Abstract
Mammalian sex chromosomes arose from an ordinary pair of autosomes. Over hundreds of millions of years, they have evolved into highly divergent X and Y chromosomes and have become increasingly specialized for male reproduction. Both sex chromosomes have acquired and amplified testis-specific genes, suggestive of roles in spermatogenesis. To understand how the sex chromosome genes participate in the regulation of spermatogenesis, we review genes, including single-copy, multi-copy, and ampliconic genes, whose spermatogenic functions have been demonstrated in mouse genetic studies. Sex chromosomes are subject to chromosome-wide transcriptional silencing in meiotic and postmeiotic stages of spermatogenesis. We also discuss particular sex-linked genes that escape postmeiotic silencing and their evolutionary implications. The unique gene contents and genomic structures of the sex chromosomes reflect their strategies to express genes at various stages of spermatogenesis and reveal the driving forces that shape their evolution.
Introduction
Mammalian sex chromosomes arose from an ordinary pair of autosomes about 200–300 million years ago (Bellott, et al. 2014, Lahn and Page 1999, Ohno 1967, Ross, et al. 2005). Since then, the X has preserved most of the ancestral autosomal genes, while the Y has lost most of them and only kept a selective group of critical genes (Bellott, et al. 2014, Hughes, et al. 2012, Hughes, et al. 2010, Ross, et al. 2005, Skaletsky, et al. 2003, Soh, et al. 2014). Remarkably, both chromosomes have acquired a substantial amount of ampliconic sequences, from which amplified genes are expressed predominantly in the testicular germ cells (Bellott, et al. 2010, Mueller, et al. 2013, Soh, et al. 2014). Several ancestral genes on the human and mouse Y chromosomes are also found to undergo amplification and change their broad expression pattern to a testis-specific one (Bellott, et al. 2014, Skaletsky, et al. 2003, Soh, et al. 2014). These findings suggest an evolutionary trend of increasing specialization for male reproduction for the sex chromosomes.
Male reproduction relies on functional spermatogenesis, which consists of cells of four major differentiation stages: spermatogonia (mitotic), spermatocyte (meiotic), spermatid (postmeiotic; spermiogenic), and spermatozoa (sperm) (Figure 1). Spermatogenesis genes are abundant in sex chromosomes. Based on the genomic structures, these genes can be divided into two major groups: single-copy genes and ampliconic/multi-copy genes. The majority of single-copy genes involved in spermatogenesis are ancestral genes and tend to be broadly expressed in the body (examples in Table 1). There are also a small group of single-copy genes that are specifically expressed in the spermatogenic cells, some of which have been shown to exert critical functions in spermatogenesis (Wang, et al. 2001, Zheng, et al. 2010). In contrast, most of the ampliconic/multi-copy genes were acquired following the divergence of the X and Y chromosomes and are expressed predominantly in the male germline. Interestingly, a few newly acquired genes on the sex chromosomes that have not been amplified, for instance, Prssly and Teyorf1 on the mouse Y chromosome (Soh, et al. 2014), also show a germline-specific expression pattern.
Figure 1. Schematic of spermatogenesis and events on the sex chromosomes.
Due to sex chromosome inactivation, sex chromosomes function in two critical phases before meiosis in mitotically proliferating spermatogonia and after meiosis in haploid spermatids. Blue arrows: two stages of sex chromosome inactivation, Red arrows: Groups of genes expressed from the sex chromosomes.
Table 1.
Sex-chromosome genes shown to contribute to male fertility in mouse genetic studies
| Gene name | Biological function | Expression | Human homolog on the X; associated diseases | Fertility phenotype of genetically altered XY mice | References |
|---|---|---|---|---|---|
| X-linked genes | |||||
| Abcd1 | Peroxisomal transporter of very long-chain fatty acids | Multiple tissues | Yes; Adrenoleukodystrophy | Reduced fertility; increased number of interstitial cells with accumulated lipid inclusions | (Forss-Petter, et al. 1997) |
| Akap4 | A-kinase anchoring protein | Spermatids, spermatozoa | Yes | Infertility; abnormal sperm flagellum morphology and motility | (Miki, et al. 2002a) |
| Ar | Androgen receptor | Multiple tissues | Yes; Androgen Insensitivity syndrome | Infertility; testicular feminization; impaired spermatogenesis; tissue-specific KOs indicate that AR signaling in Sertoli, Leydig, and peritubular myoid cells is crucial for spermatogenesis | (Chang, et al. 2004, De Gendt, et al. 2004, Holdcraft and Braun 2004, Lyon and Hawkes 1970, Xu, et al. 2007, Yeh, et al. 2002, Zhang, et al. 2006) |
| Arx | Homeodomain transcription factor | Multiple tissues | Yes; Early-onset epileptic encephalopathy 1 and Partington syndrome | Small testis; fetal Leydig cell differentiation defect | (Kitamura, et al. 2002, Miyabayashi, et al. 2013) |
| Atp7a | Copper transpoting ATPase | Multiple tissues | Yes; Menkes disease | Infertility; spermatogenic cell death; poor sperm morphology and motility | (Kotula-Balak, et al. 2007, Kowal, et al. 2010, Llanos, et al. 2006) |
| Cdk16 | Cyclin-dependent kinase | Multiple tissues | Yes | Infertility; abnormal sperm morphology and motility | (Mikolcevic, et al. 2012) |
| Dmd | Dystrophin | Multiple tissues | Yes; Duchenne and Becker muscular dystrophy | Reduced fertility; abnormal sperm flagella morphology and motility | (Hernandez-Gonzalez, et al. 2005, Kudoh, et al. 2005) |
| Fmr1 | Fragile X mental retardation protein | Multiple tissues | Yes; Fragile X syndrome | Macroorchidism-associated increase in Sertoli cell proliferation; defective synapsis and crossover formation in spermatocytes | (Alpatov, et al. 2014, Slegtenhorst-Eegdeman, et al. 1998) |
| Foxp3 | Forkhead transcription factor | Multiple | Yes; Immunodysregulation, polyendocrinopathy, enteropathy | Infertility; hypogonadism and arrested spermatogenesis caused by insufficient pituitary gonadotropins | (Jasurda, et al. 2014) |
| Gpr64 | G protein-coupled receptor | Proximal epididymis, efferent ductules | Yes | Age-dependent infertility; accumulation of spermatozoa in the efferent ductules caused by defective fluid reabsorption | (Davies, et al. 2004) |
| Gria3 | Ionotropic glutamate receptor | Multiple tissues | Yes; Mental retardation X-linked 94 | Reduced fertility with unknown causes | (Meng, et al. 2003) |
| L1cam | Neural cell adhesion molecule | Multiple tissues | Yes; Hydrocephalus, CRASH and MASA syndromes | Infertility at high frequency with unknown causes | (Cohen, et al. 1998) |
| Mecp2 | Methyl CpG binding protein | Multiple tissues | Yes; Rett Syndrome | Infertility; cryptorchidism | (Guy, et al. 2001) |
| Nr0b1 | Nuclear receptor | Multiple tissues | Yes; 46XY sex reversal; Adrenal hypoplasia | Infertility; impaired spermatogenesis; multiple defects in Sertoli, Leydig, and peritubular myoid cells | (Jeffs, et al. 2001, Meeks, et al. 2003, Yu, et al. 1998) |
| Nxf2 | Nuclear RNA export factor | Spermatogonia | Yes | Reduced fertility; age-dependent depletion of spermatogonial cells; meiotic arrest; reduced sperm count and motility | (Pan, et al. 2009) |
| Pcyt1b | CTP: phosphocholine cytidylyltransferase | Multiple tissues | Yes | Reduced fertility; age-dependent depletion of spermatogonial cells | (Jackowski, et al. 2004) |
| Porcn | Membrane-bound O- acyl transferase | Multiple tissues | Yes; Focal dermal hypoplasia | Small testis; abnormal vas deferens morphology | (Liu, et al. 2012) |
| Prdx4 | Peroxiredoxin | Multiple tissues | Yes | Reduced testis weight; increased spermatogenic cell death by oxidative damage; reduced sperm counts | (Iuchi, et al. 2009) |
| Rhox5 | Reproductive homeodomain transcription factor | Multiple tissues | Yes* | Reduced fertility caused by defective Sertoli cells; increased spermatocyte death; reduced sperm counts and motility | (Maclean, et al. 2005) |
| Scml2 | Epigenetic regulator, Polycomb protein | Spermatogenic cells | Yes; Medulloblastomas | Infertility; suppression of somatic/progenitor genes during spermatogenesis; suppression of histone H2A ubiquitination on the sex chromosomes | (Hasegawa, et al. 2015, Luo, et al. 2015) |
| Slx/Slxl-1 | Sycp3-like X-linked | Spermatogenic cells | No | Reduced fertility in single knockdowns; infertility in double knockdowns; impaired spermiogenesis leading to cell death; abnormal sperm morphology; reduced sperm counts and motility** | (Cocquet, et al. 2012, Cocquet, et al. 2010) |
| Sms | Spermine synthase | Multiple tissues | Yes; Snyder-Robinson syndrome | Infertility with unknown causes | (White, et al. 2013) |
| Sox3 | High-mobility group box transcription factor | Multiple tissues | Yes; Mental retardation and panhypopituitarism | Reduced testis weight; spermatogonial differentiation defects; reduced sperm counts | (Laronda and Jameson 2011, Rizzoti, et al. 2004, Weiss, et al. 2003) |
| Taf7l | TATA box binding protein (TBP)- associated factor | Spermatogenic cells | Yes | Reduced fertility; reduced sperm counts; abnormal sperm morphology and motility | (Cheng, et al. 2007) |
| Tex11 | Meiotic protein | Spermatogenic cells | Yes | Depletion of spermatocytes caused by meiotic defects | (Adelman and Petrini 2008, Yang, et al. 2008) |
| Tsc22d3 | Glucocorticoid- induced leucine zipper protein | Spermatogenic cells | Yes | Infertility; defects in proliferation and differentiation of undifferentiated spermatogonia; massive cell death during meiotic prophase | (Bruscoli, et al. 2012) |
| Tsx | Non-coding RNA | Multiple tissues | No | Grossly undisturbed fertility, albeit smaller testis and partial germ cell death during meiotic prophase | (Anguera, et al. 2011) |
| Zfx | Zinc finger protein | Multiple tissues | Yes | Grossly undisturbed fertility, albeit smaller testis and reduced sperm counts | (Luoh, et al. 1997) |
| Y-linked genes | |||||
| Sry | High-mobility group box transcription factor | Fetal gonad and brain | Yes; 46XY sex reversal | Infertility; male-to-female sex reversal | (Koopman, et al. 1991b, Lovell-Badge and Robertson 1990) |
| Eif2s3y | Translation initiation factor | Multiple tissues | No | Essential for spermatogonial proliferation** | (Mazeyrat, et al. 2001) |
| Sly | Sycp3-like Y-linked | Spermatogenic cells | No | Reduced fertility; impaired spermiogenesis; sperm DNA damage; abnormal sperm chromatin packaging and morphology | (Cocquet, et al. 2012, Cocquet, et al. 2009, Riel, et al. 2013) |
| Zfy1 | Zinc finger transcription factor | Spermatogenic cells | Yes | Functions in promoting 2nd meiotic divisions in spermatocytes*** | (Vernet, et al. 2011, Vernet, et al. 2014) |
| Zfy2 | Zinc finger transcription factor | Spermatogenic cells | Yes | Functions in meiotic checkpoints and divisions in spermatocytes*** | (Vernet, et al. 2011, Vernet, et al. 2014) |
Gene belongs to a Rhox homeobox gene cluster. Due to a rapid evolution of this gene cluster, a human homolog cannot be precisely defined.
Genes belong to multi-copy gene families. The phenotypes were demonstrated in male transgenic mice carrying specific siRNA transgenes.
Gene function was demonstrated by transgene rescue experiments in male mice lacking various portions of the Y chromosome.
Spermatogenic cells undergo meiosis to generate haploid gametes (Figure. 1). Unlike autosomes that undergo synapsis along their entire lengths during meiosis, large regions of the X and Y chromosomes remain unsynapsed and trigger transcriptional silencing, called meiotic sex chromosome inactivation (MSCI: reviewed elsewhere (Ichijima, et al. 2012, Turner 2007, van der Heijden, et al. 2011)). MSCI is accompanied by the formation of distinct heterochromatin called the XY body (also known as the sex body). This chromosome-wide silencing is maintained into round spermatids by postmeiotic sex chromatin (PMSC) (Greaves, et al. 2006, Namekawa, et al. 2006, Turner, et al. 2006). Importantly, there is a group of sex-linked genes that escape postmeiotic silencing and become expressed in postmeiotic spermatids (reviewed elsewhere (Sin and Namekawa 2013)). The regulatory mechanisms by which sex-linked genes are inactivated by MSCI and activated to escape postmeiotic silencing were identified as DNA damage response pathways adopted from somatic machinery to recognize damaged DNA (Broering, et al. 2014, Ichijima, et al. 2011, Sin, et al. 2012a, Turner, et al. 2004). The genes that escape postmeiotic silencing, termed escape genes, include most of the ampliconic/multi-copy genes, as well as many single-copy genes, on both X and Y chromosomes (Cocquet, et al. 2009, Mueller, et al. 2008, Sin, et al. 2012b, Toure, et al. 2004a). Therefore, the unique gene contents and genomic structures of the sex chromosomes reflect their strategies to express genes during critical stages of spermatogenesis. In this review article, we summarize the genomic structure of the sex chromosomes and the functions of single-copy and ampliconic/multi-copy genes in the regulation of spermatogenesis, and try to shed light on the evolution of their unique genomic properties.
Genomic structure of the sex chromosomes
The differentiation of the sex chromosomes from ancestral autosomes was initiated when one of the autosome pairs acquired a sex-determining gene and underwent chromosomal inversion that prevented recombination in the region (Bellott, et al. 2014, Lahn and Page 1999, Ohno 1967). Since then, the X and Y chromosomes have evolved in their own way (described below), but share a striking common feature: gaining specialization for male reproduction through acquisition and amplification of testis-specific genes (Bellott, et al. 2014, Bellott, et al. 2010, Hughes, et al. 2012, Hughes, et al. 2010, Mueller, et al. 2008, Mueller, et al. 2013, Ross, et al. 2005, Skaletsky, et al. 2003, Soh, et al. 2014, Warburton, et al. 2004). The recent improvement in the accuracy of sex chromosome sequence assemblies, by means of the Single-Haplotype Iterative Mapping and Sequencing (SHIMS) technique, has allowed for detailed annotation of large palindromic and/or ampliconic sequences and for comparisons of sex chromosomes among species (Bellott, et al. 2014, Bellott, et al. 2010, Hughes, et al. 2012, Hughes, et al. 2010, Mueller, et al. 2013, Skaletsky, et al. 2003, Soh, et al. 2014).
X chromosome
The present-day human X chromosome has preserved 98% of genes from the ancestral autosome; however, due to an intergenic expansion of non-coding sequences (particularly long interspersed repeat elements and retroviral sequences) during evolution, the gene density on the X chromosome is about half of the average of all autosomes (Bailey, et al. 2000, Bellott, et al. 2010, Lander, et al. 2001, Ross, et al. 2005). Despite the low gene density, the X chromosome has acquired a substantial number of protein-coding genes, the majority of which are members of multi-copy gene families, including cancer/testis antigen genes, and exhibit testis-predominant expression patterns (Bellott, et al. 2010, Mueller, et al. 2013, Scanlan, et al. 2004). These features are also seen in the sex chromosomes of other mammals, such as chimpanzees, rhesus monkeys and mice (Soh, et al. 2014). In the avian ZW system, the Z chromosome that is shared by both sexes (equivalent to the X chromosome in the XY system, though males are the homogametic sex) also has a lower gene density than the autosomes due to increased intergenic distances, and contains a massive tandem amplification of genes that are expressed in the testis (Bellott, et al. 2010). Thus, the convergent acquisition and amplification of testis-expressed genes biases the X, as well as Z, chromosomes toward male reproduction functions.
Y chromosome
The human Y chromosome retains only 3% of its ancestral genes, due to a lack of a crossover partner in the male-specific region, leading to genetic decay (Bellott, et al. 2010, Charlesworth and Charlesworth 2000, Skaletsky, et al. 2003). Since the start of its differentiation, the human Y chromosome has undergone evolutionary decay at least four times in a step-wise fashion, as evidenced by the presence of the “evolutionary strata” of the X-Y pairs of ancestral genes (Lahn and Page 1999). After each event that created a stratum (e.g., chromosomal inversions), genes in the newly non-recombining region of the Y chromosome decayed rapidly at first, followed by a stable phase, leaving a constant set of surviving genes (Hughes, et al. 2012). These surviving ancestral genes were selected during evolution for their critical functions in males: male sex determination, sperm production, and viability (Bellott, et al. 2014). In addition to surviving ancestral genes, the Y chromosome acquired and amplified genes that are predominantly expressed in the testis (Hughes, et al. 2012, Hughes, et al. 2010, Skaletsky, et al. 2003, Soh, et al. 2014). Interestingly, ancestral genes on the human Y chromosome, such as Rbmy, Tspy, and Hsfy, evolved into multi-copy and testis-specific genes, in contrast to other Y-linked ancestral genes, which are broadly expressed (Bellott, et al. 2014). Two ancestral genes on the mouse Y chromosome, Rbmy and Zfy, have also undergone amplification and turned into germ cell-specific genes (Mahadevaiah, et al. 1998, Mardon, et al. 1989, Soh, et al. 2014, Vernet, et al. 2014).
Lineage-specific acquisition and amplification
The amplified testis-specific genes on the X and Y chromosomes are rapidly evolving and demonstrate relatively recent and lineage-specific acquisition (Hughes, et al. 2010, Mueller, et al. 2013, Soh, et al. 2014). The recent improvement in the accuracy of the human and mouse sex chromosome sequence assemblies allows for a detailed comparison of the ampliconic regions between the two species (Mueller, et al. 2013, Soh, et al. 2014). Different from the single-copy genes that are highly conserved among placental mammals, most of the ampliconic genes are independently acquired and amplified between the human and mouse lineages (Mueller, et al. 2013, Soh, et al. 2014). Similarly, a comparison of the Y chromosome sequence between human and a closely related species, chimpanzee, also suggests a substantial change in the gene content and genomic structures in the ampliconic regions (Hughes, et al. 2010). Therefore, both X and Y chromosomes are subject to selective evolutionary forces that increase specialization for lineage-specific male reproductive functions.
Functions of single-copy genes vs. ampliconic/multi-copy genes during spermatogenesis
Spermatogenesis is a biological process where self-renewing mitotic spermatogonia give rise to mature spermatozoa through two major differentiation steps: meiosis and spermiogenesis. Spermatogenic cells reside in the testis, which provides a somatic environment to support spermatogenesis. Two major somatic cell types are Sertoli cells (which nourish germ cells through stages of spermatogenesis within the seminiferous tubule) and Leydig cells (which produce and regulate hormones, such as androgens). According to currently available mouse genetic studies, genes on the sex chromosomes regulate spermatogenesis in many different ways (Table 1). Most X-linked single-copy genes that exhibit function in spermatogenesis are expressed in multiple tissues and control the process through either somatic and/or germ cells, but a small group of spermatogonially expressed, germ cell-specific genes have also been identified (Wang, et al. 2001). With the exception of the male sex-determining gene, Sry, surviving ancestral Y-linked single-copy genes are broadly expressed, both in the adult and throughout development (Bellott, et al. 2014). In contrast, ampliconic/multi-copy genes on the sex chromosomes tend to be expressed specifically in male germ cells, with potential roles in postmeiotic spermatids (Cocquet, et al. 2010, Comptour, et al. 2014, Mueller, et al. 2008, Mueller, et al. 2013, Reynard, et al. 2009, Riel, et al. 2013, Sin, et al. 2012b). In this section, we provide examples of single-copy and ampliconic/multi-copy genes from mouse genetic studies in the regulation of spermatogenesis.
X-linked spermatogonial genes
A systematic genomic screen for spermatogonially expressed, germ cell-specific genes in mice has identified 36 genes, 11 of which are on the X chromosome and 3 of which are on the Y chromosome (Wang, et al. 2001). Further genetic studies on 4 out of the 11 X-linked genes (Tex11, Taf7l, Nxf2, Tktl1) in mice have been reported. Tex11 is critical for meiotic prophase by promoting synapsis and crossover formation in spermatocytes. Deletion of Tex11 spanning from exon 3 to exon 29 leads to apoptosis of spermatocytes at the pachytene stage due to meiotic failure (Yang, et al. 2008). However, a different Tex11 mutant mouse line, with a deletion at only exon 3, leads to a premature termination of translation and exhibits milder meiotic defects and normal fertility (Adelman and Petrini 2008). Taf7l encodes a germ cell-specific subunit of the TFIID complex that is essential for polymerase II-mediated transcription (Cheng, et al. 2007, Pointud, et al. 2003). Although Taf7l is expressed in spermatogonia, spermatocytes, and spermatids, its targeted deletion causes a relatively mild phenotype associated with a reduction in sperm counts and sperm motility. The mild phenotype may be due to a functional compensation by its ubiquitously expressed autosomal paralog, Taf7, which was retrotransposed from an mRNA of the Taf7l gene (Cheng, et al. 2007, Pointud, et al. 2003). Interestingly, Tex11 Taf7l double knockout mice show a much more severe meiotic phenotype than either single mutant: an earlier spermatocyte death at the zygotene stage, suggesting a synergistic regulation of both genes in meiotic prophase (Zheng, et al. 2010). Nxf2, a nuclear mRNA export factor, has implicated roles in mRNA stability and trafficking (Lai, et al. 2006, Takano, et al. 2007, Tretyakova, et al. 2005). Male mice lacking Nxf2 show reduced fertility, resulting from defects in spermatogonial proliferation and meiotic chromosome segregation (Pan, et al. 2009). In contrast to the other 3 genes, targeted deletion of Tktl1 in mice did not yield any obvious phenotype in reproduction (Bentz, et al. 2011).
Scml2, another critical X-linked gene in spermatogenesis was recently identified in proteomics screens. SCML2, a germline-specific Polycomb protein, was identified as a component of γH2AX (histone variant H2AX phosphorylated at serine 139, an essential histone modification of the XY body)-containing nucleosomes from testes (Hasegawa, et al. 2015). Another proteomics study independently identified SCML2 as a meiosis-specific protein (Luo, et al. 2013). Scml2 is highly transcribed in spermatogonia, but then subject to MSCI in spermatocytes due to its X-linkage (Hasegawa, et al. 2015), consistent with a previous report using RNA FISH (fluorescence in situ hybridization) analysis (Mueller, et al. 2008). SCML2 protein is detected in the entire nuclei of undifferentiated spermatogonia and persists until meiosis, where it becomes accumulated on the XY body despite its transcriptional silencing (Hasegawa, et al. 2015, Luo, et al. 2015). Scml2 knockout mice are infertile (Hasegawa, et al. 2015), while an independent Scml2 knockout mouse line with a distinct genetic background exhibits severely compromised fertility (Luo, et al. 2015). SCML2 has two distinct functions between autosomes and sex chromosomes in the establishment of the epigenome during spermatogenesis. On autosomes in spermatogonia, SCML2 positively regulates mono-ubiquitinated histone H2A at lysine 119 (H2AK119ub) and, during spermatogenic differentiation, suppresses genes commonly expressed among somatic cells as well as spermatogenesis-progenitor genes (Hasegawa, et al. 2015). Paradoxically, on sex chromosomes during meiosis, SCML2 prevents H2AK119ub (Hasegawa, et al. 2015, Luo, et al. 2015), thereby enabling unique epigenetic programming of sex chromosomes for male reproduction. Therefore, the X chromosome carries the critical regulator SCML2 for its own gene regulation during spermatogenesis—as is the case for the X-linked Xist non-coding RNA, and the proteins RLIM and ATRX in the regulation of female X chromosome inactivation (Lee and Bartolomei 2013, Sarma, et al. 2014, Shin, et al. 2010).
Ancestral single-copy and multi-copy genes
Male reproduction relies on successful male sex differentiation and development of an individual. Two major genes, Sry and Ar, which drive male differentiation, are located on the Y and X chromosomes, respectively (Gubbay, et al. 1990, Sinclair, et al. 1990, Wang, et al. 2009). Sry initiates testis differentiation of the bipotential gonad in mid-gestation (Koopman, et al. 1991a). The testis then secretes androgens whose functions are mediated through an androgen receptor, encoded from Ar, to trigger the differentiation of male sex organs (e.g., external genitalia and internal accessory organs) and secondary characteristics (Wang, et al. 2009). While androgen receptor signaling in germ cells is not required for spermatogenesis, androgen receptor signaling in the surrounding Sertoli, Leydig, and peritubular myoid cells plays critical roles in the completion of sperm production (Chang, et al. 2004, De Gendt, et al. 2004, Holdcraft and Braun 2004, Lyon and Hawkes 1970, Xu, et al. 2007, Yeh, et al. 2002, Zhang, et al. 2006). Other X- and Y-linked genes that have been shown to regulate spermatogenesis, based on mouse genetic studies, are listed in Table 1. In particular, targeted deletion of either Sox3 or Tsc22d3 impairs spermatogonial differentiation (Bruscoli, et al. 2012, Laronda and Jameson 2011). Similar to the function of the spermatogonia-specific gene Nxf2, disruption of Pcyt1b causes an age-dependent depletion of spermatogenic cells, indicating a role in the maintenance of spermatogonial stem cells (Jackowski, et al. 2004).
The function of Y-linked genes has been mainly studied in mice carrying spontaneous deletions (see review in (Burgoyne 1998)), due to the difficulty of generating targeted mutations on the Y chromosome using conventional homologous recombination methods in embryonic stem (ES) cells. Nevertheless, these studies have revealed the critical functions of the Y chromosome in spermatogenesis. The Y chromosome is the only chromosome that is acrocentric in the mouse genome, while all others are telocentric (Mouse Genome Sequencing, et al. 2002, Soh, et al. 2014). Its long arm contains the highly amplified germ cell-specific gene families Sly, Srsy, and Ssty, all of which were acquired during the evolution of the rodent lineage (Soh, et al. 2014). Mice lacking part or all of the long-arm genes show mild-to-severe defects in sperm morphology, fertilization, and fertility (Burgoyne, et al. 1992, Conway, et al. 1994, Riel, et al. 2013, Suh, et al. 1989, Toure, et al. 2004b). On the short arm, Eif2s3y is the sole factor essential for spermatogonial proliferation (Mazeyrat, et al. 2001). In mice lacking the entire Y chromosome (XO), introduction of two transgenes expressing Sry and Eif2s3y are sufficient to initiate testis differentiation and drive spermatogenesis through meiosis up to the round spermatid stage (Yamauchi, et al. 2014). Using similar transgene rescue strategies to study ancestral multi-copy genes on the mouse Y chromosome, Zfy2 is found to play an important function in meiotic checkpoints that remove spermatocytes with synaptic errors (Royo, et al. 2010). Zfy2, together with Zfy1 and Zfx (an X homolog), has been discovered to exert a major role in promoting the completion of the 2nd division of meiosis (Vernet, et al. 2011, Vernet, et al. 2014).
Acquired ampliconic/multi-copy genes that escape postmeiotic silencing
During meiosis in spermatocytes, MSCI results in an almost complete shut-down of sex-linked genes, besides a few exceptions, such as the non-coding RNA Tsx (Anguera, et al. 2011) and some microRNA genes (Song, et al. 2009). On the other hand, many more sex-linked genes escape from postmeiotic silencing in round spermatids. Based on microarray analysis, 13% of mouse X-linked genes escape postmeiotic silencing while 87% remain repressed in round spermatids (Namekawa, et al. 2006). Subsequent studies demonstrated that most ampliconic/multi-copy genes on the mouse X chromosome (33 gene families, representing ~273 genes in ampliconic regions and non-ampliconic regions) are highly expressed in the round spermatids (Mueller, et al. 2008, Sin, et al. 2012b). Because conventional targeting knockout strategies cannot be applied to multicopy genes, their function has been investigated through the generation of transgenic mice carrying small interfering RNAs. For instance, using shRNAs to knock down Slx (~25 copies), Slxl1 (~14 copies), or both, mice showed impairments in sperm motility and counts (Cocquet, et al. 2010). While Slx or Slxl1 knockdown mice were subfertile, double Slx/Slxl1 transgenic mice were sterile due to more apparent defects in spermatid elongation and sperm release (Cocquet, et al. 2010). These data suggest that X-linked ampliconic/multi-copy genes escape postmeiotic silencing for a functional role in spermiogenesis.
Additionally, Y-linked ampliconic/multi-copy genes are also known to escape postmeiotic silencing (Toure, et al. 2005). For instance, Sly (126 copies) is located on the long arm of the Y chromosome and expressed in postmeiotic spermatids (Reynard, et al. 2009, Soh, et al. 2014). Using a shRNA transgene to knock down Sly, mice exhibited sperm head abnormalities and reduced fertility (Cocquet, et al. 2009). In addition, knockdown of Sly also leads to sperm DNA damage and defective chromatin packaging (Riel, et al. 2013). Interestingly, SLY protein accumulates on PMSC, and Sly knockdown causes derepression of sex-linked genes in spermatids, suggesting that Sly escapes postmeiotic silencing to regulate PMSC (Cocquet, et al. 2009).
Single-copy genes that escape postmeiotic silencing
In addition to ampliconic/multi-copy genes, there are also many single-copy genes that escape postmeiotic silencing. One example of a single-copy escape gene from mouse genetic studies is Akap4, a gene critical for sperm motility function by encoding a protein anchoring a critical enzyme, cyclic AMP-dependent protein kinase, in the fibrous sheath of the sperm flagellum (Carrera, et al. 1994, Miki, et al. 2002b). Akap4 belongs to a group of genes that commonly escape postmeiotic silencing in both human and mouse, and is also involved in sperm function in humans (Luconi, et al. 2011). Expectedly, deletion of Akap4 in male mice leads to infertility due to abnormal sperm morphology and motility (Miki, et al. 2002a). On the other hand, not all of the escape genes are essential for fertility. Rlim (also known as Rnf12) escapes postmeiotic silencing in mice; however, Rlim is not required for male fertility, although maternal RLIM is a critical regulator of imprinted X chromosome inactivation in embryos (Shin, et al. 2010).
Human sex-linked genes that escape postmeiotic silencing
In humans, although the involvement of the X chromosome in male fertility is in debate (Stouffs, et al. 2009), the Y chromosome microdeletions clearly contribute to spermatogenic defects (Krausz, et al. 2011, Stouffs and Lissens 2012). Escape genes in humans have also been implicated in male infertility. In addition to AKAP4, the X-linked single-copy escape gene TEX13A may be associated with azoospermia and abnormal sperm maturation (Lee, et al. 2003, Luconi, et al. 2011). Mutations of other X-linked escape genes, such as CUL4B and AFF2, are associated with hypergonadism and developmental delay, suggesting a possible function in male reproduction (Isidor, et al. 2010, Sahoo, et al. 2011). Further investigation of escape genes is warranted for a better understanding of human male infertility.
Evolutionary implications of genes that escape postmeiotic silencing
Recent advances in the biology of sex chromosomes, summarized above, enable us to revisit old evolutionary theories of sex chromosomes. In this section, we discuss the driving forces and strategies that have shaped the evolution of the sex chromosomes. MSCI is common between eutherians and marsupials (Hornecker, et al. 2007, Namekawa, et al. 2007) and suggested to contribute to the emergence of the X and Y chromosomes in therian ancestors (Potrzebowski, et al. 2008). With this history, gene reactivation within the context of sex chromosome inactivation was a necessary step to acquire the spermiogenesis functions of the sex chromosomes.
The mammalian X chromosome is enriched with male reproductive genes (Sin, et al. 2012b, Wang, et al. 2001, Zhang, et al. 2010). It has been shown that these male-biased genes tend to be those recently acquired on the X chromosome and expressed in round spermatids (Zhang, et al. 2010). Acquired genes include both single-copy and ampliconic/multi-copy genes. While genes subject to postmeiotic silencing are highly conserved between humans and mice, male reproductive genes that escape postmeiotic silencing are significantly diverged (Sin, et al. 2012b). Compared with non-escape genes, escape genes exhibit higher rates of amino acid changes calculated by Ka/Ks values (Ka/Ks value: the ratio of the number of non-synonymous substitutions per non-synonymous site (Ka) to the number of synonymous substitutions per synonymous site (Ks)). In addition, although these escape genes are considered important to sperm function, different species tend to have different sets of escape genes (Sin, et al. 2012b). In the study looking at X-linked escape genes, of 54 mouse and 66 human genes, only 12 of them were common between the two species. For example, Rlim escapes postmeiotic silencing in mice but is subject to postmeiotic silencing in humans (Sin, et al. 2012b). It remains unknown how the expression change of Rlim between mice and humans affects reproductive fitness. Such expression change in the round spermatids was also observed between two closely related mouse species, suggesting that the expression of escape genes could also be an evolutionary constraint, separating the species (Homolka, et al. 2011). Furthermore, 25 out of 54 mouse escape gene are rodent-specific, and 8 out of 66 human escape genes are primate/ape-specific. Such specificities suggest that escape genes evolved rapidly in both sequence and expression pattern, and are likely to contribute to reproductive isolation between species (Sin, et al. 2012b).
Rice’s hypothesis
These findings are in accord with long-standing evolutionary theories of sex chromosomes. Due to sexual antagonisms (i.e., conflicting fitness between males and females during sexual reproduction), the sex chromosomes accumulated sexually antagonistic alleles that are favored in one sex but detrimental to the other (Rice 1984). Rice hypothesized that the X chromosome is enriched for genes benefiting males due to the selection of hemizygously expressed favorable effects in males, while their deleterious effects in females would initially be hidden by heterozygosity (Rice 1984). Similarly, any recessive mutation that is advantageous for male fitness can more likely spread on the X chromosome rather than an autosome, where the effects are masked by heterozygosity and likely to be lost. The Y chromosome is also predicted to be enriched for male-beneficial genes because of the hemizygous expression of Y-linked genes and selection that only occurs in males.
Ohno’s law
A subsequent study compared the sequences of human and mouse X chromosomes and showed that species-specific sequences apparently reside in the ampliconic regions, suggesting that these genes are significantly diverged and are, for the most part, independently acquired between these two species (Mueller, et al. 2013). This study challenged Ohno’s law (Ohno 1967), stating that the gene contents of the X chromosome is conserved across placental mammals due to the somatic dosage compensation between the X chromosome and autosomes (i.e., any translocations between the X chromosome and autosomes would disturb the gene dosage). Genes that are expressed before meiosis and subject to postmeiotic silencing are found to be highly conserved (Sin, et al. 2012b) and thus follow Ohno’s law. However, ampliconic genes that are predominantly expressed in postmeiotic spermatids escape not only postmeiotic silencing, but also Ohno’s law. Therefore, Ohno’s law pertains mainly to the genes subject to postmeiotic silencing, whereas sexual antagonism underlies escape gene activation in postmeiotic spermatids, consistent with Rice’s hypothesis (Figure 2). These findings suggest that sex chromosomes face at least two antagonistic evolutionary driving forces: (1) sex chromosome inactivation, which preserves the gene content (mostly ancestral genes) among species; and (2) sexual antagonism, which facilitates escape gene activation and the acquisition of new genes.
Figure 2. Model of conflict between two evolutionary driving forces on the X chromosome.
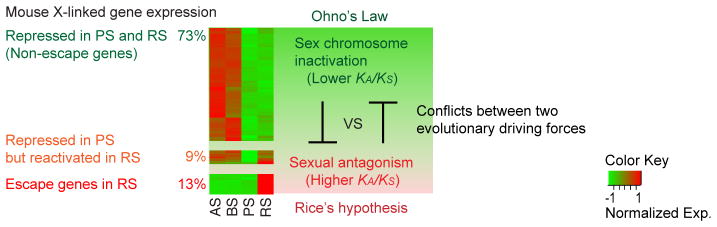
Microarray heatmap adapted from Namekawa et al. 2006. AS: type A spermatogonia, BS: type B spermatogonia, PS: pachytene spermatocytes, RS: round spermatids.
Intragenomic conflict between X and Y chromosomes
The human and mouse sex chromosomes contain X-Y pairs of ampliconic gene families that were co-acquired and amplified during their evolution (Skaletsky, et al. 2003, Soh, et al. 2014). Due to the high divergence in sequence between these pairs of gene families, it has been thought that they may have resulted from intragenomic conflict, which may affect transmission disorders (or segregation distortion) by the action of selfish genetic elements transmitted to the next generation at a frequency higher than the expected Mendelian inheritance ratio. This hypothesis has been tested in mice by knocking down the ampliconic gene family pair: for instance, Slx/Slxl1 on the X chromosome and Sly on the Y chromosome (Cocquet, et al. 2012). Slx/Slxl1 deficiency leads to male-biased litters, and Sly deficiency leads to female-biased litters (Cocquet, et al. 2009, Cocquet, et al. 2010). However, double knockdown of Slx/Slxl1 and Sly rescues the sex ratio distortion phenotype and the fertility problem caused by either knockdown (Cocquet, et al. 2012). Based on this result, it was proposed that SLX/SLXL1 and SLY are responsible for intragenomic conflict between X - and Y-bearing sperm by differentially regulating sperm function (Cocquet, et al. 2012, Good 2012). Further, Y-linked SSTY also localizes on PMSC, interacts with SLX/SLX1 and SLY, and is proposed to be involved in the intragenomic conflict (Comptour, et al. 2014). Similar to Slx/Slxl1-Sly, other acquired ampliconic gene family pairs on the mouse sex chromosomes, including Sstx-Ssty1/Ssty2 and Srsx-Srsy, are rodent-specific and show significant divergence in copy number and sequence (Soh, et al. 2014). Taken together, the intragenomic conflict has an important impact on the evolution of sex chromosomes by co-acquiring and amplifying genes that escape postmeiotic silencing and regulate spermatid differentiation, which is in line with Rice’s hypothesis. Given that these genes are rapidly evolving in a lineage-specific manner, they may also contribute to the speciation process.
Concluding remark
Mammalian sex chromosomes possess unique genomic features and gene expression that might have conferred an advantage in reproductive fitness of the species. Sequence analysis has revealed that both the X and Y chromosomes contain testis-specific ampliconic/multi-copy gene families, most of which were acquired during recent evolution, and some of which were derived from ancestral single-copy genes. Many of these genes escape postmeiotic silencing, suggesting that they exert important functions in spermatid differentiation and maturation. Because they are rapidly evolving and are highly divergent in gene content among species, it is thought that they contribute to the speciation process. Similar to ampliconic/multi-copy genes, single-copy escape genes are found to evolve rapidly in sequence and vary markedly between human and mouse (Sin, et al. 2012b). Therefore, the accumulation of ampliconic/multi-copy genes and genes escaping postmeiotic silencing on the sex chromosomes is likely to benefit male reproduction in a lineage-specific manner, thus supporting Rice’s hypothesis (Figure 2). In contrast, genes that are subject to postmeiotic silencing tend to be more conserved and thus follow Ohno’s law (Figure 2). Therefore, these antagonistic driving forces and strategies have shaped the evolution of the sex chromosomes.
Looking forward
In spite of accumulating evidence, the mechanisms that drive unique genomic arrangements and expression, such as amplification of genomic elements and escape from postmeiotic silencing, remain unknown. A promising path for future investigation is to identify the potential mechanisms that induce gene amplification and regulate gene expression during the postmeiotic period of spermatogenesis. Curiously, ampliconic DNA regions exhibit unique epigenetic signatures, such as DNA hypomethylation, throughout the germline prior to gene activation (Ikeda, et al. 2013). It suggests that the uniquely evolved ampliconic sequences are distinctly regulated during spermatogenesis.
Furthermore, in humans, sex chromosome abnormalities such as Kleinfelter (XXY) and Turner (XO) syndromes are associated with infertility, suggesting that the gene dosage of the sex chromosomes is also critical for fertility both in males and females (Heard and Turner 2011). Thus, in addition to the function of individual genes, the chromosome-wide regulation of gene dosage of the sex chromosomes may be warranted for further investigation.
Another interesting direction for future investigation is the relationship between the function of sex-chromosome genes and some types of cancer. Large palindromic and/or repetitive sequences of the sex chromosomes are enriched with cancer/testis (CT) antigens that are commonly expressed in testes and cancer (Simpson, et al. 2005). Unique genomic and epigenomic features likely underlie germline-specific gene activation on the sex chromosomes, as well as ectopic expression in some types of cancers. It would be intriguing to investigate how the germline program recapitulates in the case of abnormal somatic cells such as cancer cells, and how to safeguard and maintain our genome through the germline. The extraordinary genomic structure and genetic complement of sex chromosomes are likely to be key players in such processes.
Acknowledgments
We thank Tyler J. Broering and Kris G. Alavattam for editing, and Daniel W. Bellott for critical comments on the manuscript. This work was supported by the endowment from the Perinatal Institute and Cincinnati Children’s Hospital Medical Center to Y.-C.H, the Developmental Fund at Cincinnati Children’s Hospital Medical Center to S.H.N., the Research Grant from the March of Dimes Foundation to S.H.N., and NIH Grant GM098605 to S.H.N.
Footnotes
Declaration of Interests
We declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpatov R, Lesch BJ, Nakamoto-Kinoshita M, Blanco A, Chen S, Stutzer A, Armache KJ, Simon MD, Xu C, Ali M, Murn J, Prisic S, Kutateladze TG, Vakoc CR, Min J, Kingston RE, Fischle W, Warren ST, Page DC, Shi Y. A chromatin-dependent role of the fragile X mental retardation protein FMRP in the DNA damage response. Cell. 2014;157:869–881. doi: 10.1016/j.cell.2014.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera MC, Ma W, Clift D, Namekawa S, Kelleher RJ, 3rd, Lee JT. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011;7:e1002248. doi: 10.1371/journal.pgen.1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A. 2000;97:6634–6639. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, Kremitzki C, Fulton RS, Dugan S, Ding Y, Morton D, Khan Z, Lewis L, Buhay C, Wang Q, Watt J, Holder M, Lee S, Nazareth L, Alfoldi J, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK, Page DC. Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature. 2010;466:612–616. doi: 10.1038/nature09172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentz S, Pesch T, Wolfram L, de Valliere C, Leucht K, Fried M, Coy JF, Hausmann M, Rogler G. Lack of transketolase-like (TKTL) 1 aggravates murine experimental colitis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G598–607. doi: 10.1152/ajpgi.00323.2010. [DOI] [PubMed] [Google Scholar]
- Broering TJ, Alavattam KG, Sadreyev RI, Ichijima Y, Kato Y, Hasegawa K, Camerini-Otero RD, Lee JT, Andreassen PR, Namekawa SH. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J Cell Biol. 2014;205:663–675. doi: 10.1083/jcb.201311050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruscoli S, Velardi E, Di Sante M, Bereshchenko O, Venanzi A, Coppo M, Berno V, Mameli MG, Colella R, Cavaliere A, Riccardi C. Long glucocorticoid-induced leucine zipper (L-GILZ) protein interacts with ras protein pathway and contributes to spermatogenesis control. J Biol Chem. 2012;287:1242–1251. doi: 10.1074/jbc.M111.316372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS. The role of Y-encoded genes in mammalian spermatogenesis. Semin Cell Dev Biol. 1998;9:423–432. doi: 10.1006/scdb.1998.0228. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Sutcliffe MJ, Palmer SJ. Fertility in mice requires X-Y pairing and a Y-chromosomal “spermiogenesis” gene mapping to the long arm. Cell. 1992;71:391–398. doi: 10.1016/0092-8674(92)90509-b. [DOI] [PubMed] [Google Scholar]
- Carrera A, Gerton GL, Moss SB. The major fibrous sheath polypeptide of mouse sperm: structural and functional similarities to the A-kinase anchoring proteins. Dev Biol. 1994;165:272–284. doi: 10.1006/dbio.1994.1252. [DOI] [PubMed] [Google Scholar]
- Chang C, Chen YT, Yeh SD, Xu Q, Wang RS, Guillou F, Lardy H, Yeh S. Infertility with defective spermatogenesis and hypotestosteronemia in male mice lacking the androgen receptor in Sertoli cells. Proc Natl Acad Sci U S A. 2004;101:6876–6881. doi: 10.1073/pnas.0307306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol. 2007;27:2582–2589. doi: 10.1128/MCB.01722-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet. 2012;8:e1002900. doi: 10.1371/journal.pgen.1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol. 2009;7:e1000244. doi: 10.1371/journal.pbio.1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Riel JM, Karacs TP, Rattigan A, Ojarikre OA, Affara NA, Ward MA, Burgoyne PS. Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol Biol Cell. 2010;21:3497–3505. doi: 10.1091/mbc.E10-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NR, Taylor JS, Scott LB, Guillery RW, Soriano P, Furley AJ. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr Biol. 1998;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- Comptour A, Moretti C, Serrentino ME, Auer J, Ialy-Radio C, Ward MA, Toure A, Vaiman D, Cocquet J. SSTY proteins co-localize with the post-meiotic sex chromatin and interact with regulators of its expression. Febs j. 2014;281:1571–1584. doi: 10.1111/febs.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Mahadevaiah SK, Darling SM, Capel B, Rattigan AM, Burgoyne PS. Y353/B: a candidate multiple-copy spermiogenesis gene on the mouse Y chromosome. Mamm Genome. 1994;5:203–210. doi: 10.1007/BF00360546. [DOI] [PubMed] [Google Scholar]
- Davies B, Baumann C, Kirchhoff C, Ivell R, Nubbemeyer R, Habenicht UF, Theuring F, Gottwald U. Targeted deletion of the epididymal receptor HE6 results in fluid dysregulation and male infertility. Mol Cell Biol. 2004;24:8642–8648. doi: 10.1128/MCB.24.19.8642-8648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gendt K, Swinnen JV, Saunders PT, Schoonjans L, Dewerchin M, Devos A, Tan K, Atanassova N, Claessens F, Lecureuil C, Heyns W, Carmeliet P, Guillou F, Sharpe RM, Verhoeven G. A Sertoli cell-selective knockout of the androgen receptor causes spermatogenic arrest in meiosis. Proc Natl Acad Sci U S A. 2004;101:1327–1332. doi: 10.1073/pnas.0308114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forss-Petter S, Werner H, Berger J, Lassmann H, Molzer B, Schwab MH, Bernheimer H, Zimmermann F, Nave KA. Targeted inactivation of the X-linked adrenoleukodystrophy gene in mice. J Neurosci Res. 1997;50:829–843. doi: 10.1002/(SICI)1097-4547(19971201)50:5<829::AID-JNR19>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Good JM. The conflict within and the escalating war between the sex chromosomes. PLoS Genet. 2012;8:e1002955. doi: 10.1371/journal.pgen.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves IK, Rangasamy D, Devoy M, Marshall Graves JA, Tremethick DJ. The X and Y chromosomes assemble into H2A.Z-containing [corrected] facultative heterochromatin [corrected] following meiosis. Mol Cell Biol. 2006;26:5394–5405. doi: 10.1128/MCB.00519-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. doi: 10.1038/85899. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, Andreassen PR, Barski A, Namekawa SH. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell. 2015;32:547–548. doi: 10.1016/j.devcel.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Turner J. Function of the sex chromosomes in mammalian fertility. Cold Spring Harb Perspect Biol. 2011;3:a002675. doi: 10.1101/cshperspect.a002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Gonzalez EO, Mornet D, Rendon A, Martinez-Rojas D. Absence of Dp71 in mdx3cv mouse spermatozoa alters flagellar morphology and the distribution of ion channels and nNOS. J Cell Sci. 2005;118:137–145. doi: 10.1242/jcs.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdcraft RW, Braun RE. Androgen receptor function is required in Sertoli cells for the terminal differentiation of haploid spermatids. Development. 2004;131:459–467. doi: 10.1242/dev.00957. [DOI] [PubMed] [Google Scholar]
- Homolka D, Ivanek R, Forejt J, Jansa P. Differential expression of non-coding RNAs and continuous evolution of the X chromosome in testicular transcriptome of two mouse species. PLoS One. 2011;6:e17198. doi: 10.1371/journal.pone.0017198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornecker JL, Samollow PB, Robinson ES, Vandeberg JL, McCarrey JR. Meiotic sex chromosome inactivation in the marsupial Monodelphis domestica. Genesis. 2007;45:696–708. doi: 10.1002/dvg.20345. [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, Wang Q, Shen H, Holder M, Villasana D, Nazareth LV, Cree A, Courtney L, Veizer J, Kotkiewicz H, Cho TJ, Koutseva N, Rozen S, Muzny DM, Warren WC, Gibbs RA, Wilson RK, Page DC. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, Trask BJ, Mardis ER, Warren WC, Repping S, Rozen S, Wilson RK, Page DC. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Ichijima M, Lou Z, Nussenzweig A, Camerini-Otero RD, Chen J, Andreassen PR, Namekawa SH. MDC1 directs chromosome-wide silencing of the sex chromosomes in male germ cells. Genes Dev. 2011;25:959–971. doi: 10.1101/gad.2030811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichijima Y, Sin HS, Namekawa SH. Sex chromosome inactivation in germ cells: emerging roles of DNA damage response pathways. Cell Mol Life Sci. 2012;69:2559–2572. doi: 10.1007/s00018-012-0941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R, Shiura H, Numata K, Sugimoto M, Kondo M, Mise N, Suzuki M, Greally JM, Abe K. Large, male germ cell-specific hypomethylated DNA domains with unique genomic and epigenomic features on the mouse X chromosome. DNA Res. 2013;20:549–565. doi: 10.1093/dnares/dst030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidor B, Pichon O, Baron S, David A, Le Caignec C. Deletion of the CUL4B gene in a boy with mental retardation, minor facial anomalies, short stature, hypogonadism, and ataxia. Am J Med Genet A. 2010;152A:175–180. doi: 10.1002/ajmg.a.33152. [DOI] [PubMed] [Google Scholar]
- Iuchi Y, Okada F, Tsunoda S, Kibe N, Shirasawa N, Ikawa M, Okabe M, Ikeda Y, Fujii J. Peroxiredoxin 4 knockout results in elevated spermatogenic cell death via oxidative stress. Biochem J. 2009;419:149–158. doi: 10.1042/BJ20081526. [DOI] [PubMed] [Google Scholar]
- Jackowski S, Rehg JE, Zhang YM, Wang J, Miller K, Jackson P, Karim MA. Disruption of CCTbeta2 expression leads to gonadal dysfunction. Mol Cell Biol. 2004;24:4720–4733. doi: 10.1128/MCB.24.11.4720-4733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasurda JS, Jung DO, Froeter ED, Schwartz DB, Hopkins TD, Farris CL, McGee S, Narayan P, Ellsworth BS. The forkhead transcription factor, FOXP3: a critical role in male fertility in mice. Biol Reprod. 2014;90:4. doi: 10.1095/biolreprod.113.112375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs B, Meeks JJ, Ito M, Martinson FA, Matzuk MM, Jameson JL, Russell LD. Blockage of the rete testis and efferent ductules by ectopic Sertoli and Leydig cells causes infertility in Dax1-deficient male mice. Endocrinology. 2001;142:4486–4495. doi: 10.1210/endo.142.10.8447. [DOI] [PubMed] [Google Scholar]
- Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kusaka M, Omichi K, Suzuki R, Kato-Fukui Y, Kamiirisa K, Matsuo M, Kamijo S, Kasahara M, Yoshioka H, Ogata T, Fukuda T, Kondo I, Kato M, Dobyns WB, Yokoyama M, Morohashi K. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nat Genet. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991a;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991b;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]
- Kotula-Balak M, Lenartowicz M, Kowal M, Styrna J, Bilinska B. Testicular morphology and expression of aromatase in testes of mice with the mosaic mutation (Atp7a mo-ms) Theriogenology. 2007;67:423–434. doi: 10.1016/j.theriogenology.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kowal M, Lenartowicz M, Pecio A, Golas A, Blaszkiewicz T, Styrna J. Copper metabolism disorders affect testes structure and gamete quality in male mice. Syst Biol Reprod Med. 2010;56:431–444. doi: 10.3109/19396361003734624. [DOI] [PubMed] [Google Scholar]
- Krausz C, Chianese C, Giachini C, Guarducci E, Laface I, Forti G. The Y chromosome-linked copy number variations and male fertility. J Endocrinol Invest. 2011;34:376–382. doi: 10.1007/BF03347463. [DOI] [PubMed] [Google Scholar]
- Kudoh H, Ikeda H, Kakitani M, Ueda A, Hayasaka M, Tomizuka K, Hanaoka K. A new model mouse for Duchenne muscular dystrophy produced by 2.4 Mb deletion of dystrophin gene using Cre-loxP recombination system. Biochem Biophys Res Commun. 2005;328:507–516. doi: 10.1016/j.bbrc.2004.12.191. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lai D, Sakkas D, Huang Y. The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. RNA. 2006;12:1446–1449. doi: 10.1261/rna.94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Laronda MM, Jameson JL. Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology. 2011;152:1606–1615. doi: 10.1210/en.2010-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152:1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee SH, Chung TG, Kim HJ, Yoon TK, Kwak IP, Park SH, Cha WT, Cho SW, Cha KY. Molecular and cytogenetic characterization of two azoospermic patients with X-autosome translocation. J Assist Reprod Genet. 2003;20:385–389. doi: 10.1023/A:1025437329427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Shaver TM, Balasa A, Ljungberg MC, Wang X, Wen S, Nguyen H, Van den Veyver IB. Deletion of Porcn in mice leads to multiple developmental defects and models human focal dermal hypoplasia (Goltz syndrome) PLoS One. 2012;7:e32331. doi: 10.1371/journal.pone.0032331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llanos RM, Ke BX, Wright M, Deal Y, Monty F, Kramer DR, Mercer JF. Correction of a mouse model of Menkes disease by the human Menkes gene. Biochim Biophys Acta. 2006;1762:485–493. doi: 10.1016/j.bbadis.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Development. 1990;109:635–646. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- Luconi M, Cantini G, Baldi E, Forti G. Role of a-kinase anchoring proteins (AKAPs) in reproduction. Front Biosci. 2011;16:1315–1330. doi: 10.2741/3791. [DOI] [PubMed] [Google Scholar]
- Luo M, Yang F, Leu NA, Landaiche J, Handel MA, Benavente R, La Salle S, Wang PJ. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat Commun. 2013;4:2788. doi: 10.1038/ncomms3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Zhou J, Leu NA, Abreu CM, Wang J, Anguera MC, de Rooij DG, Jasin M, Wang PJ. Polycomb Protein SCML2 Associates with USP7 and Counteracts Histone H2A Ubiquitination in the XY Chromatin during Male Meiosis. PLoS Genet. 2015;11:e1004954. doi: 10.1371/journal.pgen.1004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoh SW, Bain PA, Polakiewicz RD, Goodheart ML, Gardner H, Jaenisch R, Page DC. Zfx mutation results in small animal size and reduced germ cell number in male and female mice. Development. 1997;124:2275–2284. doi: 10.1242/dev.124.11.2275. [DOI] [PubMed] [Google Scholar]
- Lyon MF, Hawkes SG. X-linked gene for testicular feminization in the mouse. Nature. 1970;227:1217–1219. doi: 10.1038/2271217a0. [DOI] [PubMed] [Google Scholar]
- Maclean JA, 2nd, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn LL, McCarrey JR, Cattanach BM, Lovell-Badge R, Burgoyne PS. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- Mardon G, Mosher R, Disteche CM, Nishioka Y, McLaren A, Page DC. Duplication, deletion, and polymorphism in the sex-determining region of the mouse Y chromosome. Science. 1989;243:78–80. doi: 10.1126/science.2563173. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- Meeks JJ, Crawford SE, Russell TA, Morohashi K, Weiss J, Jameson JL. Dax1 regulates testis cord organization during gonadal differentiation. Development. 2003;130:1029–1036. doi: 10.1242/dev.00316. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Jia Z. Synaptic transmission and plasticity in the absence of AMPA glutamate receptor GluR2 and GluR3. Neuron. 2003;39:163–176. doi: 10.1016/s0896-6273(03)00368-4. [DOI] [PubMed] [Google Scholar]
- Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002a;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- Miki K, Willis WD, Brown PR, Goulding EH, Fulcher KD, Eddy EM. Targeted disruption of the Akap4 gene causes defects in sperm flagellum and motility. Dev Biol. 2002b;248:331–342. doi: 10.1006/dbio.2002.0728. [DOI] [PubMed] [Google Scholar]
- Mikolcevic P, Sigl R, Rauch V, Hess MW, Pfaller K, Barisic M, Pelliniemi LJ, Boesl M, Geley S. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol Cell Biol. 2012;32:868–879. doi: 10.1128/MCB.06261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabayashi K, Katoh-Fukui Y, Ogawa H, Baba T, Shima Y, Sugiyama N, Kitamura K, Morohashi K. Aristaless related homeobox gene, Arx, is implicated in mouse fetal Leydig cell differentiation possibly through expressing in the progenitor cells. PLoS One. 2013;8:e68050. doi: 10.1371/journal.pone.0068050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing C. Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, VandeBerg JL, McCarrey JR, Lee JT. Sex chromosome silencing in the marsupial male germ line. Proc Natl Acad Sci U S A. 2007;104:9730–9735. doi: 10.1073/pnas.0700323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Berlin, New York [etc.]: Springer-Verlag; 1967. [Google Scholar]
- Pan J, Eckardt S, Leu NA, Buffone MG, Zhou J, Gerton GL, McLaughlin KJ, Wang PJ. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol. 2009;330:167–174. doi: 10.1016/j.ydbio.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointud JC, Mengus G, Brancorsini S, Monaco L, Parvinen M, Sassone-Corsi P, Davidson I. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J Cell Sci. 2003;116:1847–1858. doi: 10.1242/jcs.00391. [DOI] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jégov B, Kaessmann H. Chromosomal Gene Movements Reflect the Recent Origin and Biology of Therian Sex Chromosomes. PLoS Biology. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynard LN, Cocquet J, Burgoyne PS. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol Reprod. 2009;81:250–257. doi: 10.1095/biolreprod.108.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Riel JM, Yamauchi Y, Sugawara A, Li HY, Ruthig V, Stoytcheva Z, Ellis PJ, Cocquet J, Ward MA. Deficiency of the multi-copy mouse Y gene Sly causes sperm DNA damage and abnormal chromatin packaging. J Cell Sci. 2013;126:803–813. doi: 10.1242/jcs.114488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzoti K, Brunelli S, Carmignac D, Thomas PQ, Robinson IC, Lovell-Badge R. SOX3 is required during the formation of the hypothalamo-pituitary axis. Nat Genet. 2004;36:247–255. doi: 10.1038/ng1309. [DOI] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RI, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol. 2010;20:2117–2123. doi: 10.1016/j.cub.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Sahoo T, Theisen A, Marble M, Tervo R, Rosenfeld JA, Torchia BS, Shaffer LG. Microdeletion of Xq28 involving the AFF2 (FMR2) gene in two unrelated males with developmental delay. Am J Med Genet A. 2011;155A:3110–3115. doi: 10.1002/ajmg.a.34345. [DOI] [PubMed] [Google Scholar]
- Sarma K, Cifuentes-Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell. 2014;159:869–883. doi: 10.1016/j.cell.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. [PubMed] [Google Scholar]
- Shin J, Bossenz M, Chung Y, Ma H, Byron M, Taniguchi-Ishigaki N, Zhu X, Jiao B, Hall LL, Green MR, Jones SN, Hermans-Borgmeyer I, Lawrence JB, Bach I. Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature. 2010;467:977–981. doi: 10.1038/nature09457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, Andreassen PR, Namekawa SH. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012a;26:2737–2748. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin HS, Ichijima Y, Koh E, Namiki M, Namekawa SH. Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res. 2012b;22:827–836. doi: 10.1101/gr.135046.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin HS, Namekawa SH. The great escape: Active genes on inactive sex chromosomes and their evolutionary implications. Epigenetics. 2013;8:887–892. doi: 10.4161/epi.25672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Slegtenhorst-Eegdeman KE, de Rooij DG, Verhoef-Post M, van de Kant HJ, Bakker CE, Oostra BA, Grootegoed JA, Themmen AP. Macroorchidism in FMR1 knockout mice is caused by increased Sertoli cell proliferation during testicular development. Endocrinology. 1998;139:156–162. doi: 10.1210/endo.139.1.5706. [DOI] [PubMed] [Google Scholar]
- Soh YQS, Alföldi J, Pyntikova T, Brown Laura G, Graves T, Minx Patrick J, Fulton Robert S, Kremitzki C, Koutseva N, Mueller Jacob L, Rozen S, Hughes Jennifer F, Owens E, Womack James E, Murphy William J, Cao Q, de Jong P, Warren Wesley C, Wilson Richard K, Skaletsky H, Page David C. Sequencing the Mouse Y Chromosome Reveals Convergent Gene Acquisition and Amplification on Both Sex Chromosomes. Cell. 2014;159:800–813. doi: 10.1016/j.cell.2014.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffs K, Lissens W. X chromosomal mutations and spermatogenic failure. Biochim Biophys Acta. 2012;1822:1864–1872. doi: 10.1016/j.bbadis.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update. 2009;15:623–637. doi: 10.1093/humupd/dmp023. [DOI] [PubMed] [Google Scholar]
- Suh DS, Styrna J, Moriwaki K. Effect of Y chromosome and H-2 complex derived from Japanese wild mouse on sperm morphology. Genet Res. 1989;53:17–19. doi: 10.1017/s0016672300027816. [DOI] [PubMed] [Google Scholar]
- Takano K, Miki T, Katahira J, Yoneda Y. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res. 2007;35:2513–2521. doi: 10.1093/nar/gkm125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure A, Clemente EJ, Ellis P, Mahadevaiah SK, Ojarikre OA, Ball PA, Reynard L, Loveland KL, Burgoyne PS, Affara NA. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 2005;6:R102. doi: 10.1186/gb-2005-6-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure A, Grigoriev V, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A protein encoded by a member of the multicopy Ssty gene family located on the long arm of the mouse Y chromosome is expressed during sperm development. Genomics. 2004a;83:140–147. doi: 10.1016/s0888-7543(03)00216-7. [DOI] [PubMed] [Google Scholar]
- Toure A, Szot M, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. A new deletion of the mouse Y chromosome long arm associated with the loss of Ssty expression, abnormal sperm development and sterility. Genetics. 2004b;166:901–912. doi: 10.1534/genetics.166.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova I, Zolotukhin AS, Tan W, Bear J, Propst F, Ruthel G, Felber BK. Nuclear export factor family protein participates in cytoplasmic mRNA trafficking. J Biol Chem. 2005;280:31981–31990. doi: 10.1074/jbc.M502736200. [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, Wang R, Kim S, Chandramouli GV, Barrett JC, Burgoyne PS, Deng CX. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr Biol. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Eijpe M, Baarends WM. The X and Y chromosome in meiosis: how and why they keep silent. Asian J Androl. 2011;13:779–780. doi: 10.1038/aja.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Ojarikre OA, Longepied G, Prosser HM, Bradley A, Mitchell MJ, Burgoyne PS. The Y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr Biol. 2011;21:787–793. doi: 10.1016/j.cub.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Yamauchi Y, Decarpentrie F, Mitchell MJ, Ward MA, Burgoyne PS. Mouse Y-linked Zfy1 and Zfy2 are expressed during the male-specific interphase between meiosis I and meiosis II and promote the 2nd meiotic division. PLoS Genet. 2014;10:e1004444. doi: 10.1371/journal.pgen.1004444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Wang RS, Yeh S, Tzeng CR, Chang C. Androgen receptor roles in spermatogenesis and fertility: lessons from testicular cell-specific androgen receptor knockout mice. Endocr Rev. 2009;30:119–132. doi: 10.1210/er.2008-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14:1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J, Meeks JJ, Hurley L, Raverot G, Frassetto A, Jameson JL. Sox3 is required for gonadal function, but not sex determination, in males and females. Mol Cell Biol. 2003;23:8084–8091. doi: 10.1128/MCB.23.22.8084-8091.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Gerdin AK, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP Sanger Institute Mouse Genetics P. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Lin HY, Yeh SD, Yu IC, Wang RS, Chen YT, Zhang C, Altuwaijri S, Chen LM, Chuang KH, Chiang HS, Yeh S, Chang C. Infertility with defective spermatogenesis and steroidogenesis in male mice lacking androgen receptor in Leydig cells. Endocrine. 2007;32:96–106. doi: 10.1007/s12020-007-9015-0. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science. 2014;343:69–72. doi: 10.1126/science.1242544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 2008;22:682–691. doi: 10.1101/gad.1613608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh S, Tsai MY, Xu Q, Mu XM, Lardy H, Huang KE, Lin H, Yeh SD, Altuwaijri S, Zhou X, Xing L, Boyce BF, Hung MC, Zhang S, Gan L, Chang C. Generation and characterization of androgen receptor knockout (ARKO) mice: an in vivo model for the study of androgen functions in selective tissues. Proc Natl Acad Sci U S A. 2002;99:13498–13503. doi: 10.1073/pnas.212474399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RN, Ito M, Saunders TL, Camper SA, Jameson JL. Role of Ahch in gonadal development and gametogenesis. Nat Genet. 1998;20:353–357. doi: 10.1038/3822. [DOI] [PubMed] [Google Scholar]
- Zhang C, Yeh S, Chen YT, Wu CC, Chuang KH, Lin HY, Wang RS, Chang YJ, Mendis-Handagama C, Hu L, Lardy H, Chang C. Oligozoospermia with normal fertility in male mice lacking the androgen receptor in testis peritubular myoid cells. Proc Natl Acad Sci U S A. 2006;103:17718–17723. doi: 10.1073/pnas.0608556103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M. Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Yang F, Wang PJ. Regulation of male fertility by X-linked genes. J Androl. 2010;31:79–85. doi: 10.2164/jandrol.109.008193. [DOI] [PMC free article] [PubMed] [Google Scholar]



