 140, 4499-4509. 10.1242/dev.100081 [DOI] [PubMed] [Google Scholar]
- Molina M. D., Neto A., Maeso I., Gómez-Skarmeta J. L., Saló E. and Cebrià F. (2011). Noggin and noggin-like genes control dorsoventral axis regeneration in planarians. Curr. Biol. 21, 300-305. 10.1016/j.cub.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Nateri A. S., Spencer-Dene B. and Behrens A. (2005). Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437, 281-285. 10.1038/nature03914 [DOI] [PubMed] [Google Scholar]
- Niehrs C. (2010). On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137, 845-857. 10.1242/dev.039651 [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Newmark P. A. and Sánchez Alvarado A. (2003). Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev. Dyn. 226, 326-333. 10.1002/dvdy.10228 [DOI] [PubMed] [Google Scholar]
- Pellettieri J., Fitzgerald P., Watanabe S., Mancuso J., Green D. R. and Sánchez Alvarado A. (2010). Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338, 76-85. 10.1016/j.ydbio.2009.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2008). Smed-betacatenin-1 is required for anteroposterior blastema polarity in planarian regeneration. Science 319, 327-330. 10.1126/science.1149943 [DOI] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2009a). Wnt signaling and the polarity of the primary body axis. Cell 139, 1056-1068. 10.1016/j.cell.2009.11.035 [DOI] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2009b). A wound-induced Wnt expression program controls planarian regeneration polarity. Proc. Natl. Acad. Sci. USA 106, 17061-17066. 10.1073/pnas.0906823106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C. P. and Reddien P. W. (2011). Polarized notum activation at wounds inhibits Wnt function to promote planarian head regeneration. Science 332, 852-855. 10.1126/science.1202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss K. D. (2010). Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 11, 710-722. 10.1038/nrg2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P. W. (2011). Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 27, 277-285. 10.1016/j.tig.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C. (2013). Stem cell systems and regeneration in planaria. Dev. Genes Evol. 223, 67-84. 10.1007/s00427-012-0426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C., Gurley K. A., Elliott S. A. and Sanchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406-1410. 10.1126/science.1178712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S. M. C., Ross E. and Sanchez Alvarado A. (2008). SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res. 36, D599-D606. 10.1093/nar/gkm684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadeddin A., Babaei-Jadidi R., Spencer-Dene B. and Nateri A. S. (2009). The links between transcription, beta-catenin/JNK signaling, and carcinogenesis. Mol. Cancer Res. 7, 1189-1196. 10.1158/1541-7786.MCR-09-0027 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M. C. and Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665. 10.1242/dev.00167 [DOI] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B. et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone M. L., Lapan S. W. and Reddien P. W. (2014). A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS Genet. 10, e1003999 10.1371/journal.pgen.1003999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Kao D., Mihaylova Y., Jaber-Hijazi F., Malla S., Wilson R. and Aboobaker A. (2012). Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNA-seq, RNA interference and irradiation approach. Genome Biol. 13, R19 10.1186/gb-2012-13-3-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J., Gamberi C., Mihaylova Y., Grosswendt S., Chen C., Lasko P., Rajewsky N. and Aboobaker A. A. (2013). The CCR4-NOT complex mediates deadenylation and degradation of stem cell mRNAs and promotes planarian stem cell differentiation. PLoS Genet. 9, e1004003 10.1371/journal.pgen.1004003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Kinoshita I., Shimizu Y., Ohba Y., Itoh T., Matsuno Y., Shichinohe T. and Dosaka-Akita H. (2008). Clinicopathological significance of expression of p-c-Jun, TCF4 and beta-Catenin in colorectal tumors. BMC Cancer 8, 328 10.1186/1471-2407-8-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka E. M. and Reddien P. W. (2011). The cellular basis for animal regeneration. Dev. Cell 21, 172-185. 10.1016/j.devcel.2011.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanemura S., Momose H., Shimizu N., Kitagawa D., Seo J., Yamasaki T., Nakagawa K., Kajiho H., Penninger J. M., Katada T. et al. (2009). Blockage by SP600125 of Fcepsilon receptor-induced degranulation and cytokine gene expression in mast cells is mediated through inhibition of phosphatidylinositol 3-kinase signalling pathway. J. Biochem. 145, 345-354. 10.1093/jb/mvn172 [DOI] [PubMed] [Google Scholar]
- Tasaki J., Shibata N., Sakurai T., Agata K. and Umesono Y. (2011). Role of c-Jun N-terminal kinase activation in blastema formation during planarian regeneration. Dev. Growth Differ. 53, 389-400. 10.1111/j.1440-169X.2011.01254.x [DOI] [PubMed] [Google Scholar]
- Toualbi K., Güller M. C., Mauriz J.-L., Labalette C., Buendia M.-A., Mauviel A. and Bernuau D. (2007). Physical and functional cooperation between AP-1 and beta-catenin for the regulation of TCF-dependent genes. Oncogene 26, 3492-3502. 10.1038/sj.onc.1210133 [DOI] [PubMed] [Google Scholar]
- Valesio E. G., Zhang H. and Zhang C. (2013). Exposure to the JNK inhibitor SP600125 (anthrapyrazolone) during early zebrafish development results in morphological defects. J. Appl. Toxicol. 33, 32-40. 10.1002/jat.1708 [DOI] [PubMed] [Google Scholar]
- Vásquez-Doorman C. and Petersen C. P. (2014). zic-1 Expression in Planarian neoblasts after injury controls anterior pole regeneration. PLoS Genet. 10, e1004452 10.1371/journal.pgen.1004452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg M. C., Owlarn S., Pérez Rico Y. A., Xie J., Suzuki Y., Gentile L., Wu W. and Bartscherer K. (2014). Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Dev. Biol. 390, 136-148. 10.1016/j.ydbio.2014.03.016 [DOI] [PubMed] [Google Scholar]
- Wagner D. E., Ho J. J. and Reddien P. W. (2012). Genetic regulators of a pluripotent adult stem cell system in planarians identified by RNAi and clonal analysis. Cell Stem Cell 10, 299-311. 10.1016/j.stem.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D., Lapan S. W., Wilkinson A. W., Bell G. W. and Reddien P. W. (2012). A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 26, 988-1002. 10.1101/gad.187377.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston C. R. and Davis R. J. (2002). The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12, 14-21. 10.1016/S0959-437X(01)00258-1 [DOI] [PubMed] [Google Scholar]
- Weston C. R. and Davis R. J. (2007). The JNK signal transduction pathway. Curr. Opin. Cell Biol. 19, 142-149. 10.1016/j.ceb.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Witchley J. N., Mayer M., Wagner D. E., Owen J. H. and Reddien P. W. (2013). Muscle cells provide instructions for planarian regeneration. Cell Rep. 4, 633-641. 10.1016/j.celrep.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa S., Umesono Y., Hayashi T., Tarui H. and Agata K. (2009). Planarian Hedgehog/Patched establishes anterior-posterior polarity by regulating Wnt signaling. Proc. Natl. Acad. Sci. USA 106, 22329-22334. 10.1073/pnas.0907464106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas R. M., Hernandez A., Habermann B., Wang Y., Stary J. M. and Newmark P. A. (2005). The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc. Natl. Acad. Sci. USA 102, 18491-18496. 10.1073/pnas.0509507102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.