Abstract
The paternally expressed imprinted retrotransposon-like 1 (Rtl1) is a retrotransposon-derived gene that has evolved a function in eutherian placentation. Seven miRNAs, including miR-127, are processed from a maternally expressed antisense Rtl1 transcript (Rtl1as) and regulate Rtl1 levels through RNAi-mediated post-transcriptional degradation. To determine the relative functional role of Rtl1as miRNAs in Rtl1 dosage, we generated a mouse specifically deleted for miR-127. The miR-127 knockout mice exhibit placentomegaly with specific defects within the labyrinthine zone involved in maternal-fetal nutrient transfer. Although fetal weight is unaltered, specific Rtl1 transcripts and protein levels are increased in both the fetus and placenta. Phenotypic analysis of single (ΔmiR-127/Rtl1 or miR-127/ΔRtl1) and double (ΔmiR-127/ΔRtl1) heterozygous miR-127- and Rtl1-deficient mice indicate that Rtl1 is the main target gene of miR-127 in placental development. Our results demonstrate that miR-127 is an essential regulator of Rtl1, mediated by a trans-homologue interaction between reciprocally imprinted genes on the maternally and paternally inherited chromosomes.
KEY WORDS: Genomic imprinting, Rtl1 (Peg11), miR-127, Mir127, Placenta development
Summary: MiR-127 is an essential regulator of the paternally expressed imprinted gene Rtl1 and acts via trans-homologue interactions to regulate Rtl1 dosage and placental growth.
INTRODUCTION
Mammalian genomic imprinting is an epigenetic process whereby genes are mono-allelically expressed in a parent-of-origin-specific manner (Ferguson-Smith, 2011). The imprinted gene cluster on mouse chromosome 12 contains four paternally expressed protein-coding genes and maternally expressed non-coding RNAs (Fig. 1A) (da Rocha et al., 2008). One of these paternally expressed genes, retrotransposon-like 1 (Rtl1; also known as Peg11), is derived from a Ty3/Gypsy-type retrotransposon that in eutherians has evolved a large conserved open reading frame (ORF) but has lost its long terminal repeats (LTRs), resulting in loss of the original retroviral promoter activity (Brandt et al., 2005; Youngson et al., 2005; Edwards et al., 2008).
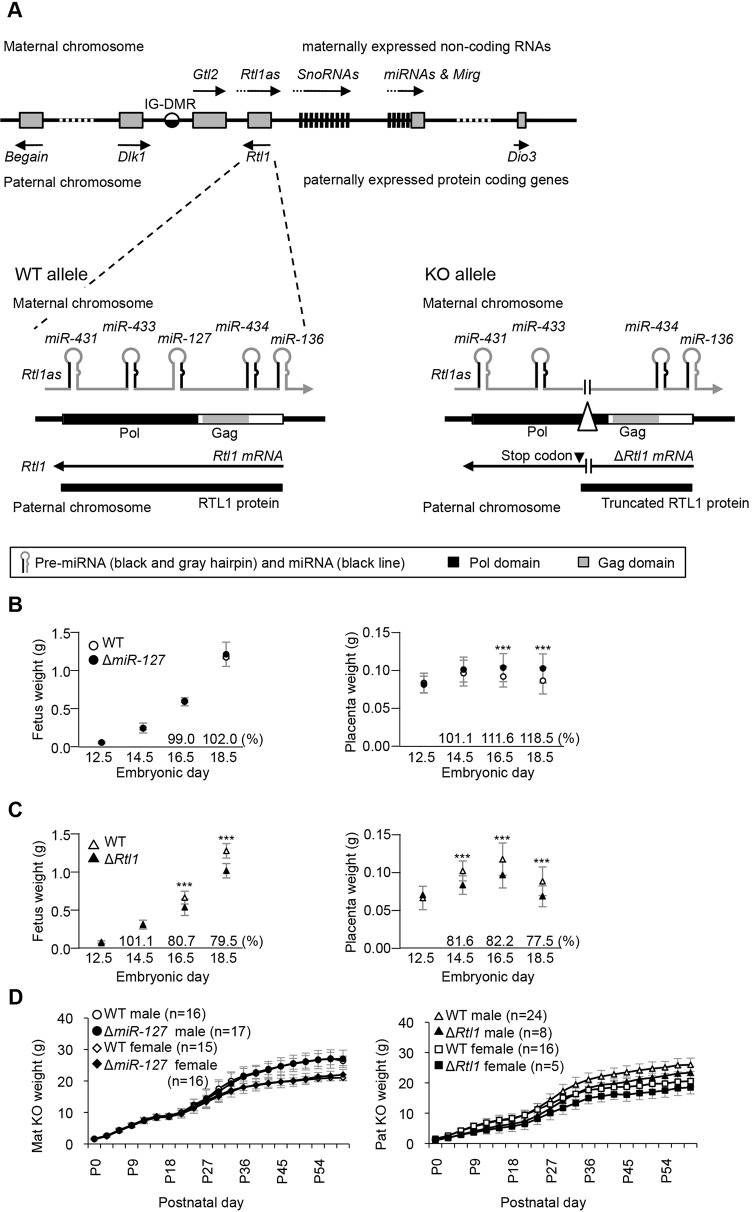
Fig. 1.
Structure of the Rtl1 locus and pre- and postnatal growth of miR-127 and Rtl1 knockout mice. (A) Schematic presentation of the mouse Dlk1-Dio3 cluster. (Lower left) The WT Rtl1 locus (exon 3). Rtl1 is expressed from the paternal chromosome and Rtl1as is exclusively transcribed from the maternal chromosome. (Lower right) The knockout (KO) allele. The paternally transmitted deletion introduces an in-frame stop codon that results in premature termination of RTL1. The maternally transmitted deletion lacks miR-127 expression. (B,C) Prenatal growth of ΔmiR-127 mice and ΔRtl1 mice, respectively. Left and right panels show embryonic and placental growth curves in mutant and WT littermates from E12.5 to E18.5. All embryos and placentas were collected from the N6 and N7 generation. (D) Postnatal growth curve of ΔmiR-127 (left) and ΔRtl1 (right) from birth to 2 months. Weights were measured every 3 days. ΔRtl1 mice were significantly smaller than WT. ***P<0.005 (Student's t-test). Error bars indicate s.d.
The primary antisense Rtl1 transcript (Rtl1as) is exclusively expressed from the maternally inherited Rtl1 locus but in the opposite direction to Rtl1 (Fig. 1A) (Seitz et al., 2003). At least seven microRNAs (miRNAs) processed from Rtl1as are therefore perfectly complementary in sequence to Rtl1 (Davis et al., 2005). Maternally inherited deletion of the differentially methylated imprinting control region for the locus (IG-DMR) causes a maternal-to-paternal epigenotype switch across the whole imprinted gene cluster (Lin et al., 2003). This is associated with repression of all the maternally expressed non-coding RNAs, including the miRNAs, and inappropriate activation of the usually paternally expressed protein-coding genes on the maternally inherited chromosome, resulting in a double dose. However, Rtl1 mRNA levels increase 4.5-fold from both alleles, instead of the double dose expected from loss of imprinting (LOI). This suggests that the increase in Rtl1 dosage in the mutant is the cumulative effect of both LOI and a failure to destabilise the now biallelically expressed transcript by the antisense miRNAs (Lin et al., 2003). Further evidence that these miRNAs can degrade Rtl1 transcripts by the RNAi machinery in vivo came from the identification of both DROSHA and DICER cleavage products for each of the miRNAs (Davis et al., 2005). Previous work has shown that Rtl1 gene deletion causes growth retardation of both the fetus and placenta and that removal of six of the seven miRNAs on Rtl1as leads to Rtl1 overproduction and placentomegaly (Sekita et al., 2008).
Further findings indicate that miR-127 on Rtl1as can be independently regulated in human cancer (Iorio et al., 2005; Lu et al., 2005), and that on its own miR-127 may be the major contributor to Rtl1 silencing in differentiating mouse embryonic stem cells (ESCs) (Ciaudo et al., 2009). These findings suggest that miR-127 might play a prominent role in controlling Rtl1 dosage during normal development. In order to clarify the biological significance of miR-127, we generated miR-127 (Mir127) knockout mice and studied its impact on Rtl1 transcript and protein levels and consequences for placental development.
RESULTS AND DISCUSSION
Maternal miR-127 deletion induces placentomegaly
The schematic organisation of the imprinted Rtl1 sense and antisense transcripts is shown in Fig. 1A. A 134 bp deletion removed miR-127 upon maternal transmission (ΔmiR-127), while the same deletion when paternally transmitted (ΔRtl1) introduces a nonsense mutation in the third exon of Rtl1, resulting in premature translation termination of a normally transcribed mutant transcript (Fig. 1A; supplementary material Fig. S1B). Western blotting data showed no detectable RTL1 protein in ΔRtl1 conceptuses (supplementary material Fig. S1G), although Rtl1 mRNA was stable (supplementary material Fig. S3A). All phenotypic analyses were carried out on the C57BL/6J background unless otherwise indicated.
Placentae were significantly overgrown in ΔmiR-127 mutants, which was first apparent at E16.5; placental weights were 111.6% and 118.5% compared with wild type (WT) at E16.5 and E18.5, respectively (Fig. 1B). By contrast, there was no effect of ΔmiR-127 on fetal weight during development (Fig. 1B). Previous work had shown that when six miRNAs, including miR-127, are deleted, mutant placental weights are 156% of WT values at E18.5, although fetal weights are not different (Sekita et al., 2008). These data suggest that miR-127 functions to suppress placental growth in pregnancy, although placentomegaly in ΔmiR-127 was milder than with the larger deletion encompassing six miRNAs. After birth, the ΔmiR-127 mice grew at comparable rates to WT and no lethality was observed either pre- or postnatally in these mice (Fig. 1D; supplementary material Tables S1 and S2).
ΔRtl1 mice showed prenatal growth retardation starting at E16.5; fetal weights were ∼80% of WT (Fig. 1C). Mice have reduced wet weight at birth (∼70% of WT) and remain growth retarded into adulthood (Fig. 1D). Prenatally, the placenta is growth restricted from E14.5, prior to the onset of fetal growth restriction, suggesting a causal role for the placenta in the fetal growth phenotype (Fig. 1C). Prenatal lethality was not observed in ΔRtl1 but the majority of neonates died within 1 day of birth (supplementary material Tables S1 and S2). In situations in which ΔRtl1 newborns survived more than 2 days, animals survived to adulthood. The lethality of ΔRtl1 was not evident on a mixed 129aa and C57BL/6J background (supplementary material Table S1). The embryonic lethality we report differs from that associated with the previously reported larger deletion, where lethality occurred during gestation upon paternal transmission (Sekita et al., 2008), despite both mutants lacking the RTL1 protein.
ΔmiR-127 causes defects in the placental labyrinthine zone
Placental structure was analysed stereologically (Gundersen et al., 1988; Mandarim-de-Lacerda, 2003; Coan et al., 2004) upon both maternal and paternal transmission of the deletion at E18.5. In ΔmiR-127 the labyrinthine zone (Lz), which is the site of nutrient and gaseous exchange between the maternal and fetal blood supplies, was expanded (142.3% of WT; Fig. 2A,C). Conversely, the volume of the Lz was reduced in ΔRtl1 (64.7% of WT; Fig. 2B,D). In contrast to the Lz, the junctional zone, decidual basalis and chorion were all unaffected by miR-127 or Rtl1 deficiency.
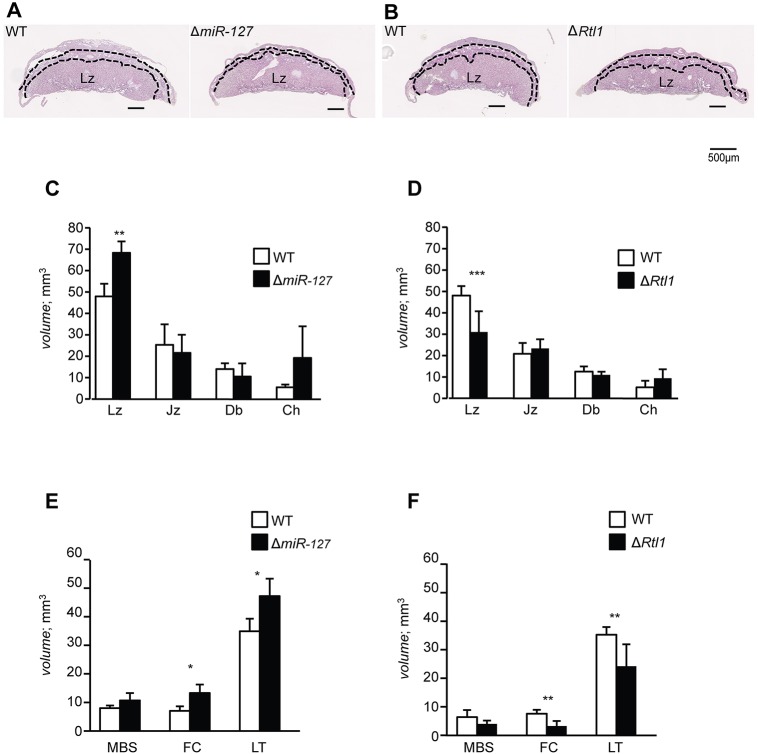
Fig. 2.
Histological analysis shows abnormality in the labyrinthine zone in ΔmiR-127 and ΔRtl1. (A,B) Histological analysis of WT littermate and ΔmiR-127 or ΔRtl1, respectively. H&E-stained paraffin sections of E18.5 placentae. Dashed lines demarcate Lz (bottom layer), Jz (middle layer) and Db (top layer). Scale bars: 500 μm. (C-F) The volumes of placental and labyrinthine compartments in WT, ΔmiR-127 and ΔRtl1. Lz, labyrinthine zone; Jz, junctional zone; Db, decidual basalis; Ch, chorion; MBS, maternal blood spaces; FC, fetal capillaries; LT, labyrinthine trophoblast. *P<0.05, **P<0.01, ***P<0.005 (Student's t-test). Error bars indicate s.d.
Detailed structural analysis of the Lz showed that both the fetal capillaries (FCs) and the labyrinthine trophoblast (LT) were significantly increased in ΔmiR-127, with a non-significant trend for expanded maternal blood spaces (MBSs) (Fig. 2E; supplementary material Fig. S2). Similar to the volume differences, the surface areas of FCs and MBSs were also extended in ΔmiR-127 (supplementary material Table S3). Moreover, the average length of FCs in the Lz was increased in ΔmiR-127, without a change in diameter. There was no effect of miR-127 deficiency on the thickness of the interhemal trophoblast membrane, where nutritional exchange takes place. These results suggest that miR-127 suppresses fetal capillarisation of the placental exchange region.
In ΔRtl1, placental abnormalities were observed in the same compartments affected by miR-127 deficiency, but with opposite phenotypes (Fig. 2F; supplementary material Fig. S2). These results suggest that Rtl1 supports FC elongation and that the two genes interact to regulate the same placental processes. The alterations in MBS and FC surface area would affect nutrient and oxygen supply to the fetus and contribute to the observed fetal growth restriction. The theoretical diffusion capacity (TDC) and specific diffusion capacity (SDC) are barometers for the potential ability of small molecules such as oxygen to transfer by passive diffusion from mother to fetus (Laga et al., 1973). The TDC and SDC values of the mutant placentae indicate that ΔmiR-127 mice have a higher diffusive capacity than WT and, conversely, that ΔRtl1 placentae have a reduced capacity (supplementary material Table S3). Although this is likely to contribute to the growth retardation of the ΔRtl1 fetuses, it is noteworthy that the ΔmiR-127 mutants are not growth enhanced. Previous work has proposed that Rtl1 cleaves an extracellular matrix (ECM) component resulting in a release of growth factors to promote hepatocarcinogenesis (Riordan et al., 2013). During angiogenesis, the degradation of the basement membrane and ECM facilitates migration into the interstitial matrix and the formation of new capillaries (Jain, 2003). Since placental Lz RTL1 protein is expressed in the capillary endothelial cells (Sekita et al., 2008), we propose that RTL1 promotes cleavage of the basement membrane to progress the vascularisation of FCs.
All Rtl1 isoforms are regulated by miR-127
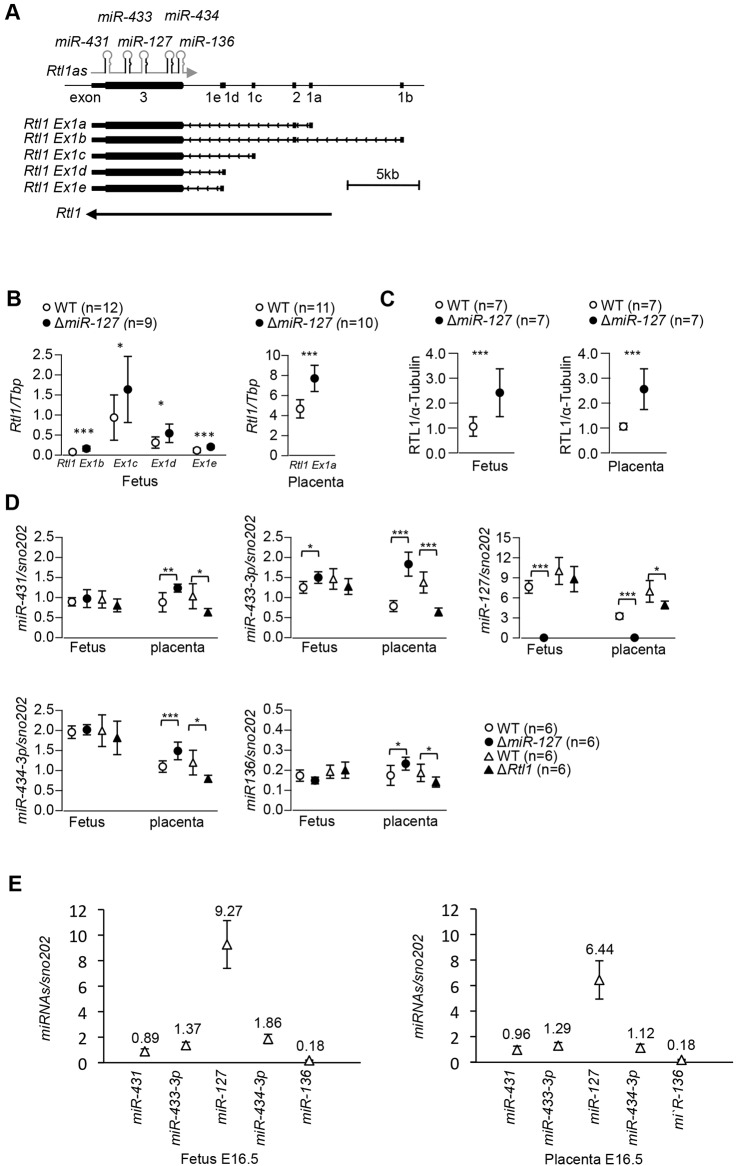
cDNA screening previously revealed that Rtl1 has two exons and a transcription start site located 5 kb upstream of the retrotransposon-like sequences (Hagan et al., 2009), suggesting that Rtl1 might be regulated by a host-derived promoter outside the retrotransposon. In order to clarify Rtl1 transcript structure, we identified further Rtl1 transcription start sites by 5′RACE. One alternative leader exon was identified in E15.5 placenta (Rtl Ex1a) and three alternatives were identified in the E11 embryo (Rtl1 Ex1b, Ex1d and Ex1e) (Fig. 3A; supplementary material Fig. S4). All five Rtl1 alternative transcripts, including the known Rtl1 Ex1c (GenBank: EU434918), contain a common large exon, namely exon 3, which contains the retrotransposon-derived ORF, and different small exons. All alternative exon 1s are spread over a 12 kb region, suggesting that they might be transcribed from different promoters. To address this, real-time RT-PCR was performed using alternative transcript-specific forward primers and a common reverse primer in exon 3. This showed that Rtl1 Ex1c is the most abundant transcript in E16.5 whole embryos (supplementary material Fig. S3B). The other Rtl1 transcripts were also detectable in E16.5 embryos, but the Rtl1 Ex1a expression level was much lower (0.6% of total) than for the other four. Conversely, the most abundant mRNA in the placenta was Rtl1 Ex1a, which contributed more than 97% of total Rtl1 expression compared with the others (supplementary material Fig. S3B).
Fig. 3.
Expression of Rtl1 alternative transcripts and miRNAs in embryo and placenta at E16.5. (A) Structure of the mouse Rtl1 locus. Alternative transcripts are transcribed from different leader exons. Exon 1s are named a-e. All alternative transcripts have a common exon 3 that has a conserved retrotransposon sequence. Rtl1 Ex1a and Ex1b also have a common exon 2. Exons are represented by a solid box. (B) Quantitative expression analysis for each alternative Rtl1 transcript in ΔmiR-127 embryo and placenta at E16.5. (C) Western blotting for RTL1 normalised to α-tubulin in ΔmiR-127 embryo and placenta at E16.5. (D) miRNA expression is shown normalised to snoRNA202. (E) The relative expression of miRNAs. miR-127 is the most abundant of the miRNAs in Rtl1as. *P<0.05, **P<0.01, ***P<0.005 (Student's t-test). Error bars indicate s.d.
In order to address whether all Rtl1 transcripts were equivalently modulated by miR-127, we quantified Rtl1 transcript levels in ΔmiR-127 embryos and placentae. Results showed that all alternative transcripts were significantly overexpressed (∼1.7-fold of control) in E16.5 ΔmiR-127 embryos (Fig. 3B). Rtl1 Ex1a was significantly increased (1.7-fold) in ΔmiR-127 placentae (Fig. 3B). This is not an indirect effect caused by a disproportionate increase in the number of endothelial cells, since there is a similar increase of 70% in Ex1a expression when normalised to the endothelial cell marker Pecam1 (CD31) (supplementary material Fig. S3C). Analysis of hybrid fetuses and placentae showed that all alternative transcripts are exclusively transcribed from the paternal chromosome in ΔmiR-127 (supplementary material Fig. S3D), indicating that the overexpression is not associated with LOI. Western blotting showed that RTL1 protein was significantly increased and in proportion to the increased level of the transcript in E16.5 ΔmiR-127 embryos and placentae (Fig. 3C; supplementary material Fig. S1H). Since deletion of six of the seven miRNAs results in only a 2.5-fold increase in Rtl1 mRNA (supplementary material Fig. S3E), our findings indicate that, compared with the other miRNAs in the cluster, miR-127 contributes a proportionately greater effect on Rtl1 levels in placentae, and disruption of this repression causes placental overgrowth.
Consistent with its impact on Rtl1 levels, miR-127 is the most abundant miRNA generated from Rtl1as (Fig. 3E). We next determined whether the deletion of miR-127 influences expression of the neighbouring miRNAs to potentially impact Rtl1 expression. As expected, miR-127 was not detected in ΔmiR-127 embryos and placentae (Fig. 3D). In ΔmiR-127 fetuses, only miR-433-3p was upregulated, with no change in miR-431, miR-434-3p and miR-136 expression (Fig. 3D). By contrast, all four miRNAs were significantly upregulated, with miR-433-3p the most induced, in ΔmiR-127 placenta. The same miRNAs that were upregulated in the placenta in the ΔmiR-127 mutant were downregulated in ΔRtl1 (Fig. 3D). Together, these results suggest that there might be a compensatory feedback mechanism involving RTL1 that acts specifically in the placenta to minimise the impact on Rtl1 transcript levels. miR-433 has its own promoter and thus might be more sensitive to this feedback mechanism (Song and Wang, 2008).
Rtl1 is the main target gene of miR-127 for placenta development
Our data suggest miR-127 can regulate placental growth through Rtl1 repression. However, to address the possibility that other target genes of miR-127 might also contribute to placental development, we generated double-heterozygous mice (ΔmiR-127/ΔRtl1) lacking both Rtl1 and miR-127. If Rtl1 is the main target of miR-127 leading to repressed placental growth then the ΔmiR-127/ΔRtl1 mutant should show a similar phenotype to ΔRtl1. However, if miR-127 has other targets contributing to this phenotype then the ΔmiR-127/ΔRtl1 mutant would be expected to have an intermediate phenotype between that seen in ΔRtl1 and that seen in ΔmiR-127. The ΔmiR-127/ΔRtl1 mutant mouse embryo and placental weight data show that they are similar to ΔRtl1 at E18.5 rather than to ΔmiR-127 (Fig. 4A). Histological analysis also showed that the extent and volume reduction of the placental Lz was the same in both ΔmiR-127/ΔRtl1 and ΔRtl1 (Fig. 2C and Fig. 4B). Detailed analysis of the Lz also determined that volumes and surface areas of the MBS, FC and LT and the TDC and SDC were similarly decreased in ΔRtl1 and ΔmiR-127/ΔRtl1 compared with WT (supplementary material Table S3). By contrast, these volumes were increased in ΔmiR-127. These striking similarities between ΔRtl1 and ΔmiR-127/ΔRtl1 placentae suggest that miR-127 specifically acts upstream of Rtl1 during placental development.
Fig. 4.
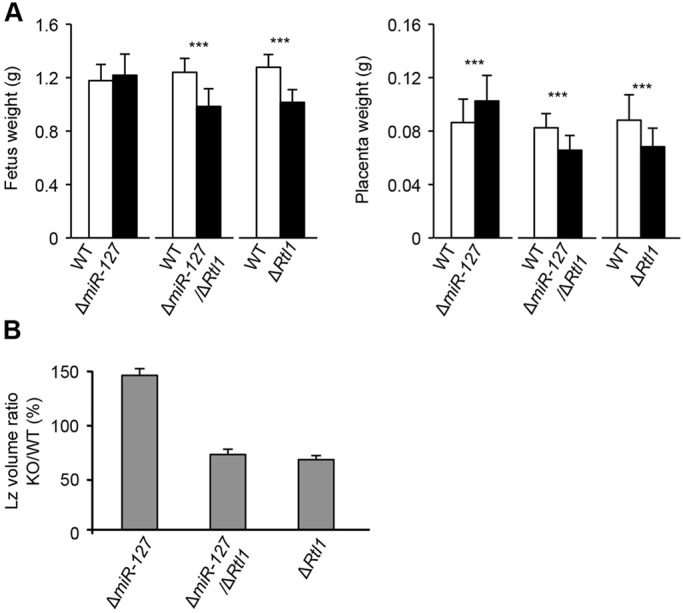
ΔmiR-127/ΔRtl1 knockout mice are comparable to ΔRtl1. Double-heterozygous mice carrying both ΔmiR-127 and ΔRtl1 were born from heterozygous parents. (A) Fetal and placental weights at E18.5. (B) Volume ratio of the placental Lz at E18.5. ***P<0.005 (Student's t-test). Error bars indicate s.d.
Comparative analysis of the genomic locus between eutherian, metatherian and protherian mammals suggests that miRNAs on Rtl1as evolved in eutherians along with the neofunctionalisation of RTL1 (Edwards et al., 2008). Marsupial mammals lack the miRNAs and have retained only remnants of the Ty3/Gypsy retrotransposon that evolved into Rtl1 in eutherians. Hence, it is likely that Rtl1as miRNAs evolved as a host defence mechanism to suppress the activity of this retrotransposon-derived gene (Edwards et al., 2008). In particular, the reciprocally imprinted miR-127 and Rtl1, which interact so effectively in trans, co-evolved to regulate placenta development.
MATERIALS AND METHODS
Generation of ΔmiR-127/ΔRtl1 mice
We generated a miR-127 deletion construct that lacks 134 bp incorporating miR-127 (chr12:109,592,803-109,592,936) (supplementary material Fig. S1A). The miR-127 targeting construct was transfected into female 129SV ESCs and clones containing the targeting vector were selected (supplementary material Fig. S1C-E). After deletion of the neomycin resistance gene (supplementary material Fig. S1F), targeted ESCs were injected into blastocysts to make chimaeras and germline transmission confirmed. Animals were backcrossed to C57BL/6J for ten generations with consistent growth and viability phenotypes noted after N5 on this genetic background (supplementary material Table S1). Mice were subsequently maintained on a C57BL/6J genetic background. For further details, see the supplementary Materials and Methods.
Placental histology
Placentae from embryonic day (E) 18.5 conceptuses were dissected free of fetal membranes, weighed and bisected mid-sagittally. One half was fixed in 4% paraformaldehyde, paraffin embedded, sectioned, stained with Hematoxylin and Eosin (H&E) and the gross placental structure analysed. The other half was fixed in 4% glutaraldehyde, resin embedded, stained with Toluidine Blue and the structure stereologically assessed. Analyses were performed using the Computer Assisted Stereological Toolbox (CAST v2.0) program as previously described (Coan et al., 2004). Further details of placental histology are given in the supplementary Materials and Methods.
Rapid amplification of cDNA ends (5′RACE) and quantitative RT-PCR
5′RACE was performed using the First Choice RLM-RACE Kit (Ambion) following the manufacturer's protocol; 10 µg of total RNA from E11 fetus and E15.5 placenta was used as the starting material.
For real-time PCR, total RNA (10 µg) from whole embryos and placenta at E16.5 was treated with RQ1 RNase-free DNase (Promega). cDNA was synthesized using RevertAid H Minus First Strand cDNA Synthesis Kit with random hexamers (Fermentas). Real-time RT-PCR assay for Rtl1 was performed using alternative exon 1-specific forward primers and a common reverse primer on exon 3. TATA box binding protein (Tbp) and Pecam1 (CD31; Wang et al., 2005) expression was used as an internal control.
For mature miRNA expression, we carried out real-time RT-PCR using TaqMan microRNA assays (Applied Biosystems). Additional details of 5′RACE and real-time RT-PCR are provided in the supplementary Materials and Methods.
Western blotting
Proteins were extracted from E16.5 embryos and placentae using RIPA buffer containing protease inhibitors (Complete, EDTA-free, Roche). RTL1 was detected by rabbit anti-RTL1 antibody (YZ2844) created in the C.L.S. laboratory, and then normalised by anti-α-tubulin (Sigma-Aldrich, T6199). Further details are given in the supplementary Materials and Methods.
Supplementary Material
Acknowledgements
We thank members of the A.C.F.-S. laboratory for discussions; and Neil Youngson, Simao Teixeira da Rocha, Sylvia Kocialkowski, Marika Charalambous and Marie Watkins for assistance and discussions during the course of this work.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) and UK Medical Research Council (MRC) [MR/J001597/1] and EU FP7 Marie Curie Action 290123 (INGENIUM). This work was partly funded by a Australian Government National Health and Medical Research Council (NHMRC) CJ Martin Biomedical Fellowship to A.N.S.-P. Deposited in PMC for release after 6 months.
Author contributions
M.I. and A.C.F.-S. designed the study. M.I., A.N.S.-P., C.A.E., T.-H.L. and M.K. performed experiments. M.I., A.N.S.-P., S.E.A., T.K.-I., F.I., C.L.S. and A.C.F.-S. analysed and discussed the data. M.I., A.N.S.-P., B.T.A. and A.C.F.-S. wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.121996/-/DC1
References
- Brandt J., Schrauth S., Veith A.-M., Froschauer A., Haneke T., Schultheis C., Gessler M. Leimeister C. and Volff J.-N. (2005). Transposable elements as a source of genetic innovation: expression and evolution of a family of retrotransposon-derived neogenes in mammals. Gene 345, 101-111. 10.1016/j.gene.2004.11.022 [DOI] [PubMed] [Google Scholar]
- Ciaudo C., Servant N., Cognat V., Sarazin A., Kieffer E., Viville S., Colot V., Barillot E., Heard E. and Voinnet O. (2009). Highly dynamic and sex-specific expression of microRNAs during early ES cell differentiation. PLoS Genet. 5, e1000620 10.1371/journal.pgen.1000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coan P. M., Ferguson-Smith A. C. and Burton G. J. (2004). Developmental dynamics of the definitive mouse placenta assessed by stereology. Biol. Reprod. 70, 1806-1813. 10.1095/biolreprod.103.024166 [DOI] [PubMed] [Google Scholar]
- da Rocha S. T., Edwards C. A., Ito M., Ogata T. and Ferguson-Smith A. C. (2008). Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306-316. 10.1016/j.tig.2008.03.011 [DOI] [PubMed] [Google Scholar]
- Davis E., Caiment F., Tordoir X., Cavaillé J., Ferguson-Smith A., Cockett N., Georges M. and Charlier C. (2005). RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743-749. 10.1016/j.cub.2005.02.060 [DOI] [PubMed] [Google Scholar]
- Edwards C. A., Mungall A. J., Matthews L., Ryder E., Gray D. J., Pask A. J., Shaw G., Graves J. A. M., Rogers J.; SAVOIR consortium, et al. (2008). The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6, e135 10.1371/journal.pbio.0060135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith A. C. (2011). Genomic imprinting: the emergence of an epigenetic paradigm. Nat. Rev. Genet. 12, 565-575. 10.1038/nrg3032 [DOI] [PubMed] [Google Scholar]
- Gundersen H. J. G., Bendtsen T. F., Korbo L., Marcussen N., Møller A., Nielsen K., Nyengaard J. R., Pakkenberg B., Sørensen F. B., Vesterby A. et al. (1988). Some new, simple and efficient stereological methods and their use in pathological research and diagnosis. APMIS 96, 379-394. 10.1111/j.1699-0463.1988.tb05320.x [DOI] [PubMed] [Google Scholar]
- Hagan J. P., O'Neill B. L., Stewart C. L., Kozlov S. V. and Croce C. M. (2009). At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4, e4352 10.1371/journal.pone.0004352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio M. V., Ferracin M., Liu C.-G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. et al. (2005). MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65, 7065-7070. 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- Jain R. K. (2003). Molecular regulation of vessel maturation. Nat Med. 9, 685-693. 10.1038/nm0603-685 [DOI] [PubMed] [Google Scholar]
- Laga E. M., Driscoll S. G. and Munro H. N. (1973). Quantitative studies of human placenta. I. Morphometry. Biol. Neonate. 23, 231-259. 10.1159/000240605 [DOI] [PubMed] [Google Scholar]
- Lin S.-P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J. and Ferguson-Smith A. C. (2003). Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 35, 97-102. 10.1038/ng1233 [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A. et al. (2005). MicroRNA expression profiles classify human cancers. Nature 435, 834-838. 10.1038/nature03702 [DOI] [PubMed] [Google Scholar]
- Mandarim-de-Lacerda C. A. (2003). Stereological tools in biomedical research. An. Acad. Bras. Cienc. 75, 469-486. 10.1590/s0001-37652003000400006 [DOI] [PubMed] [Google Scholar]
- Riordan J. D., Keng V. W., Tschida B. R., Scheetz T. E., Bell J. B., Podetz-Pedersen K. M., Moser C. D., Copeland N. G., Jenkins N. A., Roberts L. R. et al. (2013). Identification of Rtl1, a retrotransposon-derived imprinted gene, as a novel driver of hepatocarcinogenesis. PLoS Genet. 9, e1003441 10.1371/journal.pgen.1003441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H., Youngson N., Lin S.-P., Dalbert S., Paulsen M., Bachellerie J.-P., Ferguson-Smith A. C. and Cavaillé J. (2003). Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261-262. 10.1038/ng1171 [DOI] [PubMed] [Google Scholar]
- Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., Hino T., Suzuki-Migishima R., Kohda T., Ogura A. et al. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243-248. 10.1038/ng.2007.51 [DOI] [PubMed] [Google Scholar]
- Song G. and Wang L. (2008). MiR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433–127 locus. PLoS ONE 3, e3574 10.1371/journal.pone.0003574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Riha G. M., Yan S., Li M., Chai H., Yang H., Yao Q. and Chen C. (2005). Shear stress induces endothelial differentiation from a murine embryonic mesenchymal progenitor cell line. Arterioscler. Thromb. Vasc. Biol. 25, 1817-1823. 10.1161/01.ATV.0000175840.90510.a8 [DOI] [PubMed] [Google Scholar]
- Youngson N. A., Kocialkowski S., Peel N. and Ferguson-Smith A. C. (2005). A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481-490. 10.1007/s00239-004-0332-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.