Abstract
Neurogenesis involves deeply conserved patterning molecules, such as the proneural basic helix-loop-helix transcription factors. Sox proteins and specifically members of the SoxB and SoxC groups are another class of conserved transcription factors with an important role in neuronal fate commitment and differentiation in various species. In this study, we examine the expression of all five Sox genes of the nematode C. elegans and analyze the effect of null mutant alleles of all members of the SoxB and SoxC groups on nervous system development. Surprisingly, we find that, unlike in other systems, neither of the two C. elegans SoxB genes sox-2 (SoxB1) and sox-3 (SoxB2), nor the sole C. elegans SoxC gene sem-2, is broadly expressed throughout the embryonic or adult nervous system and that all three genes are mostly dispensable for embryonic neurogenesis. Instead, sox-2 is required to maintain the developmental potential of blast cells that are generated in the embryo but divide only postembryonically to give rise to differentiated neuronal cell types. Moreover, sox-2 and sox-3 have selective roles in the terminal differentiation of specific neuronal cell types. Our findings suggest that the common themes of SoxB gene function across phylogeny lie in specifying developmental potential and, later on, in selectively controlling terminal differentiation programs of specific neuron types, but not in broadly controlling neurogenesis.
KEY WORDS: Caenorhabditis elegans, Sox genes, Neurogenesis
Summary: Unlike in other organisms, SoxB and SoxC genes in C. elegans have no broad function in embryonic nervous system development; instead, SoxB genes control the terminal differentiation of restricted neuron types.
INTRODUCTION
Comparing the function of specific molecules in diverse organisms has revealed core principles of nervous system development. One such core principle is constituted by proneural basic helix-loop-helix (bHLH)-type transcription factors that act in species as diverse as Hydra, nematodes, flies and mice to convert ectodermal cells into neuroblasts (Bertrand et al., 2002; Galliot et al., 2009). Sox proteins, which contain an HMG box-type DNA-binding domain, are another family of deeply conserved transcription factors present in all metazoans (Phochanukul and Russell, 2010). Sox proteins fall into eight main groups based on sequence identity and have been implicated in various developmental processes, with members of the same group often showing functional redundancy (Table 1) (Guth and Wegner, 2008). Members of the SoxB and SoxC groups are important regulators of different steps of nervous system development in diverse species (Reiprich and Wegner, 2014).
Table 1.
Overview of Sox genes
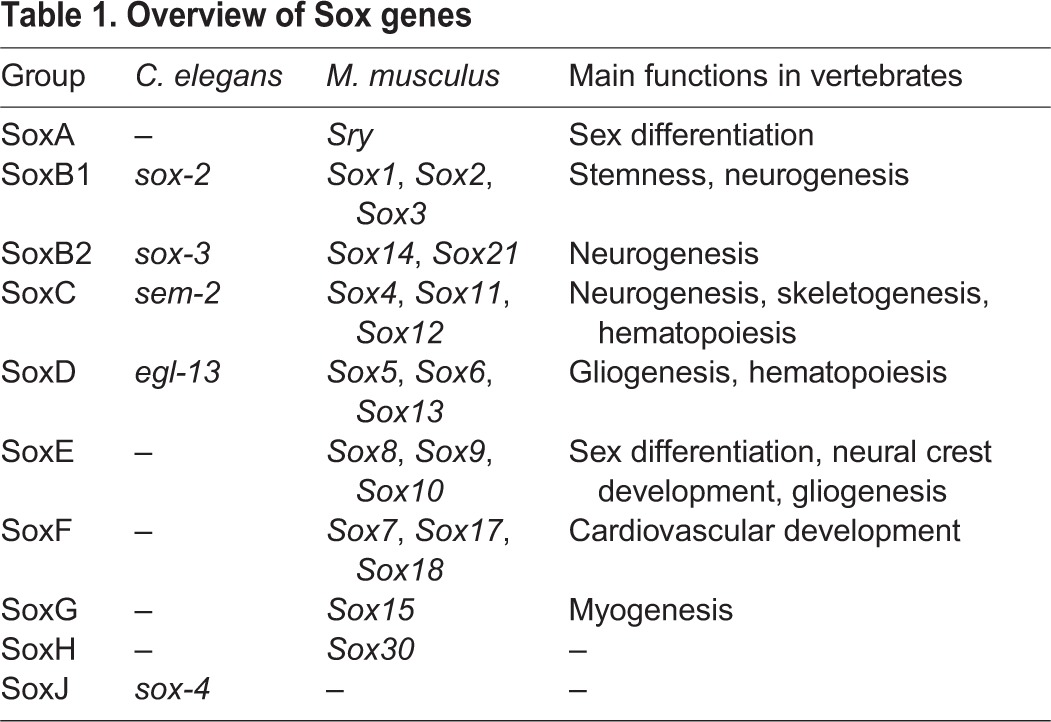
There are five SoxB genes in mammals: Sox1, Sox2, Sox3 (subgroup B1) and Sox14, Sox21 (subgroup B2). Mammalian SoxB1 genes are important to establish neuroectodermal fate and are key regulators of neural stem cell maintenance during development and in adult neurogenic regions (Pevny and Placzek, 2005; Reiprich and Wegner, 2014). Furthermore, the mammalian Sox2 gene is a key pluripotency factor (Takahashi and Yamanaka, 2006). Sox21 (SoxB2) has been shown to be important for the progression of neuronal differentiation by counteracting the action of SoxB1 proteins (Sandberg et al., 2005). Drosophila has four SoxB genes: SoxNeuro (SoxB1), Dichaete, Sox21a and Sox21b (SoxB2). SoxNeuro and Dichaete are co-expressed in the developing CNS and double-mutant embryos show severe defects in neuroblast formation (Buescher et al., 2002; Overton et al., 2002).
The SoxC group consists of three members in mammals (Sox4, Sox11, Sox12). Sox4 and Sox11 play an important role at later stages of both embryonic and adult neurogenesis, where they act downstream of SoxB genes to promote neuronal differentiation in postmitotic neurons, at least in part by directly controlling the expression of pan-neuronally expressed genes (Bergsland et al., 2006; Mu et al., 2012).
Based on sequence similarity, the genome of the nematode C. elegans contains two SoxB genes, sox-2 (SoxB1) and sox-3 (SoxB2), and one SoxC gene (sem-2) (Bowles et al., 2000; Shinzato et al., 2008) (Table 1). However, nothing is known about their potential role in embryonic nervous system development. sox-2 has only been examined in the context of a naturally occurring reprogramming event (Kagias et al., 2012), sem-2 is involved in mesoderm development (Tian et al., 2011), while sox-3 has not previously been examined at all. Here, we describe an expression pattern analysis of all C. elegans Sox genes and provide a loss-of-function analysis of the SoxB and SoxC genes in embryonic nervous system development using null mutant alleles. Surprisingly, we find that, unlike in any other system, C. elegans SoxB genes are dispensable for neuronal fate commitment during embryogenesis, while the SoxC gene has a very limited role in one specific lineage. However, SoxB genes have specific roles in the terminal differentiation of certain neuronal cell types. We also find that sox-2 is required to retain the developmental potential of distinct blast cells that are generated during embryogenesis but give rise to differentiated progeny later on during larval development. Our study therefore ascribes common and divergent themes of SoxB and SoxC gene function across phylogeny.
RESULTS
Sox genes are only selectively expressed during embryonic neurogenesis
Previous work from our laboratory has defined gene regulatory routines that operate during terminal neuronal differentiation throughout the nervous system of C. elegans (e.g. Kratsios et al., 2012; Serrano-Saiz et al., 2013). These previous studies focused on understanding how the expression of terminal effector genes in a differentiating neuron type are regulated (‘bottom-up approach’). As a complementary approach to understand nervous system development we set out to identify factors involved in early neurogenesis in C. elegans (‘top-down approach’). Given the roles of SoxB and SoxC genes in early neurogenesis in various species (Reiprich and Wegner, 2014), we examined the expression of the C. elegans SoxB1 gene sox-2, the SoxB2 gene sox-3 and the SoxC gene sem-2 using fosmid-based reporter genes (Fig. 1). These fosmid reporters rescued the mutant phenotypes associated with loss of these genes (as described below) and we confirmed their expression patterns with single-molecule fluorescent in situ hybridization (smFISH; supplementary material Fig. S1).
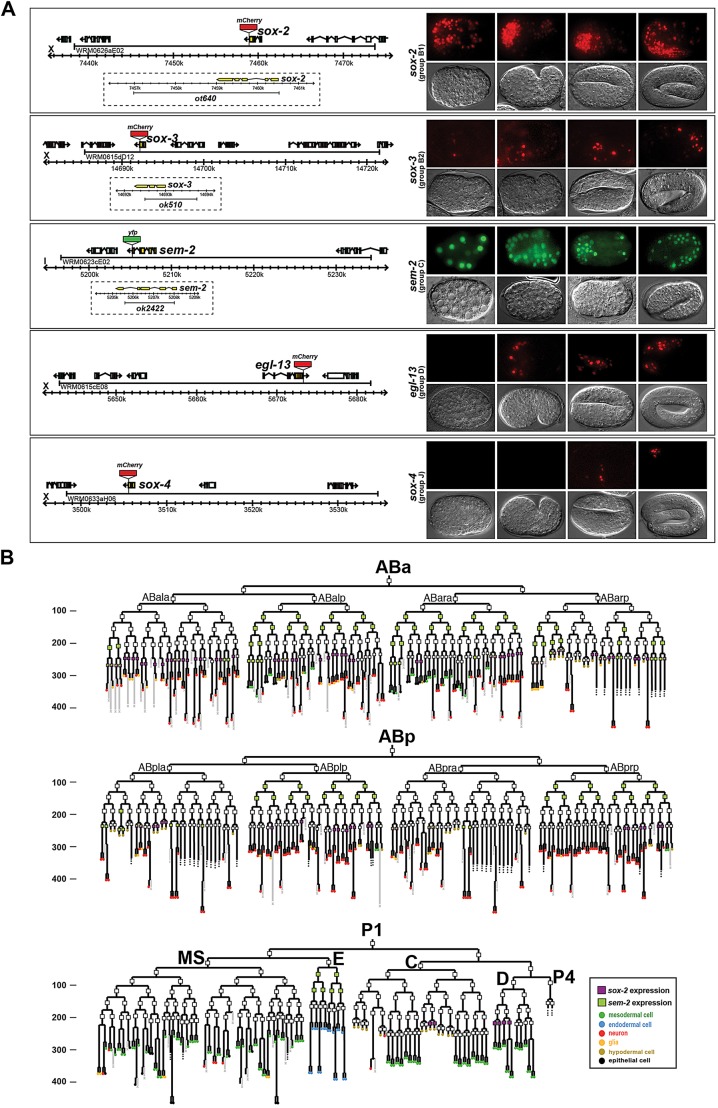
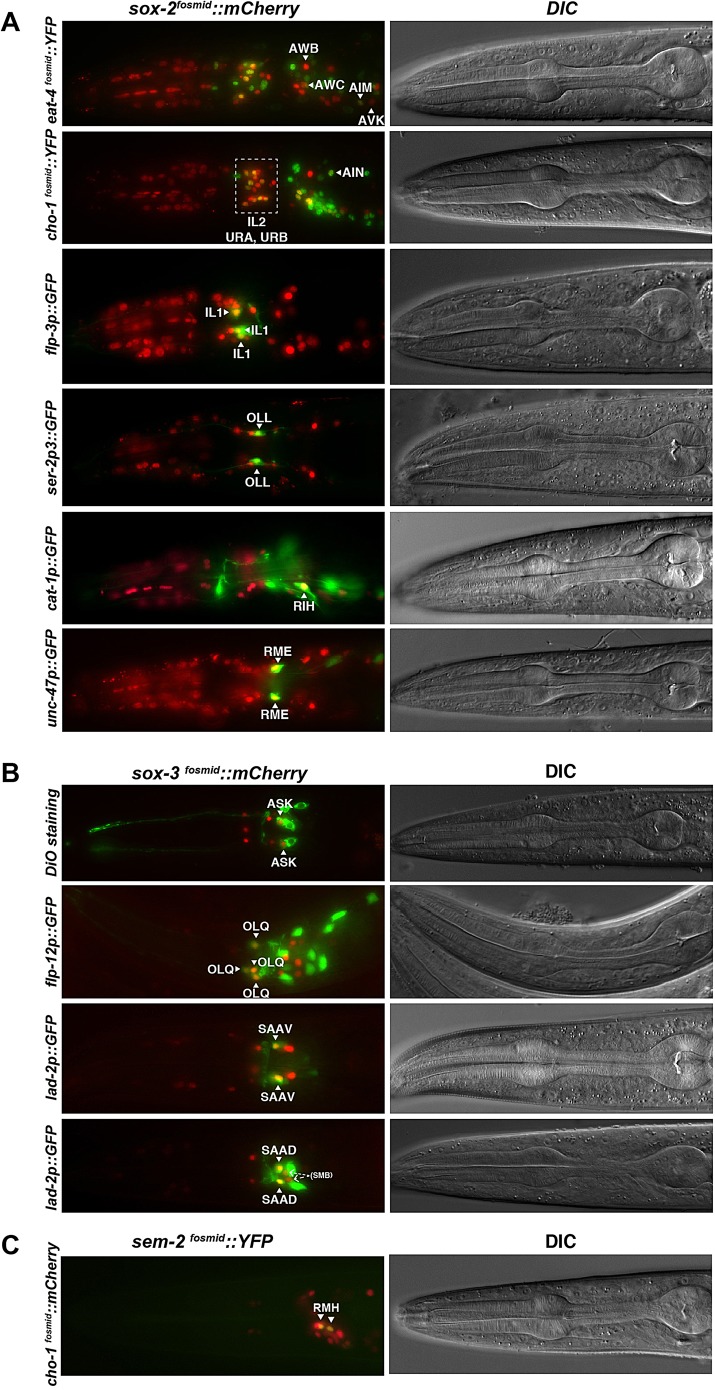
Fig. 1.
C. elegans Sox gene expression. (A) Representative images of Sox gene expression (top rows; bottom rows are DIC images) at different stages of embryonic development (right), together with fosmid reporter schematics (left). (B) Lineage diagram showing the specific cells in which sox-2 and sem-2 are expressed during embryogenesis, as determined by a combination of manual and automated lineaging using 4D microscopy and the SIMI software or the StarryNite and AceTree software, respectively. Rectangles indicate the stage at which expression was examined; a filled rectangle indicates expression, whereas no fill indicates no expression. Expression in the last cell division of the lineage is not represented in this diagram. Numbers on the y-axis represent developmental time (in minutes). N=2. See Table 2 for a summary of expression in postembryonic neuron types.
Using semi-automated four-dimensional (4D) lineage analysis (Murray et al., 2008; Schnabel et al., 1997), we found only limited expression of all SoxB and SoxC genes during C. elegans embryogenesis. sox-2 expression was restricted to subsets of neuroblasts, contrasting with the broad expression of SoxB1 in Drosophila and vertebrate neuroectoderm tissue. Moreover, again in contrast to Drosophila and vertebrate Sox2 (Pevny and Placzek, 2005), sox-2 was expressed relatively late in nervous system development, in the progenitor of differentiated neurons, but not in earlier neuroectodermal cells (Fig. 1). sox-2 was also expressed in some progenitors of non-neural tissue. Expression of the other C. elegans SoxB gene, sox-3, was not observed in neuroblasts but was observed in several differentiated postmitotic neurons (Fig. 1), as we further describe below.
sem-2 expression was identified in several early blastomeres and in some neuronal and non-neuronal progenitors (Fig. 1). Its expression colocalized with that of sox-2 in a few progenitors. As in vertebrates, sem-2 expression was observed in postmitotic neurons (Fig. 1), but in contrast to the broad expression of SoxC genes in vertebrate postmitotic neurons, only a single class of postembryonic neurons expresses sem-2 (described below).
Together, these data reveal no obvious, broad correlation between SoxB and SoxC gene expression and embryonic neurogenesis. To examine whether a broad neurogenic function might have ‘transferred’ to other Sox genes in C. elegans, we also generated fosmid-based reporters for the two remaining Sox genes in the C. elegans genome: egl-13 (SoxD) and sox-4 (SoxJ). We found that, like sox-3, the egl-13 and sox-4 fosmid reporters are expressed only in a few neurons during late embryogenesis, when all embryonic neurons are already born (Fig. 1). A role of these genes in early neurogenesis therefore also seems unlikely.
Embryonic neurogenesis is unaffected in SoxB mutants
We next sought to examine the impact of genetic removal of C. elegans SoxB genes on embryonic neurogenesis. We generated a deletion allele of sox-2, ot640, using a targeted gene deletion method based on transposon excision (MosDEL) (Frøkjaer-Jensen et al., 2010). The deletion eliminates the entire sox-2 coding region (Fig. 1) and is therefore a null allele. sox-2(ot640) null mutant animals arrest at the first larval stage and show a 100% penetrant Pun (pharynx unattached) phenotype (Fig. 2A), which is at least one of the reasons for lethality. sox-2 is expressed in the head hypodermis and the arcade cells, two cell types that have been shown to be important for the proper attachment of the pharynx to the mouth cavity (Mango, 2007). The fate of these cells is not obviously affected in sox-2(ot640) mutants since the expression of hypodermis (dpy-7) or arcade cell (ajm-1, bath-15) identity markers appears normal in the absence of sox-2 (data not shown). However, more subtle differentiation defects in these cells that could account for the Pun phenotype cannot be excluded. The Pun phenotype and lethality of the sox-2(ot640) mutant can be rescued with the sox-2 fosmid-based reporter gene described above.
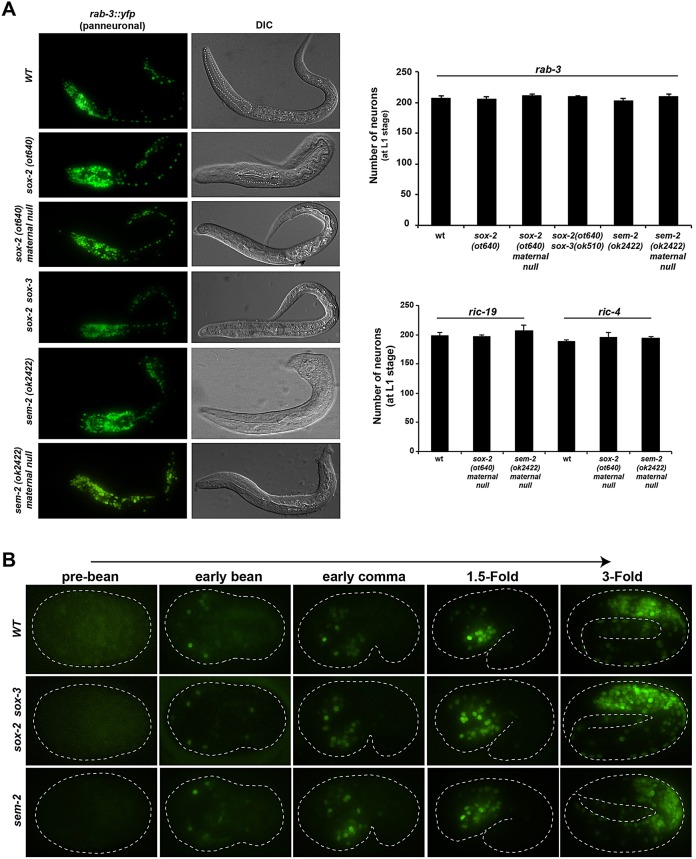
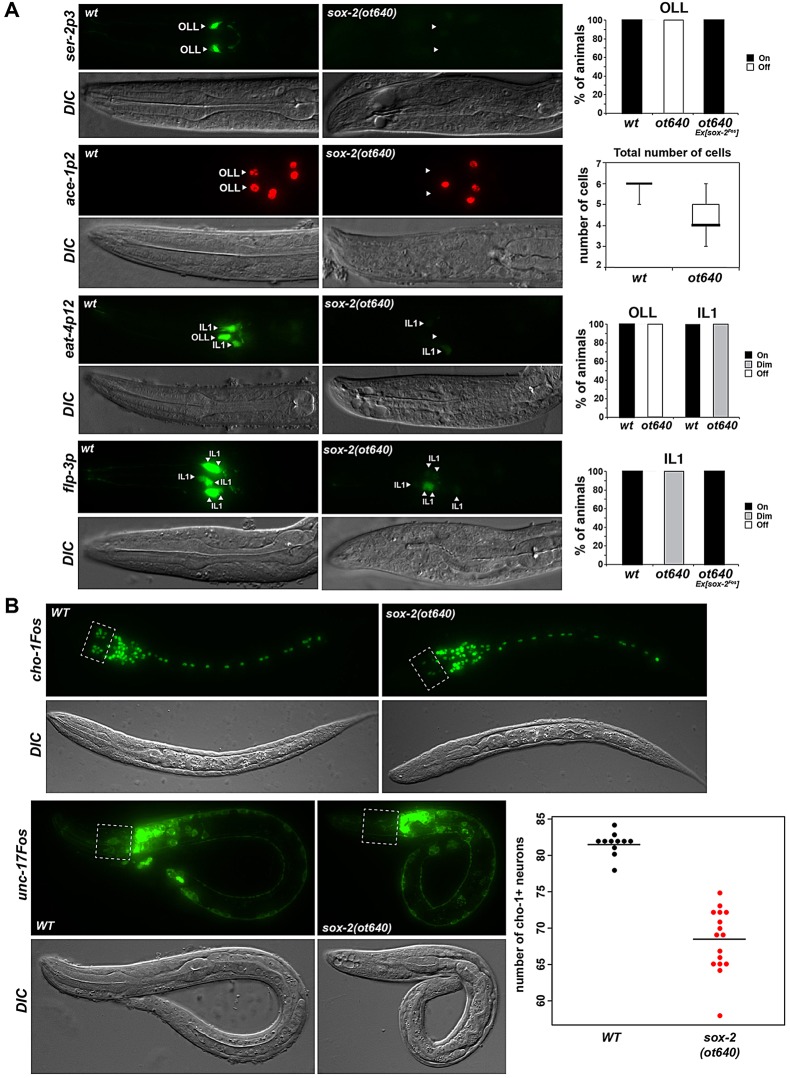
Fig. 2.
Embryonic neurogenesis is unaffected in SoxB and SoxC mutants. (A) Expression of the pan-neuronal genes rab-3, ric-19 and ric-4 is not affected in SoxB and SoxC mutants at the L1 stage. The total number of rab-3-, ric-19- and ric-4-expressing neurons was counted at the L1 stage; n=5 for each genotype. sox-2 mutants display a Pun (pharynx unattached) phenotype as shown in DIC. Error bars indicate s.d. (B) Timing of neurogenesis is not affected in SoxB and SoxC mutants. Representative images show the onset of rab-3 expression at the early bean stage and its increasing expression as development progresses. No differences are observed between wild-type (WT) embryos and sox-2(ot640) sox-3(ok510) double-mutant or sem-2(ok2422) mutant embryos.
Using rab-3, a synaptic vesicle Ras GTPase, as a pan-neuronal marker we examined whether sox-2 mutants display defects in neurogenesis. We did not find any changes in rab-3-expressing neuronal cell number in L1-arrested sox-2 null mutants (Fig. 2A), suggesting that sox-2 is not required during embryonic neuronal commitment and/or pan-neuronal identity specification. Because homozygous sox-2 mutants were generated from heterozygous parents, we addressed the possibility that defects could be masked by maternally supplied sox-2 gene activity. To this end, we analyzed maternal/zygotic null mutants derived from mosaic mothers that had lost the rescuing array in the germline-producing P lineage (see Materials and Methods). These maternal/zygotic null mutant animals were, like the zygotic nulls, viable until the L1 stage, and they still displayed the normal number of rab-3-expressing neurons (Fig. 2A). To further examine the notion that neuronal fate is not affected in sox-2 mutants, we analyzed the expression of additional pan-neuronal markers in sox-2 maternal/zygotic null animals: ric-4 (a SNARE), ric-19 (which is important for neuroendocrine secretion and DCV maturation) and rgef-1 (a Ras nucleotide exchange factor). No major defects were observed with these additional pan-neuronal markers (Fig. 2A; supplementary material Fig. S2). Also, in agreement with our functional results showing that sox-2 is not maternally contributed, the sox-2 fosmid reporter or sox-2 smFISH did not show any expression in the germ line or oocytes of young adult worms.
The possibility that the two members of the SoxB group, sox-2 (SoxB1) and sox-3 (SoxB2), act redundantly during embryonic neurogenesis seemed unlikely, since they are not expressed in the same cells and since sox-3 is not expressed in neuroblasts (Fig. 1). However, in order to rule out potential compensatory mechanisms, we also generated sox-2 and sox-3 double mutants using the above described null allele of sox-2 and a sox-3 null allele provided by the C. elegans Knockout Consortium (Fig. 1A). In these double-null mutant animals, the total number of rab-3-expressing neurons was still unaffected (Fig. 2A). Moreover, the onset of expression of the pan-neuronal marker rab-3 was unaffected in sox-2 sox-3 double mutants (Fig. 2B), indicating that the timing of neurogenesis remains normal in SoxB mutants. Additionally, we analyzed the fate of several neurons that express sox-2 transiently at the progenitor stage using neuron type-specific identity markers. Analyzing descendants of 18 of the 52 neuroblasts that express sox-2 during embryonic development, we found no obvious defects in sox-2 null mutants (supplementary material Fig. S3A).
These findings are in striking contrast to the early neurogenic defects observed upon removal of Drosophila or vertebrate SoxB genes (Buescher et al., 2002; Bylund et al., 2003; Okuda et al., 2010; Overton et al., 2002; Pevny and Placzek, 2005). The apparent lack of requirement for C. elegans SoxB genes in overall neurogenesis is consistent with their limited and relatively late expression during embryonic nervous system development, as described above.
sox-2 (SoxB1) is required for postembryonic lineage specification
We noted that sox-2 is expressed in several postembryonic blast cells that are generated in the embryo. These sox-2-expressing blast cells display epithelial characteristics from their generation until the L1 stage, and must retain their developmental potential in order to start dividing again during larval development. The larval stage divisions generate several neurons, among other cell types (Sulston and Horvitz, 1977). These blast cells include the B, Y, F, U and K rectal epithelial cells and the seam cells along the body (Fig. 3A). Although expression of sox-2 is absent in the terminal neurons generated by these lineages, sox-2 expression extends beyond the blast cell stage [e.g. sox-2 expression is maintained in the V5 daughters (V5a and V5p) but is lost in the next division]. sox-3 is co-expressed with sox-2 in the rectal epithelial cells but its expression is undetectable in seam cells or other blast cells (data not shown).
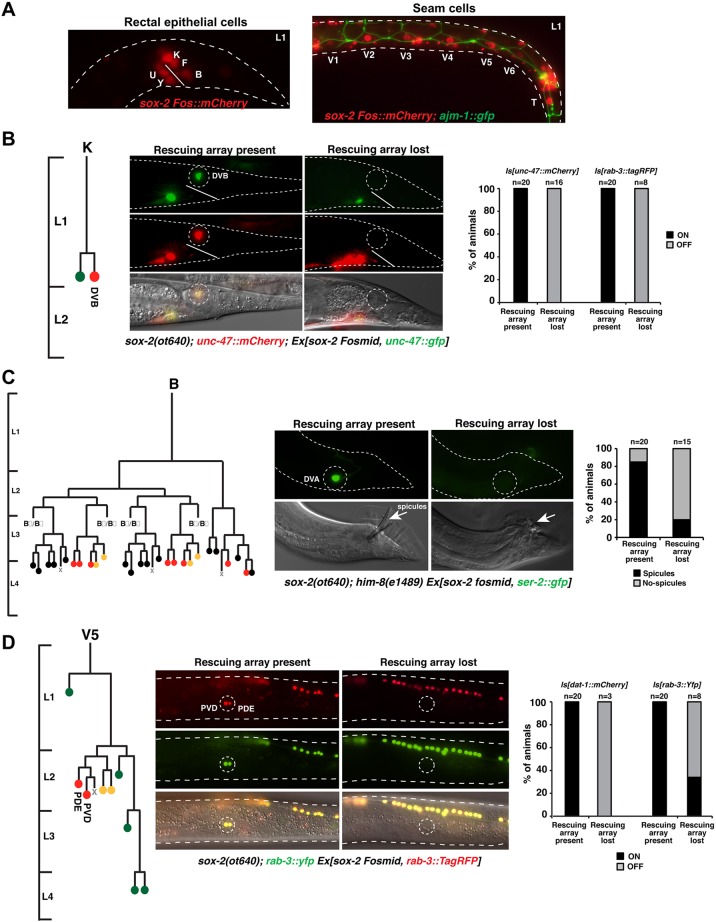
Fig. 3.
Postembryonic blast cells fail to generate certain lineages in sox-2 mutants. (A) Expression of sox-2 in rectal epithelial cells (left) and seam cells (right) at the L1 stage. (B) Mosaic analysis of sox-2(ot640) null mutants, showing loss of the DVB motor neuron. A scheme of the postembryonic K lineage is shown on the left. L1-lethal sox-2(ot640) animals were rescued with an extrachromosomal array (Ex) containing a fosmid with the sox-2 locus and the DVB reporter unc-47p::GFP in order to be able to follow the array in the K lineage. For phenotypic output, the expression of integrated (Is) fluorescent DVB (unc-47::mCherry) and pan-neuronal (rab-3::tagRFP) markers was analyzed. (C) Mosaic analysis of sox-2(ot640) null mutants, showing defective spicules in the male tail. Males were generated by crossing the sox-2(ot640) mutant into a him-8(e1489) mutant background. A scheme of the spicules-generating B lineage is shown on the left. L1-lethal sox-2(ot640) animals were rescued with an extrachromosomal array (Ex) containing a fosmid with the sox-2 locus and the DVA (sister of B) reporter ser-2p::GFP in order to infer the presence or absence of the rescuing array in the B lineage. For phenotypic output the presence of spicules in males was examined by DIC. (D) Mosaic analysis of sox-2(ot640) null mutants, showing loss of the PVD and PDE neurons. A scheme of the postembryonic V5 lineage is shown on the left. L1-lethal sox-2(ot640) animals were rescued with an extrachromosomal array (Ex) containing a fosmid with the sox-2 locus and the rab-3::TagRFP pan-neuronal reporter to assess the presence or absence of the rescuing array in the V5 lineage. For phenotypic output, expression of integrated (Is) fluorescent PVD and PDE markers (dat-1::mCherry and rab-3::yfp) was examined. Cell fate in lineage schemes: red, neurons; yellow, glial; green, hypodermal cells; black, proctodeum.
Owing to the early larval lethality of sox-2 null mutants, we used genetic mosaic analysis to address sox-2 function in these postembryonically dividing blast cells. We rescued sox-2(ot640) homozygous animals with an array containing the wild-type copy of sox-2 and analyzed mosaic animals that had specifically lost the sox-2 rescuing array in any of these blast cell lineages, as assessed with lineage-specific markers that were also present in the rescuing array. We found that in all cases examined, in the absence of sox-2 these blast cells failed to produce the neuron types that they normally produce (Fig. 3). Specifically, the K blast cell failed to produce the DVB motor neuron (Fig. 3B; supplementary material Fig. S4A), the B blast cell failed to generate male sensory structures (Fig. 3C), and the V5 blast cells failed to produce the PDE and PVD neurons in the postdeirid lineage (Fig. 3D). It is important to note that sox-2 is expressed in the blast cells from which these neurons are generated, but not in any of the neurons that fail to be generated in the sox-2 mosaic mutants. This suggests that sox-2 acts in the blast cells and not at later stages of the lineages when the terminal cells are born. In contrast to sox-2, the sox-3 mutant animals did not show any obvious defects in the generation of postembryonic lineages (not shown).
To examine the stage at which the blast cells become defective in their development, we assessed the expression of a number of genes previously shown to be expressed in these blast cells (Fig. 4A), including the Hox gene egl-5 (Wang et al., 1993), the Spalt-like transcription factor sem-4 (Jarriault et al., 2008), the POU homeobox gene ceh-6 (Burglin and Ruvkun, 2001), the Pax gene egl-38 (Johnson et al., 2001), the T-box transcription factor mab-9 (Woollard and Hodgkin, 2000) and the apical junction molecule ajm-1 (Koppen et al., 2001). We found that sox-2 regulates mab-9 expression in the B and F rectal epithelial cells (Fig. 4C). It was previously described that mab-9 expression in B and F is important for the correct specification of these cells, and in the absence of mab-9 B converts into a Y-like cell while F converts into a U-like cell (based on the lineages generated) (Chisholm and Hodgkin, 1989). Thus, the spicules lineage defect observed in sox-2 mutant males is possibly the result of a cell fate switch in which B transforms into a Y-like cell due to the absence of mab-9, which is downstream of sox-2. Since mab-9 expression is also lost in F in the absence of sox-2 we can infer that the F lineage may also be affected in sox-2 mutants. In the U cell, ceh-6 and sem-4 expression is selectively lost in sox-2 mutants. The other genes analyzed (egl-5, egl-38 and ajm-1) did not show any loss of expression in any of the rectal epithelial cells (Fig. 4C). Similarly, the seam cell markers ceh-16 (Engrailed) (Huang et al., 2009), hlh-2 (E protein transcription factor) and ajm-1 were also unaffected in the absence of sox-2 (Fig. 4D).
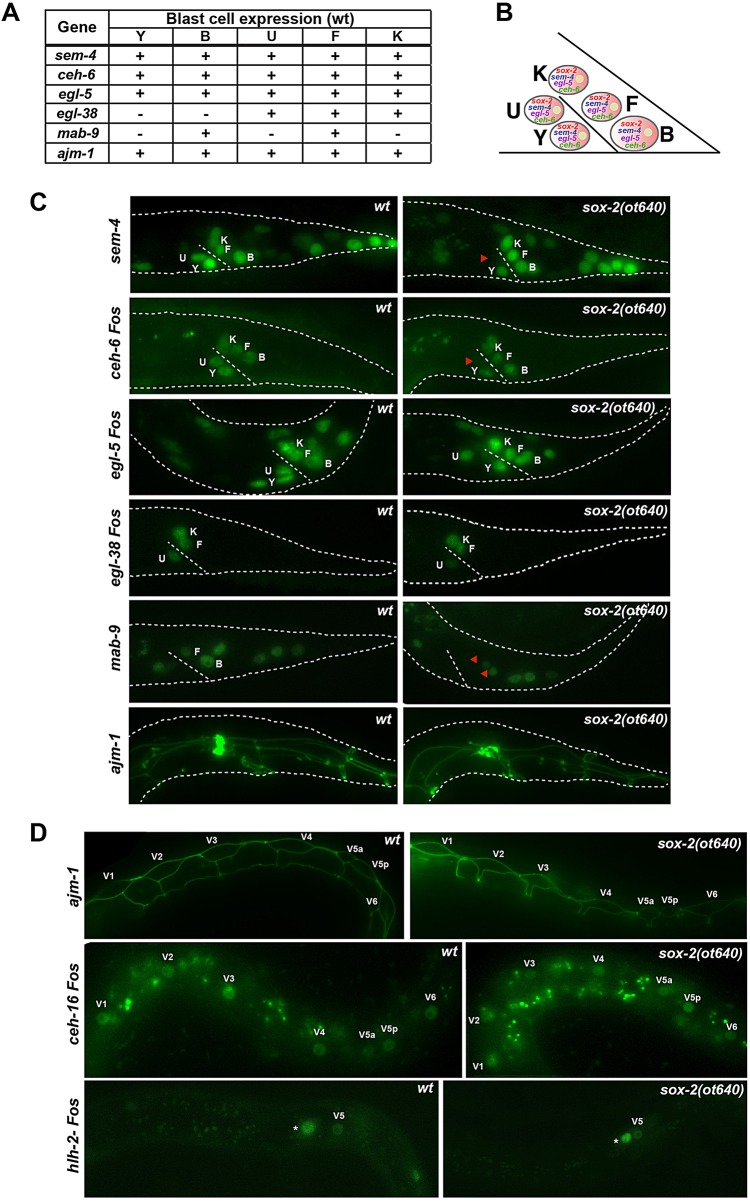
Fig. 4.
Postembryonic blast cells show selective differentiation defects in sox-2 mutants. (A) Expression summary of reporter genes expressed in rectal epithelial cells. (B) Scheme of rectal epithelial blast cells at the L1 stage, previously shown to express sem-4, egl-5 and ceh-6. sox-2 is also expressed in these cells (Fig. 3A). (C) Rectal epithelial cells show selective differentiation defects in sox-2 mutants. Reporter genes were crossed into sox-2(ot640) null mutants and animals were examined at the first larval stage. Representative images are shown; n>20. (D) Seam cell identity is unaffected in sox-2 mutants. Reporter genes were crossed into sox-2(ot640) null mutants and animals were examined at the first larval stage. Asterisk marks the Q neuroblast. Representative images are shown; n>20.
Using the ajm-1 apical marker we observed that the V5 blast cell is able to divide at least once to generate V5a and V5p in the sox-2 mutant (Fig. 4D). Moreover, in mosaic animals lacking sox-2 in the B or K rectal epithelial cells, these cells were able to enter the cell cycle, as indicated by the expression of the mcm-4 reporter, which marks dividing cells (Korzelius et al., 2011) (supplementary material Fig. S5).
In light of the failure of sox-2 mutants to generate DVB, we examined whether DVB transformed into the fate of its sister cell, i.e. a ceh-6-expressing epithelial cell. However, this is not the case (supplementary material Fig. S4B). Taken together, sox-2 expression in several postembryonic blast cells is required for the correct progression of postembryonic lineages.
SoxB genes have selective roles in the terminal differentiation of specific neuronal cell types
To investigate whether SoxB genes have later roles during neuronal differentiation we systematically examined their expression during larval and adult stages. We found that the two C. elegans SoxB genes show postmitotic and sustained expression throughout adulthood in several sensory, inter- and motor neurons (Table 2). sox-2 is more broadly expressed in the larval and adult nervous system than sox-3. sox-2 is expressed in the sensory neurons AWB, AWC, IL1, IL2, URA, URB, OLL, the interneurons AIM, AIN, AVK, RIH and the motor neuron class RME (Fig. 5A). sox-3 expression is maintained in the sensory neurons ASK, OLQ, the SMB motor neurons and the SAA neurons (Fig. 5B). The expression of the two SoxB genes does not overlap in any neuronal cell.
Table 2.
SoxB and SoxC expression in terminally differentiated neurons at larval and adult stages
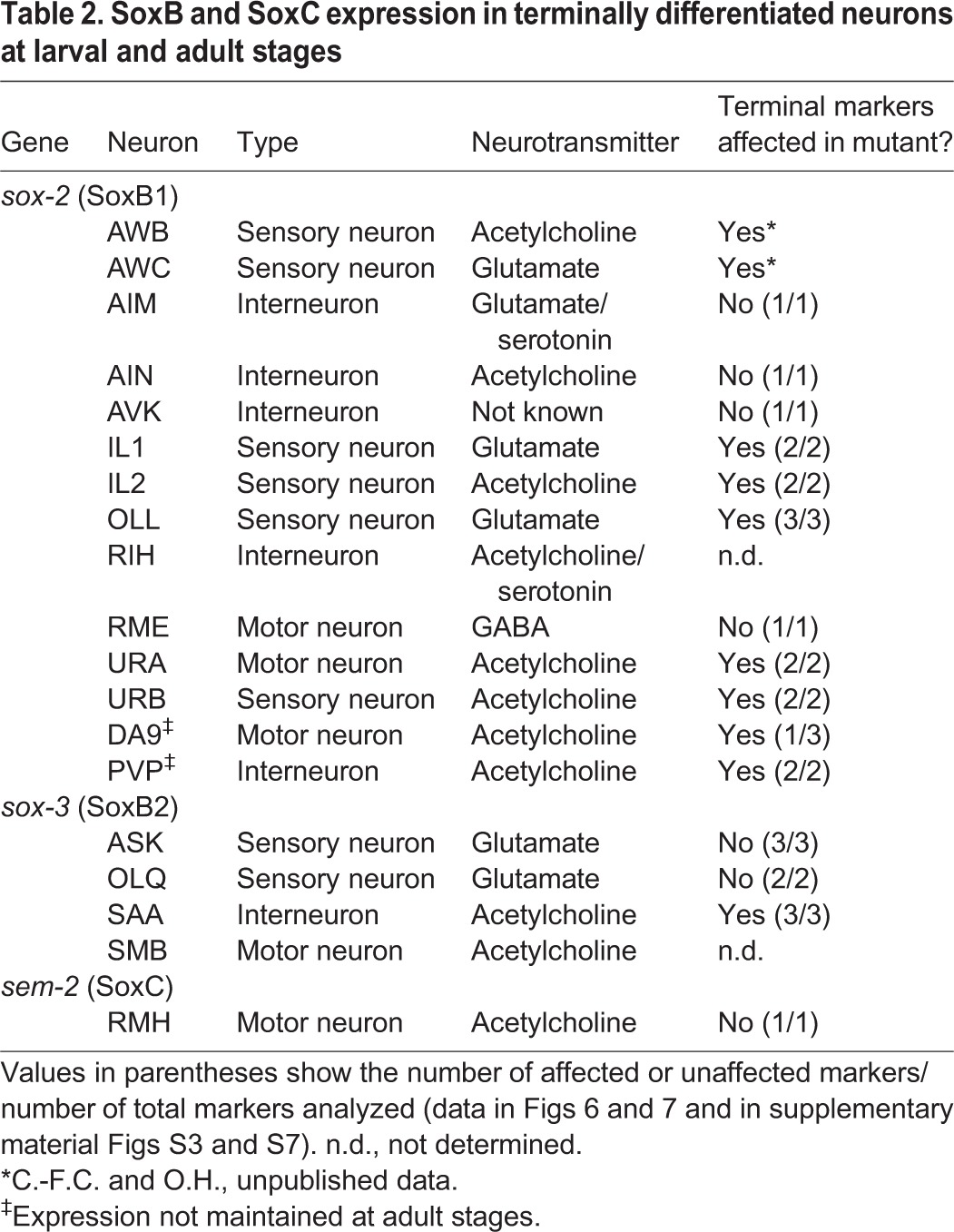
Fig. 5.
Postembryonic neuronal expression of SoxB and SoxC genes. Neurons expressing the SoxB1 gene sox-2 (A), the SoxB2 gene sox-3 (B) or the SoxC gene sem-2 (C) throughout the life of the worm were identified based on position and colocalization with several neuron type-specific markers. Representative young adult worms are shown.
To determine whether sox-2 or sox-3 has a role in neuron type-specific terminal differentiation, we then analyzed the expression in SoxB mutants of terminal identity markers that label neurons that continuously express sox-2 or sox-3 throughout their lifetime.
sox-2 (SoxB1) affects the terminal differentiation of glutamatergic and cholinergic neurons
Using a number of cell fate markers, we observed no apparent differentiation defects of the AIM, AIN, AVK or RME neurons in sox-2 null mutants (supplementary material Fig. S3A). However, the terminal identity of the glutamatergic sensory neurons OLL and IL1 was severely and partially affected, respectively (Fig. 6A). Specifically, the expression of the tyramine receptor ser-2, the acetylcholinesterase ace-1 and the vesicular glutamate transporter eat-4 was completely lost in OLL (Fig. 6A), while the expression of eat-4 and the neuropeptide flp-3 was considerably reduced in IL1 (Fig. 6A). The OLL neurons are generated in sox-2 mutants as assessed by unaffected expression of ift-20, a marker of ciliated neurons (not shown). The OLL neurons are known to also require the Pax6 homolog vab-3 for correct differentiation (Serrano-Saiz et al., 2013), and we observed that vab-3 also affects the terminal differentiation of the IL1 neurons (supplementary material Fig. S6A), suggesting that vab-3 and sox-2 might cooperate to correctly specify OLL and IL1 glutamatergic neurons. A regulatory element from the eat-4 locus that drives expression in OLL and IL1 neurons (Serrano-Saiz et al., 2013) indeed contains predicted SOX-2 and PAX-6 binding sites and deletion of either site abolishes expression driven by this regulatory element in OLL and IL1 (supplementary material Fig. S6B). These findings suggest that SOX-2 and VAB-3 cooperate to drive glutamatergic sensory neuron differentiation. Remarkably, a similar Pax6-Sox2 cooperation has been observed in vertebrate lens differentiation (Kondoh et al., 2004).
Fig. 6.
sox-2 affects the terminal differentiation of some glutamatergic and cholinergic neurons. (A) The expression of terminal differentiation markers of OLL and IL1 glutamatergic neurons is affected in sox-2 mutants. Reporter genes were crossed into sox-2(ot640) null mutants. The fraction of animals that show the indicated phenotype is indicated in the bar charts. Alternatively, standard whisker-and-box plots of total counts of rfp-expressing cells are presented. Animals were scored at the first larval stage; n>30. (B) sox-2 mutant animals show defects in cholinergic neuron differentiation. Reporter genes were crossed into sox-2(ot640) null mutants. The dotted boxes indicate the anterior ganglion where the cholinergic neurons IL2, URA and URB are located. Animals were scored at the first larval stage; n>20.
sox-2 is also necessary for the proper differentiation of several cholinergic neurons, as shown by reduced expression of the vesicular acetylcholine transporter unc-17 and the choline transporter cho-1 in sox-2 mutants (Fig. 6B). Affected cells are mainly located in the anterior ganglia (IL2, URA and URB). In the tail, cholinergic fate is affected in the PVP interneurons, although the expression of sox-2 is not maintained in these cells at adult stages (supplementary material Fig. S7A,B). By contrast, sox-2 is not required for the expression of cholinergic pathway genes (unc-17/VAChT and cho-1/ChT) in the DA9 neuron (supplementary material Fig. S7B), in which sox-2 is also transiently expressed (supplementary material Fig. S7A), but it is required for proper expression of the DA9-specific marker gene mig-13 (supplementary material Fig. S7C).
sox-3 (SoxB2) affects the terminal differentiation of a stretch receptor class
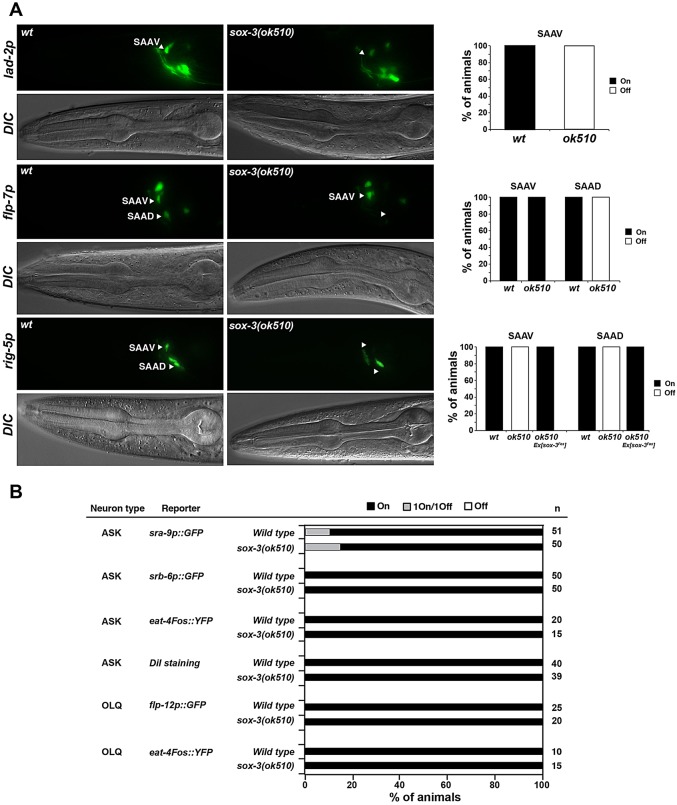
Using a panel of terminal identity markers for the neurons in which sox-3 is normally expressed, we examined potential differentiation defects in sox-3 null mutants (Fig. 7A,B). We observed defects in one specific neuron class, the SAA class, which are putative stretch/proprioceptive neurons. Expression of rig-5 and lad-2, which encode immunoglobulin superfamily adhesion molecules, is abolished in the dorsal and ventral SAA neurons, while expression of the neuropeptide flp-7 gene is lost in the dorsal SAA subtype but remains unaffected in the ventral SAA subtype (Fig. 7A).
Fig. 7.
sox-3 affects the terminal differentiation of SAA neurons. (A) The expression of certain terminal differentiation markers of SAA identity is affected in sox-3 mutants. Reporter genes were crossed into sox-3(ok510) null mutants. The fraction of animals that show the indicated phenotype is indicated in the bar charts. Animals were scored at the L4 or adult stage; n>30. (B) Expression of markers for other neuronal types is not affected in sox-3(ok510) mutants. Animals were scored at the L4 or adult stage.
Taken together, our observations demonstrate that, rather than being involved in early neurogenesis during embryonic development, C. elegans SoxB genes have roles in the terminal differentiation of specific neuron types.
Mutation of the C. elegans SoxC gene causes no broad neuronal differentiation defects
We next examined whether the function of the sole C. elegans SoxC gene, sem-2, mirrors the previously described function of vertebrate SoxC genes in neuronal differentiation throughout the nervous system (Bergsland et al., 2006; Mu et al., 2012). We used a null allele of sem-2, ok2422, provided by the C. elegans Knockout Consortium, which deletes almost the entire coding region (Fig. 1A). Most sem-2(ok2422) mutant animals die at embryonic or early larval stages showing different degrees of morphogenesis defects, as previously reported (Tian et al., 2011). This lethality can be rescued with the sem-2 fosmid-based reporter described above.
Using the pan-neuronal rab-3 marker, we examined whether sem-2 mutants display defects in neuronal differentiation. We did not observe changes in rab-3-expressing neuronal cell number in L1-arrested sem-2 null mutants (Fig. 2A), suggesting that sem-2 is not required during pan-neuronal identity specification. The onset of expression of the pan-neuronal marker rab-3 was also unaffected in sem-2 null mutants (Fig. 2B). Because homozygous sem-2 mutants were generated from heterozygous parents, we analyzed maternal/zygotic sem-2 null mutants derived from mosaic mothers that had lost the rescuing array in the germline-producing P lineage (see Materials and Methods). These maternal/zygotic null mutants still displayed the normal number of neurons as assessed by expression of the pan-neuronal markers rab-3, ric-4, ric-19 and rgef-1 (Fig. 2A; supplementary material Fig. S2). These findings are in striking contrast to the neuronal differentiation defects observed upon removal of vertebrate SoxC genes (Bergsland et al., 2006; Mu et al., 2012).
The lack of a broad effect of sem-2 mutants on embryonic neuronal differentiation is consistent with the very restricted expression of sem-2 in the terminally differentiating embryonic nervous system. We find that sem-2 is exclusively expressed in one head motor neuron class, RMH (Fig. 5C). sem-2 expression in these neurons is observed throughout larval and adult stages. The identity of these neurons is not obviously affected in sem-2 null mutants, as assessed by the intact expression of two terminal markers (supplementary material Fig. S3B).
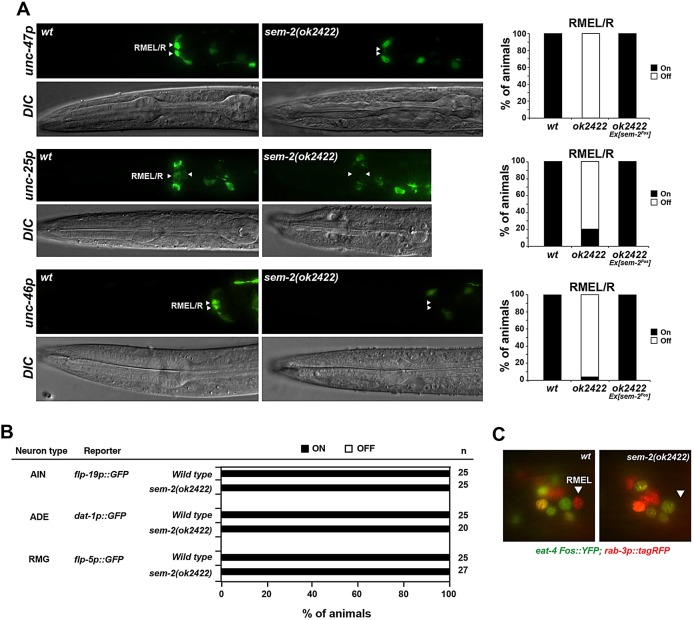
However, we found that the fate of the RME GABAergic motor neurons was affected in sem-2 mutants, as inferred from the loss of expression of the GABA biosynthetic enzyme unc-25, the vesicular GABA transporter unc-47 and the LAMP-like protein unc-46 genes (Fig. 8A). We did not detect any sem-2 expression in the RME neurons themselves, but sem-2 is instead expressed in the progenitor of RMEL/R (Fig. 1B). In sem-2 mutants, we could not detect expression of the pan-neuronal marker rab-3 in the region where the RMEL/R neurons are normally located (Fig. 8C), suggesting that these neurons are not generated in the absence of sem-2; however, it was difficult to conclude this unambiguously owing to the somewhat variable positioning of cells in the sem-2 mutant. This effect seems to be specific to this particular lineage, since the fate of other neurons derived from a sem-2-expressing progenitor is unaffected in sem-2 null mutants (Fig. 8B).
Fig. 8.
sem-2 affects the development of RME motor neurons. (A) The expression of terminal differentiation markers of RME identity is affected in sem-2 mutants. Reporter genes were crossed into sem-2(ok2422) null mutants. The fraction of animals that show the indicated phenotype is indicated in the bar charts. Animals were scored at larval stages; n>30. (B) The fate of other neurons that have a sem-2-expressing progenitor is unaffected in sem-2(ok2422) mutants. Animals were scored at larval stages. (C) RMEL/R neurons are not generated in sem-2 mutants. Wild-type and sem-2(ok2422) mutant animals were crossed with the pan-neuronal marker rab-3 (red) and the glutamatergic marker eat-4 (green). eat-4 expression was used as a landmark to assess rab-3 expression in RME neurons.
Taken together, the C. elegans SoxC gene, like the C. elegans SoxB genes, has a function that is markedly different from that of vertebrate homologs.
DISCUSSION
SoxB genes have been associated with broad neurogenic functions throughout the animal kingdom, from deuterostomes (chordates, echinoderms, hemichordates) (Burke et al., 2014; Reiprich and Wegner, 2014; Taguchi et al., 2002) and protostomes (Ecdysozoa, Lophotrochozoa) (Kerner et al., 2009; Overton et al., 2002) to even earlier diverging animal lineages such as cnidarians (Richards and Rentzsch, 2014). SoxB proteins are required for neuronal commitment broadly throughout the embryonic nervous system during the earliest stages of development, predominantly in proliferating, undifferentiated precursors (Pevny and Placzek, 2005; Pevny and Nicolis, 2010; Reiprich and Wegner, 2014; Sarkar and Hochedlinger, 2013). SoxB proteins are usually downregulated upon progression to postmitotic neural development. At this stage, SoxC proteins are induced, resulting in the promotion of neuronal differentiation (Bergsland et al., 2006). We show that, surprisingly, SoxB and SoxC genes are not broadly required for neurogenesis in the developing C. elegans embryo, but are very selectively employed to control specific neuronal differentiation events.
SoxB genes
In contrast to the role of SoxB genes in essentially all other systems examined, embryonic neurogenesis is largely unaffected in C. elegans SoxB mutants. However, we found that C. elegans sox-2 (SoxB1) determines the developmental potential of a subset of peripheral, postembryonic blast cells. These blast cells are differentiated, polarized epithelial cells at the first larval stage, but as larval development proceeds they exit this differentiated state to produce a distinct set of neuron types. In the absence of sox-2 these blast cells are generated but they lose the ability to generate distinct cell types, apparently owing to the loss of downstream-acting lineage regulators such as mab-9/Tbx20.
sox-2 is known to be involved in a natural transdifferentiation event in C. elegans, in which the epithelial cell Y converts to the motor neuron PDA in a process that does not involve cell division (Kagias et al., 2012). In this cellular context, sox-2 was found to cooperate with the POU homeobox gene ceh-6, the Sall-type transcription factor sem-4, and the HOX cluster gene egl-5 (Kagias et al., 2012). Remarkably, the K, F, B and U blast cells express the same combination of factors (sox-2, ceh-6, sem-4, egl-5) as the Y cell and, in all cases examined, differentiation defects in postembryonic lineages derived from these additional blast cells were observed in the respective mutant backgrounds (this study; Basson and Horvitz, 1996; Burglin and Ruvkun, 2001; Chamberlin et al., 1999; Chisholm, 1991). We therefore suggest the presence of a common regulatory cassette that operates in distinct cellular contexts to maintain the potency of blast cells (Fig. 4B). This might represent a primordial function of Sox2. Indeed, the Sox gene expression pattern in organisms belonging to early diverging animal lineages, such as ctenophores and cnidarians, is consistent with the hypothesis that an ancient function of Sox family genes was to regulate the developmental potential of stem cell-like cells (Jager et al., 2011; Schnitzler et al., 2014), an idea that is reinforced by our findings in C. elegans.
However, the well-established role of SoxB members as early neurogenic genes in vertebrates is not conserved in the nematode embryo. This could be explained by the notion that, during C. elegans nervous system development, presumptive neuroectodermal cells might not need to be maintained in a multipotent state through proliferative phases, but rather pass through predetermined and lineage-specified differentiation programs.
Notably, we have identified very specific roles of SoxB genes in neuron type-specific differentiation programs. We have shown that sox-2 (SoxB1) is required for the specification of several glutamatergic and cholinergic neurons, whereas sox-3 (SoxB2) is important for the correct specification of a single, putative sensory neuron class. Loss of the respective SoxB gene does not affect the generation of these neurons, but affects the adoption of their neuron type-specific identities. In vertebrates, there are few examples of SoxB genes being important for the differentiation of subsets of neurons. Mouse Sox1 has been shown to regulate certain aspects of the identity of telencephalic neurons of the ventral striatum (Ekonomou et al., 2005) and it also plays a role in the specification of V2c interneurons in the mouse ventral spinal cord (Panayi et al., 2010). Mouse Sox2 is important for the correct differentiation of cortical GABAergic neurons (Cavallaro et al., 2008) and retinal ganglion cells (Taranova et al., 2006), while mouse Sox3 has been suggested to have a role in the development of hypothalamic neurons (Rizzoti et al., 2004). There is also evidence that implicates one of the Drosophila SoxB genes, SoxNeuro, in the terminal differentiation of neurons (Ferrero et al., 2014; Girard et al., 2006). In these cases, it is not clear whether Sox2 acts only transiently during neuronal differentiation or is continuously required to maintain the differentiated state. In the cases we have described here, C. elegans sox-2 is continuously expressed throughout the life of the specific neuron types, suggesting that sox-2 might act, in combination with other factors, as a terminal selector to initiate and maintain the terminally differentiated state (Hobert, 2011).
Sox proteins are known to engage in cooperative interactions with different partner factors, which determine target gene specificity and provide a cell specification code (Kondoh and Kamachi, 2010). A well-established example of such a partnership is the cooperative role of Sox2 and Pax6 in vertebrate lens specification (Kondoh et al., 2004). Our data indicate that sox-2 and vab-3, the C. elegans Pax6 homolog, cooperate to specify the OLL and IL1 sensory neurons in C. elegans. Thus, Sox2-Pax6 might define an ancestral cooperative partnership that could be employed in distinct differentiation program events in different organisms. Moreover, we have shown that sox-2 is important for the proper specification of several cholinergic neurons. The POU homeobox transcription factor unc-86 has been shown to control the terminal differentiation of several of these cholinergic neurons (IL2, URA, URB) (Zhang et al., 2014) and it is possible that sox-2 and unc-86 cooperate in the differentiation of these neuron types in the same manner as Sox2-POU cooperate to specify stem cell identity in vertebrates (Takahashi and Yamanaka, 2006).
SoxC genes
In the vertebrate central nervous system, SoxC proteins are mostly expressed in postmitotic differentiating neurons, where they play a role in induction of pan-neuronal gene expression and differentiation (Bergsland et al., 2006; Reiprich and Wegner, 2014). By contrast, sem-2 (SoxC) gene expression can be detected very early during C. elegans embryogenesis, several cell divisions before neurons are born and start to differentiate and also considerably earlier than SoxB gene induction. During C. elegans embryonic neuronal differentiation sem-2 expression becomes sparse and, accordingly, sem-2 seems to be dispensable for correct neuronal specification throughout the nervous system. We found that sem-2 is only necessary for the specification of the GABAergic RMEL/R neuron pair. Our results suggest that, in the absence of sem-2, which is expressed in the progenitor but not in RMEL/R themselves, these neurons are not generated. These findings are in contrast to the described function of SoxC genes in vertebrate postmitotic neurons.
In Drosophila, complete removal of the single SoxC gene, Sox14, does not appear to broadly affect embryonic neuronal differentiation either, but rather has selective effects during metamorphosis downstream of the hormone ecdysone (Kirilly et al., 2009; Ritter and Beckstead, 2010). It is therefore conceivable that the broad neuronal differentiation role of vertebrate SoxC genes downstream of proneural bHLH genes might be a vertebrate-specific invention.
In conclusion, our analysis of SoxB and SoxC gene function in the nematode has revealed remarkably conserved and divergent themes in the function of this ancient class of transcription factors, revealing commonalities and differences in the mechanisms of neuronal development in vertebrates and invertebrates.
MATERIALS AND METHODS
Strains and transgenes
A list of mutants and transgenes is provided in supplementary material Table S1.
Generation of fosmid-based reporter genes
Fosmid reporter constructs for all Sox genes were created using λ-Red-mediated recombineering in bacteria as previously described (Tursun et al., 2009). The recombined fosmids were WRM0626aE02 (sox-2), WRM0615dD12 (sox-3), WRM0623cE02 (sem-2), WRM0615cE08 (egl-13) and WRM0633aH06 (sox-4). We inserted mCherry or YFP immediately preceding the STOP codon of each Sox gene, which resulted in a translational fusion. Recombineered fosmids were linearized with SdaI and injected at 15 ng/μl into N2 worms, together with ScaI-digested rol-6(d) (pRF4; 5 ng/µl) and PvuII-digested bacterial genomic DNA (100 ng/μl) to generate a complex array. Integrated transgenes were generated by γ-irradiation and outcrossed four to six times.
Generation of the sox-2(ot640) deletion allele
The sox-2 null allele ot640 was generated by transposon excision (MosDEL) as previously described (Frøkjaer-Jensen et al., 2010), using ttTi44122, a Mos1 insertion 2002 bp downstream of the sox-2 STOP codon kindly provided by the NemaGENETAG Consortium. The resulting sox-2(ot640) allele has a 3496 bp deletion, including the whole sox-2 coding region, as verified by PCR analysis and sequencing.
Mosaic analysis
In order to assess potential sox-2 or sem-2 maternal contribution in neuronal development, mosaic analysis was performed and maternal/zygotic null mutants derived from mothers that had lost the rescuing array in the germline-producing P lineage were analyzed. sox-2(ot640) or sem-2(ok2422) null mutants were rescued with an extrachromosomal array containing the sox-2 fosmid WRM0626aE02 (15 ng/µl) or sem-2 fosmid WRM0623cE02 (15 ng/µl) together with myo-3p::mCherry (3 ng/µl), which is expressed in body wall muscles. These strains were crossed with the integrated transgenes otIs291 (rab-3p::2xnlsYFP), otIs381 (ric-19p::2xnlsGFP), otIs353 (ric-4Fos::SL2::YFP::H2B) and evIs111 (rgef-1::GFP), which label all neurons. In C. elegans, 94 body wall muscles derive from the P lineage while only one arises from the AB lineage. Mosaic mothers that had lost the rescuing array in the P lineage were identified based on the loss of myo-3p::mCherry expression in all body wall muscles but one. The number of rab-3-, ric-19- and ric-4-expressing neurons was counted in the progeny of the mosaic mothers.
Similarly, in order to overcome the L1 lethality and be able to analyze postembryonic lineages in sox-2(ot640) animals, this allele was rescued with an extrachromosomal array containing the sox-2 fosmid WRM0626aE02 (15 ng/µl) and different lineage-specific markers (3 ng/µl) so as to be able to assess whether the rescuing array had been lost in a particular lineage of interest. Cell fate in the absence of the sox-2 rescuing array was analyzed with integrated reporters.
Lineage analysis
Cells expressing sox-2 or sem-2 during embryogenesis were identified by a combination of manual and semi-automated lineaging. For the manual approach, the embryonic development of the sox-2fosmid::mCherry (otIs333) or sem-2fosmid::YFP (otIs313) reporter strains was imaged using the 4D microscopy software Steuerprg (Caenotec). Briefly, gravid adults were dissected and single two-cell embryos were mounted and visualized on a Zeiss Imager Z1 compound microscope. Nomarski stacks were taken every 35 s and embryos were illuminated with LED fluorescence light (470 or 555 nm) at predetermined time points during development. Movies were lineaged using the SIMI BioCell program as previously described (Schnabel et al., 1997). For the semi-automated approach, imaging was performed with a spinning disc confocal microscope (Zeiss Axio Observer.Z1) or iSPIM light sheet scope (Wu et al., 2011). Automated lineage tracing to follow cells labeled with a fluorescent histone marker and assign Sulston names was performed and manually validated as previously described (Bao et al., 2006; Santella et al., 2010). Nuclear-localized expression was quantified for each cell over its lifetime as previously described (Murray et al., 2008).
Supplementary Material
Acknowledgements
We thank Qi Chen for expert assistance in generating transgenic strains; members of the O.H. laboratory for comments on the manuscript; Nikos Stefanakis, Inés Carrera, Marie Gendrel and Laura Pereira for sharing reporter strains; the C. elegans Knockout Consortium for generating sem-2 and sox-3 knockout alleles and the Caenorhabditis Genetics Center (University of Minnesota) and the NemaGENETAG Consortium for providing strains.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was funded by the National Institutes of Health [R01NS039996-05 and R01NS050266-03 to O.H.; R01GM098026 to C.-F.C.]. O.H. is an Investigator of the Howard Hughes Medical Institute. B.V. was supported by a Beatriu de Pinós post-doctoral fellowship (BP-DGR). Deposited in PMC for release after 6 months.
Author contributions
B.V. and O.H. designed the study and jointly wrote the paper. Further inputs into the design of the experiments were provided by C.-F.C. B.V. performed all experiments, except the following: eat-4 analysis in vab-3 mutants and mutation/analysis of the PAX-6 binding site was performed by E.S.-S.; the StarryNite-based lineaging was performed by A.S. under supervision of Z.B.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125740/-/DC1
References
- Bao Z., Murray J. I., Boyle T., Ooi S. L., Sandel M. J. and Waterston R. H. (2006). Automated cell lineage tracing in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 103, 2707-2712. 10.1073/pnas.0511111103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. and Horvitz H. R. (1996). The Caenorhabditis elegans gene sem-4 controls neuronal and mesodermal cell development and encodes a zinc finger protein. Genes Dev. 10, 1953-1965. 10.1101/gad.10.15.1953 [DOI] [PubMed] [Google Scholar]
- Bergsland M., Werme M., Malewicz M., Perlmann T. and Muhr J. (2006). The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 20, 3475-3486. 10.1101/gad.403406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N., Castro D. S. and Guillemot F. (2002). Proneural genes and the specification of neural cell types. Nat. Rev. Neurosci. 3, 517-530. 10.1038/nrn874 [DOI] [PubMed] [Google Scholar]
- Bowles J., Schepers G. and Koopman P. (2000). Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 227, 239-255. 10.1006/dbio.2000.9883 [DOI] [PubMed] [Google Scholar]
- Buescher M., Hing F. S. and Chia W. (2002). Formation of neuroblasts in the embryonic central nervous system of Drosophila melanogaster is controlled by SoxNeuro. Development 129, 4193-4203. [DOI] [PubMed] [Google Scholar]
- Burglin T. R. and Ruvkun G. (2001). Regulation of ectodermal and excretory function by the C. elegans POU homeobox gene ceh-6. Development 128, 779-790. [DOI] [PubMed] [Google Scholar]
- Burke R. D., Moller D. J., Krupke O. A. and Taylor V. J. (2014). Sea urchin neural development and the metazoan paradigm of neurogenesis. Genesis 52, 208-221. 10.1002/dvg.22750 [DOI] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B. G. and Muhr J. (2003). Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat. Neurosci. 6, 1162-1168. 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- Cavallaro M., Mariani J., Lancini C., Latorre E., Caccia R., Gullo F., Valotta M., DeBiasi S., Spinardi L., Ronchi A. et al. (2008). Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development 135, 541-557. 10.1242/dev.010801 [DOI] [PubMed] [Google Scholar]
- Chamberlin H. M., Brown K. B., Sternberg P. W. and Thomas J. H. (1999). Characterization of seven genes affecting Caenorhabditis elegans hindgut development. Genetics 153, 731-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A. (1991). Control of cell fate in the tail region of C. elegans by the gene egl-5. Development 111, 921-932. [DOI] [PubMed] [Google Scholar]
- Chisholm A. D. and Hodgkin J. (1989). The mab-9 gene controls the fate of B, the major male-specific blast cell in the tail region of Caenorhabditis elegans. Genes Dev. 3, 1413-1423. 10.1101/gad.3.9.1413 [DOI] [PubMed] [Google Scholar]
- Ekonomou A., Kazanis I., Malas S., Wood H., Alifragis P., Denaxa M., Karagogeos D., Constanti A., Lovell-Badge R. and Episkopou V. (2005). Neuronal migration and ventral subtype identity in the telencephalon depend on SOX1. PLoS Biol. 3, e186 10.1371/journal.pbio.0030186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero E., Fischer B. and Russell S. (2014). SoxNeuro orchestrates central nervous system specification and differentiation in Drosophila and is only partially redundant with Dichaete. Genome Biol. 15, R74 10.1186/gb-2014-15-5-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C., Davis M. W., Hollopeter G., Taylor J., Harris T. W., Nix P., Lofgren R., Prestgard-Duke M., Bastiani M., Moerman D. G. et al. (2010). Targeted gene deletions in C. elegans using transposon excision. Nat. Methods 7, 451-453. 10.1038/nmeth.1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliot B., Quiquand M., Ghila L., de Rosa R., Miljkovic-Licina M. and Chera S. (2009). Origins of neurogenesis, a cnidarian view. Dev. Biol. 332, 2-24. 10.1016/j.ydbio.2009.05.563 [DOI] [PubMed] [Google Scholar]
- Girard F., Joly W., Savare J., Bonneaud N., Ferraz C. and Maschat F. (2006). Chromatin immunoprecipitation reveals a novel role for the Drosophila SoxNeuro transcription factor in axonal patterning. Dev. Biol. 299, 530-542. 10.1016/j.ydbio.2006.08.014 [DOI] [PubMed] [Google Scholar]
- Guth S. I. E. and Wegner M. (2008). Having it both ways: Sox protein function between conservation and innovation. Cell. Mol. Life Sci. 65, 3000-3018. 10.1007/s00018-008-8138-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. (2011). Regulation of terminal differentiation programs in the nervous system. Annu. Rev. Cell Dev. Biol. 27, 681-696. 10.1146/annurev-cellbio-092910-154226 [DOI] [PubMed] [Google Scholar]
- Huang X., Tian E., Xu Y. and Zhang H. (2009). The C. elegans engrailed homolog ceh-16 regulates the self-renewal expansion division of stem cell-like seam cells. Dev. Biol. 333, 337-347. 10.1016/j.ydbio.2009.07.005 [DOI] [PubMed] [Google Scholar]
- Jager M., Quéinnec E., Le Guyader H. and Manuel M. (2011). Multiple Sox genes are expressed in stem cells or in differentiating neuro-sensory cells in the hydrozoan Clytia hemisphaerica. EvoDevo 2, 12 10.1186/2041-9139-2-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S., Schwab Y. and Greenwald I. (2008). A Caenorhabditis elegans model for epithelial-neuronal transdifferentiation. Proc. Natl. Acad. Sci. USA 105, 3790-3795. 10.1073/pnas.0712159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Fitzsimmons D., Hagman J. and Chamberlin H. M. (2001). EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 128, 2857-2865. [DOI] [PubMed] [Google Scholar]
- Kagias K., Ahier A., Fischer N. and Jarriault S. (2012). Members of the NODE (Nanog and Oct4-associated deacetylase) complex and SOX-2 promote the initiation of a natural cellular reprogramming event in vivo. Proc. Natl. Acad. Sci. USA 109, 6596-6601. 10.1073/pnas.1117031109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner P., Simionato E., Le Gouar M. and Vervoort M. (2009). Orthologs of key vertebrate neural genes are expressed during neurogenesis in the annelid Platynereis dumerilii. Evol. Dev. 11, 513-524. 10.1111/j.1525-142X.2009.00359.x [DOI] [PubMed] [Google Scholar]
- Kirilly D., Gu Y., Huang Y., Wu Z., Bashirullah A., Low B. C., Kolodkin A. L., Wang H. and Yu F. (2009). A genetic pathway composed of Sox14 and Mical governs severing of dendrites during pruning. Nat. Neurosci. 12, 1497-1505. 10.1038/nn.2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H. and Kamachi Y. (2010). SOX–partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 42, 391-399. 10.1016/j.biocel.2009.09.003 [DOI] [PubMed] [Google Scholar]
- Kondoh H., Uchikawa M. and Kamachi Y. (2004). Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation. Int. J. Dev. Biol. 48, 819-827. 10.1387/ijdb.041868hk [DOI] [PubMed] [Google Scholar]
- Köppen M., Simske J. S., Sims P. A., Firestein B. L., Hall D. H., Radice A. D., Rongo C. and Hardin J. D. (2001). Cooperative regulation of AJM-1 controls junctional integrity in Caenorhabditis elegans epithelia. Nat. Cell Biol. 3, 983-991. 10.1038/ncb1101-983 [DOI] [PubMed] [Google Scholar]
- Korzelius J., The I., Ruijtenberg S., Portegijs V., Xu H., Horvitz H. R. and van den Heuvel S. (2011). C. elegans MCM-4 is a general DNA replication and checkpoint component with an epidermis-specific requirement for growth and viability. Dev. Biol. 350, 358-369. 10.1016/j.ydbio.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kratsios P., Stolfi A., Levine M. and Hobert O. (2012). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15, 205-214. 10.1038/nn.2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mango S. E. (2007). The C. elegans pharynx: a model for organogenesis. WormBook, 1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L., Berti L., Masserdotti G., Covic M., Michaelidis T. M., Doberauer K., Merz K., Rehfeld F., Haslinger A., Wegner M. et al. (2012). SoxC transcription factors are required for neuronal differentiation in adult hippocampal neurogenesis. J. Neurosci. 32, 3067-3080. 10.1523/JNEUROSCI.4679-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. I., Bao Z., Boyle T. J., Boeck M. E., Mericle B. L., Nicholas T. J., Zhao Z., Sandel M. J. and Waterston R. H. (2008). Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat. Methods 5, 703-709. 10.1038/nmeth.1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y., Ogura E., Kondoh H. and Kamachi Y. (2010). B1 SOX coordinate cell specification with patterning and morphogenesis in the early zebrafish embryo. PLoS Genet. 6, e1000936 10.1371/journal.pgen.1000936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton P. M., Meadows L. A., Urban J. and Russell S. (2002). Evidence for differential and redundant function of the Sox genes Dichaete and SoxN during CNS development in Drosophila. Development 129, 4219-4228. [DOI] [PubMed] [Google Scholar]
- Panayi H., Panayiotou E., Orford M., Genethliou N., Mean R., Lapathitis G., Li S., Xiang M., Kessaris N., Richardson W. D. et al. (2010). Sox1 is required for the specification of a novel p2-derived interneuron subtype in the mouse ventral spinal cord. J. Neurosci. 30, 12274-12280. 10.1523/JNEUROSCI.2402-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L. H. and Nicolis S. K. (2010). Sox2 roles in neural stem cells. Int. J. Biochem. Cell Biol. 42, 421-424. 10.1016/j.biocel.2009.08.018 [DOI] [PubMed] [Google Scholar]
- Pevny L. and Placzek M. (2005). SOX genes and neural progenitor identity. Curr. Opin. Neurobiol. 15, 7-13. 10.1016/j.conb.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Phochanukul N. and Russell S. (2010). No backbone but lots of Sox: invertebrate Sox genes. Int. J. Biochem. Cell Biol. 42, 453-464. 10.1016/j.biocel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Reiprich S. and Wegner M. (2014). From CNS stem cells to neurons and glia: sox for everyone. Cell Tissue Res. 359, 111-124. 10.1007/s00441-014-1909-6 [DOI] [PubMed] [Google Scholar]
- Richards G. S. and Rentzsch F. (2014). Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 141, 4681-4689. 10.1242/dev.112029 [DOI] [PubMed] [Google Scholar]
- Ritter A. R. and Beckstead R. B. (2010). Sox14 is required for transcriptional and developmental responses to 20-hydroxyecdysone at the onset of drosophila metamorphosis. Dev. Dyn. 239, 2685-2694. 10.1002/dvdy.22407 [DOI] [PubMed] [Google Scholar]
- Rizzoti K., Brunelli S., Carmignac D., Thomas P. Q., Robinson I. C. and Lovell-Badge R. (2004). SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat. Genet. 36, 247-255. 10.1038/ng1309 [DOI] [PubMed] [Google Scholar]
- Sandberg M., Källström M. and Muhr J. (2005). Sox21 promotes the progression of vertebrate neurogenesis. Nat. Neurosci. 8, 995-1001. 10.1038/nn1493 [DOI] [PubMed] [Google Scholar]
- Santella A., Du Z., Nowotschin S., Hadjantonakis A.-K. and Bao Z. (2010). A hybrid blob-slice model for accurate and efficient detection of fluorescence labeled nuclei in 3D. BMC Bioinformatics 11, 580 10.1186/1471-2105-11-580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A. and Hochedlinger K. (2013). The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell 12, 15-30. 10.1016/j.stem.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel R., Hutter H., Moerman D. and Schnabel H. (1997). Assessing normal embryogenesis in Caenorhabditis elegans using a 4D microscope: variability of development and regional specification. Dev. Biol. 184, 234-265. 10.1006/dbio.1997.8509 [DOI] [PubMed] [Google Scholar]
- Schnitzler C. E., Simmons D. K., Pang K., Martindale M. Q. and Baxevanis A. D. (2014). Expression of multiple Sox genes through embryonic development in the ctenophore Mnemiopsis leidyi is spatially restricted to zones of cell proliferation. EvoDevo 5, 15 10.1186/2041-9139-5-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E., Poole R. J., Felton T., Zhang F., De La Cruz E. D. and Hobert O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659-673. 10.1016/j.cell.2013.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C., Iguchi A., Hayward D. C., Technau U., Ball E. E. and Miller D. J. (2008). Sox genes in the coral Acropora millepora: divergent expression patterns reflect differences in developmental mechanisms within the Anthozoa. BMC Evol. Biol. 8, 311 10.1186/1471-2148-8-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E. and Horvitz H. R. (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110-156. 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Taguchi S., Tagawa K., Humphreys T. and Satoh N. (2002). Group B sox genes that contribute to specification of the vertebrate brain are expressed in the apical organ and ciliary bands of hemichordate larvae. Zoolog. Sci. 19, 57-66. 10.2108/zsj.19.57 [DOI] [PubMed] [Google Scholar]
- Takahashi K. and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663-676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Taranova O. V., Magness S. T., Fagan B. M., Wu Y., Surzenko N., Hutton S. R. and Pevny L. H. (2006). SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 20, 1187-1202. 10.1101/gad.1407906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Shi H., Colledge C., Stern M., Waterston R. and Liu J. (2011). The C. elegans SoxC protein SEM-2 opposes differentiation factors to promote a proliferative blast cell fate in the postembryonic mesoderm. Development 138, 1033-1043. 10.1242/dev.062240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursun B., Cochella L., Carrera I. and Hobert O. (2009). A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4, e4625 10.1371/journal.pone.0004625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B. B., Müller-Immergluck M. M., Austin J., Robinson N. T., Chisholm A. and Kenyon C. (1993). A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell 74, 29-42. 10.1016/0092-8674(93)90292-X [DOI] [PubMed] [Google Scholar]
- Woollard A. and Hodgkin J. (2000). The caenorhabditis elegans fate-determining gene mab-9 encodes a T-box protein required to pattern the posterior hindgut. Genes Dev. 14, 596-603. [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Ghitani A., Christensen R., Santella A., Du Z., Rondeau G., Bao Z., Colon-Ramos D. and Shroff H. (2011). Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 108, 17708-17713. 10.1073/pnas.1108494108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Bhattacharya A., Nelson J. C., Abe N., Gordon P., Lloret-Fernandez C., Maicas M., Flames N., Mann R. S., Colon-Ramos D. A. et al. (2014). The LIM and POU homeobox genes ttx-3 and unc-86 act as terminal selectors in distinct cholinergic and serotonergic neuron types. Development 141, 422-435. 10.1242/dev.099721 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.