Summary
Transcription factor complexes have varied effects on cell fate and behavior, but how this diversification of function occurs is largely unknown. The Nodal signaling pathway has many biological functions that all converge on the transcription factors Smad2/3. Smad2/3 has many cofactors, and alternative usage of these may provide a mechanism for modulating Smad2/3 function. Here we investigated how perturbation of the cofactor E2a affects global patterns of Smad2/3 binding and gene expression during gastrulation. We find that E2a regulates early development in two ways. E2a changes the position of Smad2/3 binding at the Nodal inhibitor lefty, resulting in direct repression of lefty that is critical for mesendoderm specification. Separately, E2a is necessary to drive transcription of Smad2/3 target genes, including critical regulators of dorsal cell fate and morphogenesis. Overall, we find that E2a functions as both a transcriptional repressor and activator to precisely regulate Nodal signaling.
Keywords: E2a, Smad2/3, Nodal, gastrulation, lefty
Introduction
During embryogenesis, transcription factor complexes can mediate many aspects of cell fate specification by regulating diverse sets of transcriptional targets at different points in time or space. While this scenario is well-appreciated, there remains little mechanistic insight into how it is achieved: what variables direct the choice of where a transcription factor will bind, and whether the outcome of binding has an activating, repressive, or neutral effect on the expression of the target? Several models have demonstrated that transcription factors can have variable associations with other proteins and with chromatin, resulting in different effects on gene transcription. A particularly well-studied example is the Brg1 and Brm family of ATPase complexes (BAF complexes), in which alternative use of BAF subunits significantly alters the transcriptional regulatory activity of the complex during the progression from embryonic stem cells (esBAF) to neural progenitors (npBAF) and neurons (nBAF)(Yoo and Crabtree, 2009). BAF complexes have over 10,000 targets in the mouse genome (Ho et al., 2009), and can repress or activate neuronal gene trancription through differential interaction with coactivators or corepressors. Other models of transcription factor target choice include the Drosophila mesoderm factor Twist, which binds different targets at different developmental stages (Zinzen et al., 2009), the Nodal signaling transcription factor Smad2/3, which associates with different targets in human endoderm versus stem cells (Kim et al., 2011) and the bHLH factor E2a, which has different transcriptional targets in progressive stages of B-cell development (Lin et al., 2010). However, the mechanisms that govern the switch between activation and repression of target genes, or the effects of removing or adding a subunit on the global behavior of a transcription factor complex as a whole, remain poorly understood. There remains little to no insight into how subtle changes in the interacting components of complexes cause widespread transcriptional differences and drastic cellular consequences in embryos in vivo.
In early vertebrate embryogenesis, the Nodal signaling pathway, acting through the transcription factors Smad2/Smad3, is critical for many aspects of early development and cellular differentiation, including deeply conserved roles in mesendoderm development and left/right axis specification (reviewed in (Schier, 2003; Shen, 2007)). How Nodal signaling drives many aspects of embryonic development by using a simple downstream network focused primarily on a single transcription factor, Smad2/3, remains unclear. Transcriptional regulation of Smad2/3 targets is finely balanced by autoregulation: Nodal ligands are themselves transcriptional targets of Smad2/3, as is the Nodal inhibitor lefty (Cheng et al., 2000) (Branford and Yost, 2002; Meno et al., 1999). This sets up a precise gradient of Nodal signaling activity, but how this gradient is interpreted into different transcriptional outcomes at the level of Smad2/3 binding is not understood. Possible mechanisms might include Smad2/3 binding to different target genes at different signal strengths, stronger or more stable accumulation of Smad2/3 at target sites, or association with different cofactors. Several cofactors have been discovered to interact with Smad2/3 in order to drive different functional aspects of Nodal signaling, including FoxH1/Fast1(Labbe et al., 1998), Mixer (Germain et al., 2000) Eomes (Slagle et al., 2011; Teo et al., 2011) and the bHLH proteins E2a and Heb (Yoon et al., 2011). It is not known whether any of these factors can directly influence genomic target choice by Smad2/3, or how the gain or loss of these factors affects the transcriptional behavior of the Smad2/3 complex.
To understand how transcriptional cofactors regulate Smad2/3 binding and gene transcription in the developing embryo, we investigated the requirement for the bHLH transcription factor E2a in Nodal signaling. E2a is a required cofactor for Nodal signaling in mesendoderm specification, as embryos depleted for E2a fail to form mesoderm, have reduced endoderm, and fail to gastrulate (Yoon et al., 2011). Insights from E2a’s role in hematopoiesis point to several potential models for how E2a might modulate Smad2/3 binding. In B cell development, E2a can act as either a transcriptional activator or repressor through its association with coactivators and corepressors or by forming homodimers and heterodimer in association with other class I or class II bHLH proteins, which can be repressors or activators (reviewed in (Kee, 2009)). E2a can also associate with transcriptional coactivators such as p300, CBP and TAF4 through one of two AD transactivation domains (Bayly et al., 2004; Bradney et al., 2003; Chen et al., 2013), or with the ETO/MTG class of corepressors through the AD2 and DES domains (Gow et al., 2014; Guo et al., 2009; Zhang et al., 2004). E2a can therefore have potentially widespread effects on transcriptional regulation across the genome, although the effects of E2a loss of function on global transcription patterns have not been investigated.
In this study, we asked what effect perturbation of E2a has on the behavior and function of the Smad2/3 multiprotein complex in the in vivo embryo. We identify two critical roles for E2a. First, E2a is essential for proper positioning of Smad2/3 at the lefty genomic locus. This direct interaction is mechanistically responsible for repressing lefty transcription. In the absence of E2a, lefty is dramatically upregulated, leading to failure of mesendoderm fate specification. Second, a set of genes require E2a not for Smad2/3 localization, but for transcriptional activation, leading to a failure of gastrulation morphogenesis in E2a depleted embryos. E2a can directly target these genes for activation by occupying the same regulatory regions as Smad2/3. Overall, we demonstrate that the Smad2/3 transcriptional cofactor, E2a, plays two critical roles in the regulation of early development by repressing transcription of the Nodal inhibitor lefty and by activating transcription of axial mesoderm genes. Perturbation of these roles has dramatic consequences for cell fate specification and morphogenesis.
Results
To identify how E2a affects the association of Smad2/3 with chromatin and transcription of Nodal target genes, we depleted E2A in Xenopus embryos and examined them using ChIP-Seq and RNA-Seq. Specifically, we injected 2-cell stage X. tropicalis embryos with an E2a morpholino (MO) and harvested these embryos, as well as uninjected sibling controls, at early gastrula (stage 10.5). We then conducted ChIP-Seq using anti-Smad2/3 antibodies as well as RNA-Seq with the same cohorts of embryos. Each experiment was performed in duplicate except ChIP-Seq of Smad2/3 in E2a-depleted embryos, which was performed in triplicate. For ChIP-Seq, each library was made using 1000 embryos, according to published methods (Wills et al., 2014), and our existing control Smad2/3 library data was used as one replicate (Gupta et al., 2014). For RNA-Seq, each library was made using 20μg of total starting RNA, according to published methods (Tan et al., 2013).
We then determined if Smad2/3 requires E2a to associate with chromatin across the genome. We examined ChIP-Seq datasets using Bowtie software to align ChIP-Seq reads to the X. tropicalis xenTro2 (version 4.1) genome, and used MACS2 software to call peaks. This analysis revealed 1027 regions bound by Smad2/3 in control embryos, and 1671 in E2a depleted embryos. We next used BedTools software to compare the genomic coordinates of Smad2/3-associated regions in control embryos with those in E2a-depleted embryos. We classified these regions into four categories: 1) regions that maintain Smad2/3; 2) regions where the genomic position of Smad2/3 is shifted slightly; 3) regions that lose all Smad2/3 binding; and 4) regions that gain Smad2/3 binding. 48% (495/1027) of the Smad2/3 binding falls into category 1 – meaning that these regions are not affected by E2A depletion (Figure 1A). Conversely, this also means that 52% (532/1027) of Smad2/3 binding regions are mislocalized when E2A is depleted. Overall, this indicates that E2a is not required for basic association of Smad2/3 with chromatin at gastrulation, but does direct Smad2/3 positioning at many loci, while other loci are correctly targeted independently of E2a.
Figure 1. E2a is required for subsets of Smad2/3 binding, and for global patterns of Smad2/3 target gene expression.
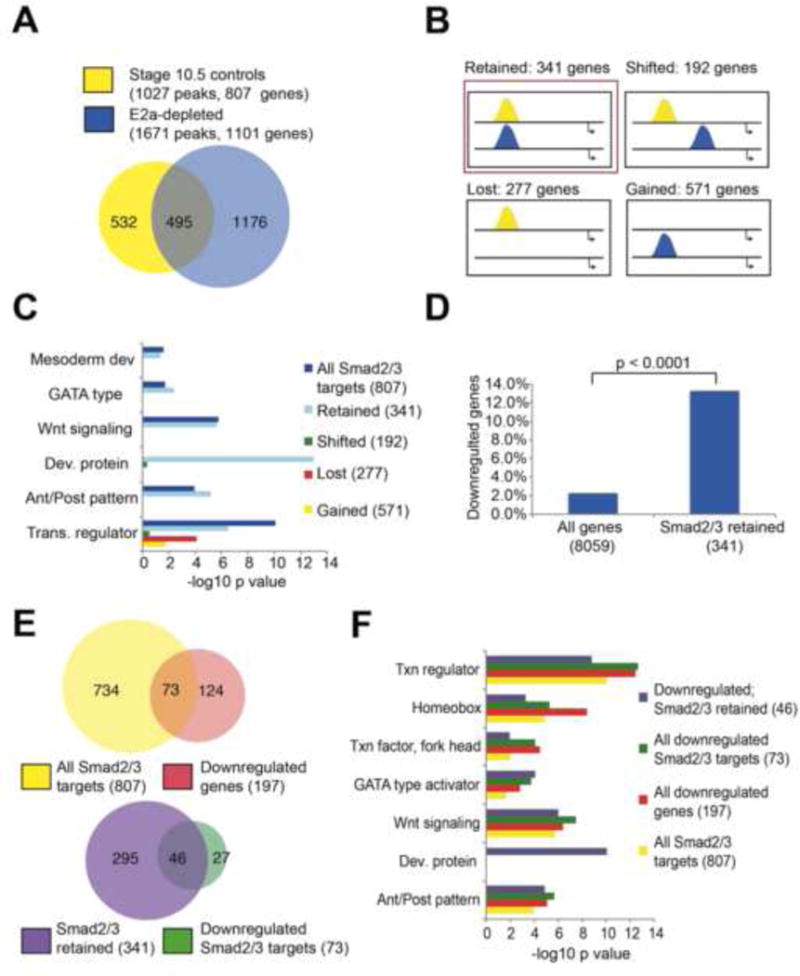
A) Smad2/3 targets 1027 distinct genomic regions in control stage 10.5 embryos (yellow), and 1671 regions in E2a-depleted embryos (blue). 495 regions are targeted in both conditions. B) Categories of Smad2/3 binding behavior in E2a-depleted embryos (blue) relative to controls (yellow). C) DAVID clustering analysis shows enrichment for developmental terms in all Smad2/3 associated genes, and for genes that have stable Smad2/3 binding when E2a is depleted (red box in B), but not other subcategories of Smad2/3 binding. D) Genes that retain Smad2/3 binding at the same genomic coordinates in E2a-depleted embryos (red box in B) are more likely to be downregulated by 2 fold or more in E2a-depleted embryos. E–F) Significant overlap exists between genes that are Smad2/3 targets and genes that are downregulated in E2a depleted embryos. Smad2/3 targets in which binding is at the same genomic position in control and E2a-depleted embryos are more likely to be downregulated in E2a-depleted embryos. These genes maintain enrichment for DAVID terms associated with early development. See also Tables S1, S2.
Next, we sought to examine the genes associated with the four categories of E2A-affected genomic regions. We used HOMER software to associate Smad2/3 bound regions to their nearest neighboring genes. We found 341 genes associated with stable Smad2/3 binding (category 1), 192 genes associated with a shift in Smad2/3 (category 2); 277 genes associated with total Smad2/3 loss (category 3) and 571 genes that gain an association with Smad2/3 (category 4)(Figure 1B, Table S1). We performed gene ontology analysis using DAVID clustering (Huang et al., 2009a, b) to identify whether any of these categories of genes was enriched for particular biological processes. We found that while all categories showed enrichment for “transcription regulation,” only genes within category 1 (stable binding) were significantly enriched for developmental processes, including anterior/posterior patterning (enrichment score 5.47; p=6.8e-6), developmental protein (enrichment score 5.4; p=1.1e-13), GATA-type transcription factor (p=3.8e-3), mesoderm development (p=4.4e-2), and Wnt signaling (score 3.76; p=2.2e-6)(Figure 1C). This category of genes also contains many well-known Nodal target genes that are critical for early development, including cer1, eomes, gsc, xbra/t, sox17β, pitx2, not, nodal1, and vegt, as well as receptors and ligands for Wnt and FGF signaling. Overall this suggests that developmental genes maintain association with a stable Smad2/3 element that is not affected by E2A depletion.
Our results suggest E2A is not required for the placement of Smad2/3 at many developmental loci. Therefore we surmised that E2a might instead act to enable transcription of these genes in response to Smad2/3 binding. To this end, we first identified the effect of E2a depletion on gene expression across the genome. Using the RNA-Seq datasets obtained from E2A depleted and control embryos, after performing FPKM normalization to account for variations in transcript length, we find that only 197 genes are significantly downregulated in E2a depleted embryos, while 3,957 genes were significantly upregulated (Table S2). Therefore we infer that, genome-wide, E2a is more frequently associated with transcriptional repression, not activation. However, the 197 genes downregulated in E2a-depleted embryos are heavily enriched for Smad2/3 target genes (73/197, 3.41 fold enrichment relative to whole gene set, p=1.091977e-27). This is particularly notable for genes in Category 1 where Smad2/3 remains stable (46/197, 6.38 fold enrichment) (Figure 1D–F). Further, if we examine the 46 downregulated genes that are associated with a stable Smad2/3 region, these represent many of the most critical and well-known Nodal target genes in early development (Table 1), and these maintain strong enrichment for DAVID terms associated with early development and patterning (Figure 1F). We were surprised to observe that these genes could be strongly downregulated at the transcriptional level while maintaining Smad2/3 binding. This suggests that E2a is essential for transcription of these genes, and that Smad2/3 binding alone is not sufficient to drive transcription. Overall, we conclude that E2a functions to repress gene transcription of many genes genome-wide, but that its role in gene activation predominates for early developmental Nodal target genes. Critically, while E2a is not required for Smad2/3 localization to these target genes, it is essential for their transcriptional activation.
Table 1.
List of all genes where Smad2/3 binding is maintained at the same position in control and E2a-depleted embryos, and that are significantly downregulated in E2a-depleted embryos. For each gene, the normalized mean expression level is shown (mean expression in E2a-depleted embryos divided by mean expression in control embryos). Gene categories are assigned as: transcription factor; cytoskeleton/transport; transmembrane receptor; signaling molecule; intracellular signaling (including secondary messenger proteins) and other/unknown (including all other major functions).
| Gene name | Expression (mean E2a-/mean Ctrl) | Function type |
|---|---|---|
| mespb | 0.008080432 | Transcription factor |
| cer1 | 0.010092776 | Signaling molecule |
| pcdh8.2 | 0.010779418 | Cytoskeleton/transport |
| t (xbra) | 0.027910383 | Transcription factor |
| frzb | 0.033973786 | Transmembrane receptor |
| gatm | 0.058238471 | Other/Unknown |
| lhx1 | 0.061176421 | Transcription factor |
| hhex | 0.075792824 | Transcription factor |
| gs17 | 0.082842955 | Other/unknown |
| foxa4 | 0.105938633 | Transcription factor |
| gsc | 0.111275047 | Transcription factor |
| gata4 | 0.112514562 | Transcription factor |
| slc38a3 | 0.135985709 | Cytoskeleton/transport |
| rspo2 | 0.155234594 | Signaling Molecule |
| nudt22 | 0.173302279 | Other/Unknown |
| foxc2 | 0.183713832 | Transcription factor |
| foxd3 | 0.195581307 | Transcription factor |
| epha4 | 0.244381447 | Transmembrane receptor |
| otx2 | 0.265712657 | Transcription factor |
| gata5 | 0.279417037 | Transcription factor |
| ngfr | 0.28420678 | Transmembrane receptor |
| ror2 | 0.298173092 | Signaling molecule |
| nr6a1 | 0.305514516 | Transcription factor |
| eomes | 0.327671011 | Transcription factor |
| cox7b | 0.381125295 | Other/Unknown |
| wnt5b | 0.389808629 | Signaling molecule |
| zic3 | 0.436294732 | Transcription factor |
| hpdl | 0.439688059 | Other/Unknown |
| efna1 | 0.459150746 | Signaling molecule |
| ventx1.1 | 0.487513979 | Transcription factor |
| snai1 | 0.506217054 | Transcription factor |
| vegt | 0.514365172 | Transcription factor |
| fgf8 | 0.53150366 | Signaling molecule |
| pkdcc.1 | 0.534870368 | Other/Unknown |
| gata6 | 0.54800291 | Transcription factor |
| tsg101 | 0.550753067 | Cytoskeleton/transport |
| crx | 0.564716113 | Transcription factor |
| map2k6 | 0.586210367 | Intracellular signaling |
| wnt11 | 0.590079259 | Signaling molecule |
| MGC76328 | 0.593841587 | Other/unknown |
| znf703 | 0.597459616 | Transcription factor |
| cdc42ep4 | 0.642916664 | Cytoskeleton/transport |
| lpar2 | 0.644983354 | Transmembrane receptor |
| stx6 | 0.64601592 | Cytoskeleton/Transport |
| LOC496805 | 0.671603875 | Unknown/Other |
| fzd8 | 0.695453455 | Transmembrane receptor |
E2a directly represses transcription of the Nodal inhibitor lefty
As E2a mostly serves to inhibit gene expression within the Xenopus embryo, we investigated whether its role as an activator for early developmental Nodal target genes might be indirect as a consequence of the ectopic upregulation of a Nodal antagonist. To this end, we examined whether any known inhibitors of Nodal signaling were highly expressed in E2a-depleted embryos. Interestingly, we find that the well-studied Nodal inhibitor Lefty is highly upregulated in E2a-depleted embryos (8.97 fold increase). We confirmed the expression of lefty by qRT-PCR in both control and E2a-depleted embryos (Figure 2A). To examine the spatial effects of this upregulation, we performed in situ hybridization for lefty in E2a-depleted and E2a-overexpressing embryos. We find that, at stage 10.5, E2a depleted embryos have stronger expression of lefty throughout the marginal zone (15/18 embryos with upregulated expression) compared to stage matched controls. (Figure 2B). Conversely, in embryos injected with E2a mRNA, lefty expression is inhibited (13/18, Figure 2B). Thus, we find multiple lines of evidence that expression of lefty is repressed by E2a. This suggests the downregulation of 46 Smad2/3 target genes observed in our RNA-Seq data could potentially arise indirectly through the upregulation of Lefty in E2a depleted embryos.
Figure 2. E2a regulates Samd2/3 positioning at lefty and represses lefty transcription.
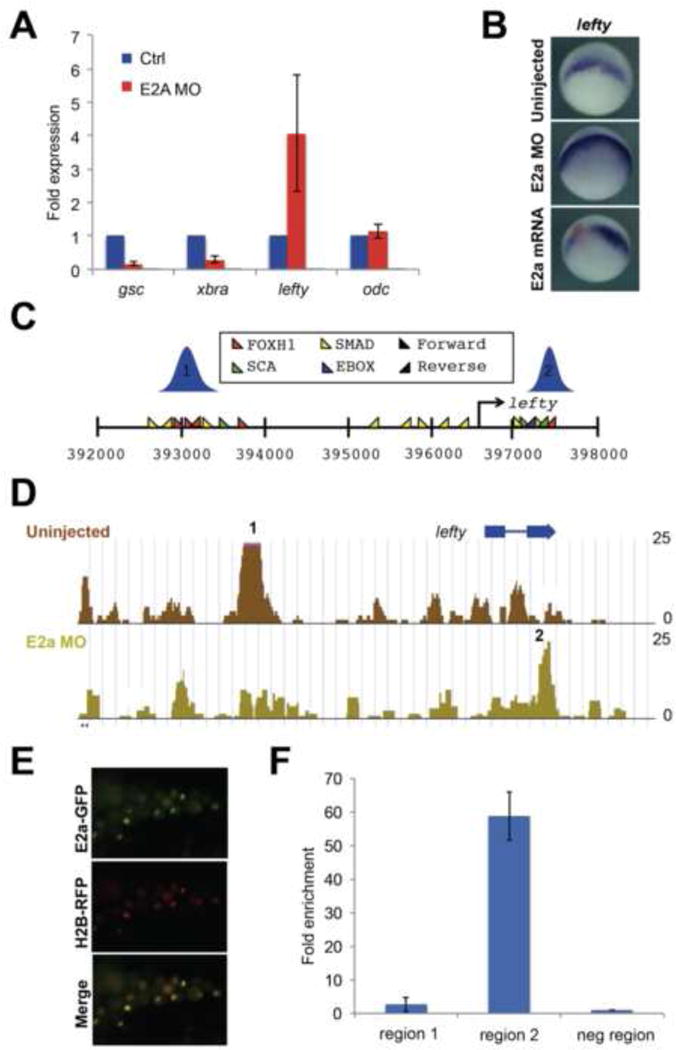
A) qRT-PCR showing upregulation of lefty in E2a depleted embryos, and downregulation of gsc and xbra. odc is shown as a loading control. Error bars represent standard deviations for three biological replicates. B) In situ hybridization for lefty expression at stage 10 in control, E2a depleted, and e2a mRNA injected embryos (red=lacZ lineage tracer). C) Distribution of key transcription factor binding sites near the lefty locus. There are two regions of observed Smad2/3 binding in control embryos, schematized in blue. D) In E2a-depleted embryos, Smad2/3 binding at region 1 is reduced, while binding at region 2 is increased. Fold enrichment over input is shown along the Y axis. E) Embryos were injected at the 2-cell stage with mE2a-GFP and RFP-tagged histone H2b mRNA and monitored for fluorescence at stage 10.5. F) ChIP-qRT-PCR using anti-GFP antibodies in stage 10.5 embryos following mE2a-GFP injection at the 2-cell stage. Y axis represents fold enrichment over a negative control region near the lefty locus. See also Figure S1.
We next investigated whether the misexpression of lefty in response to E2a depletion may be caused by changes in Smad2/3 occupancy at lefty regulatory regions. We find two regions near the lefty locus that contain densely clustered consensus sequences for Foxh1, Smad, Ebox, and SCA motifs (Yoon et al., 2011)(Figure 2C). We find that in control embryos, Smad2/3 binding is usually strongly enriched at the first of these two clusters (“region 1” in Figure 2). In E2a-depleted embryos, Smad2/3 is no longer enriched at region 1, but instead becomes more strongly enriched at region 2 (Figure 2D). We conclude that E2a regulates expression and Smad2/3 positioning at the Nodal target gene and inhibitor lefty, suggesting that E2a could serve to displace Smad2/3 from region 1 to region 2, resulting in higher levels of lefty transcription.
As our results strongly implicate E2a as a transcriptional repressor of lefty, we used a functional tagged E2A construct to examine whether E2a directly associates with these putative lefty regulatory regions. We generated an E2a-GFP fusion construct, in which GFP is fused in frame to the coding region of mouse E2a (mE2a-GFP). We confirmed the functionality of this construct in several ways. First, injection of 500pg mE2a-GFP mRNA results in GFP-positive embryos with fluorescence restricted to the nuclei of injected cells, as expected for a transcription factor (Figure 2E). Western blots using embryo lysates from mE2a-GFP injected embryos react with an anti-GFP antibody at the expected size for this fusion, and mE2a-GFP can rescue the formation of bottle cells in E2a-depleted embryos: the darkly pigmented, apically-constricted cells that form the leading edge of the blastopore (Figure S1A, B). As the mE2a-GFP construct functions similarly to wild type E2A, we used this mE2a-GFP fusion to test whether E2a was enriched at either region 1 or region 2 within the lefty locus. We injected 1000 embryos at the 2-cell stage with 500pg per cell of mE2a-GFP mRNA, raised these embryos to stage 10.5, and performed ChIP using an anti-GFP antibody, followed by quantitative PCR using primers specific for region 1, region 2, or a negative control region (Figure 2C). We found that, while there is a little to no E2a association with region 1, there is very strong enrichment of E2a association at region 2 (Figure 2F). Overall, this suggests that E2a binds directly to region 2, inhibiting the association of Smad2/3 and subsequently downregulating lefty transcription.
E2a is required in mesendoderm cells for blastopore formation
As Lefty is an important regulator of Nodal signaling in mesendoderm and dorsal midline cells, we next examined whether E2A was specifically required for the formation of these cells. Therefore, we targeted E2a morpholinos to dorsal, ventral, animal or vegetal blastomeres, and analyzed the phenotypic consequences of E2a knockdown in these regions. We find that when E2A is depleted from dorsal vegetal cells, the blastopore does not form (78%)(Figure 3A) or is incomplete (18%). Blastopore formation is inhibited or absent in fewer embryos where ventral-vegetal blastomeres (prospective ventral mesoderm and endoderm) are injected with E2a MO (20% absent, 26% incomplete), and normal in most embryos where only animal blastomeres (prospective ectoderm) are injected with E2a MO (92% normal)(Figure 3A). Thus, E2a is primarily required in regions of the embryo that have a high level of Nodal signaling, and particularly those cells that will contribute to dorsal mesendoderm.
Figure 3. Lefty is downstream of E2a in mesoderm induction.

A) Embryos were injected with E2a MO in specific blatomeres as follows: “All”= both blastomeres at the 2-cell stage (N=72); “Dorsal”=2 dorsal blastomeres at the 4-cell stage (N=51); “Ventral”=both ventral blastomeres at the 4-cell stage (N=63); “Animal”=4 animal blastomeres at the 8-cell stage (N=9). B) Embryos were injected in both blastomeres at the 2 cell stage with e2a MO, and/or at the animal pole at the 4 cell stage with xnr1 mRNA. Animal caps were harvested at stage 8 and cultured to stage 10. qPCRs were normalized to whole embryo expression and to odc expression. Error bars represent standard deviations for three biological replicates. C–J) E2a depleted embryos have reduced expression of gsc (C) and xbra (D) compared with uninjected control embryos (A,B). By contrast, Lefty-depleted embryos show increased expression of gsc (E) and xbra (F). Embryos injected with both E2a and Lefty MOs have moderate expression of gsc (G) and xbra (H). See also Figure S2.
E2A opposes Nodal activation of lefty
As E2a is required in regions of the embryo with high levels of endogenous Nodal signaling, we next determined whether E2a requires Nodal signaling to regulate lefty. To this end, we examined E2a depletion in isolated ectodermal explants, which contain no Nodal signaling. We injected E2a MO into the animal pole of the 2-cell stage embryo, isolated animal ectoderm explants at stage 8, cultured these to stage 10.5, and analyzed lefty expression by qRT-PCR. In contrast to E2a depleted whole embryos, lefty is not upregulated in E2a-depeleted ectoderm explants (Figure 3B), suggesting that E2a repression of lefty does not occur in the absence of Nodal signaling.
We then determined if E2a depletion leads to increased expression of lefty in response to Nodal signaling. To this end, we performed a double injection experiment within the prospective ectoderm, in which we first depleted E2a by injecting morpholinos at the two-cell stage, then induced Nodal expression by injecting mRNA for the Nodal ligand xnr1 at the four-cell stage. We then isolated and cultured ectoderm explants from xnr1-injected or E2a MO/xnr1 double-injected embryos, and assayed expression of mesoderm genes and lefty by qRT-PCR. As expected, lefty is induced by overexpression of xnr1 (Figure 3B). In embryos injected with both xnr1 and E2a MO, lefty expression in ectoderm is significantly increased. Thus, E2a depletion enhances lefty expression when Nodal (xnr1) is present, suggesting that Lefty is downstream of both Nodal and E2a.
Lefty is epistatic to E2a depletion in mesoderm induction
To explicitly test if Lefty is downstream of E2a, we examined whether depletion of Lefty is epistatic to depletion of E2a. We injected embryos at the two-cell stage with E2a MO, Lefty MO, or both MOs, raised them to stage 10.5, and analyzed their morphology and gene expression. In E2a depleted embryos, expression of the dorsal mesoderm gene gsc is reduced, as is expression of the pan-mesoderm marker xbra (Figure 3E, F). By contrast, in Lefty depleted embryos, the domain of gsc expression is expanded ventrally, and the xbra expression domain is expanded animally (Figure 3G,H). Surprisingly, when both morpholinos were co-injected at the two-cell stage, we observed recovery of expression for xbra (24/28) and gsc (19/24)(Figure 3I,J). Although the expression of xbra and gsc was restored in E2a/Lefty double knockdown embryos, gastrulation behaviors were still abnormal, with impaired bottle cell formation and failure of blastopore closure (data not shown). These data suggest that a substantial reason for the loss of expression of mesoderm genes in E2a depleted embryos results from upregulation of lefty, which can be rescued by a compensating depletion of lefty. However, these results further suggest that the blastopore formation defects may arise from a lefty-independent role of E2a.
E2a is required for dorsal mesoderm gene activation in ectoderm explants
We next asked whether E2a has a role in gene activation, independent from its role in repression of lefty transcription. While E2a is well known as a transcriptional activator in hematopoiesis (Bradney et al., 2003, Kee, 2009; Chen et al., 2013), two lines of evidence prompted us to investigate whether it was also acting as an activator in the early embryo. First, many well-known Nodal target genes are downregulated in the absence of E2a, but retain Smad2/3 binding (Figure 1 and Table 1). This is not explained well by upregulation of Lefty alone. Lefty acts to sequester Nodal ligands and to antagonize EGF-CFC coreceptors (Chen and Shen, 2004; Cheng et al., 2000; Cheng et al., 2004). Both these mechanisms result in an inhibition of Smad2 phosphorylation and translocation to the nucleus, and therefore, we would expect to see loss of Smad2/3 binding concurrently with downregulation of transcription of these genes. The presence of Smad2/3 in E2a depleted embryos, coupled with a strong transcriptional downregulation of these genes, suggests that E2a might also be necessary as a transcriptional activator. Second, while downregulation of Lefty is sufficient to rescue expression of xbra and gsc, it is not sufficient for rescue of gastrulation morphogenesis. We therefore investigated whether E2a might function to activate dorsal mesoderm gene expression and promote gastrulation morphogenesis through an additional mechanism independent from lefty repression.
We first asked if E2a is necessary or sufficient for gene activation in ectoderm explants, which do not express lefty. To this end, we performed qPCR for expression of representative genes from Table 1 in whole embryos, E2a-depleted embryos, control ectoderm explants, and ectoderm explants injected with e2a mRNA (Figure 4A). This confirmed that E2a is required for expression of xbra, eomes, and epha4 in whole embryos, but revealed that e2a overexpression is not sufficient to induce expression of these genes in ectoderm. As our previous work has shown that E2a binds with Smad at gene regulatory regions in hESC-derived endoderm, we hypothesized that E2a might be required downstream of Nodal to activate gene expression. We therefore tested whether eomes, xbra, or epha4 gene expression in ectoderm depends on both Nodal signaling and E2a. We therefore injected xnr1 mRNA, in the presence or absence of E2a MO, and quantified expression of xbra, eomes and epha4. All three genes are robustly activated in the ectoderm by xnr1 injection, but this activation is significantly reduced in ectoderm that is also depleted of E2a (Figure 4A and Supplemental Figure S3). This demonstrated that E2a is necessary but not sufficient for Nodal-dependent transcriptional activation of these genes. However, as lefty is also expressed in response to xnr1 (Figure 3), this experiment did not fully clarify whether this requirement for E2a was as a direct activator.
Figure 4. E2a is required for dorsal mesoderm gene expression, and binds directly to these target genes.
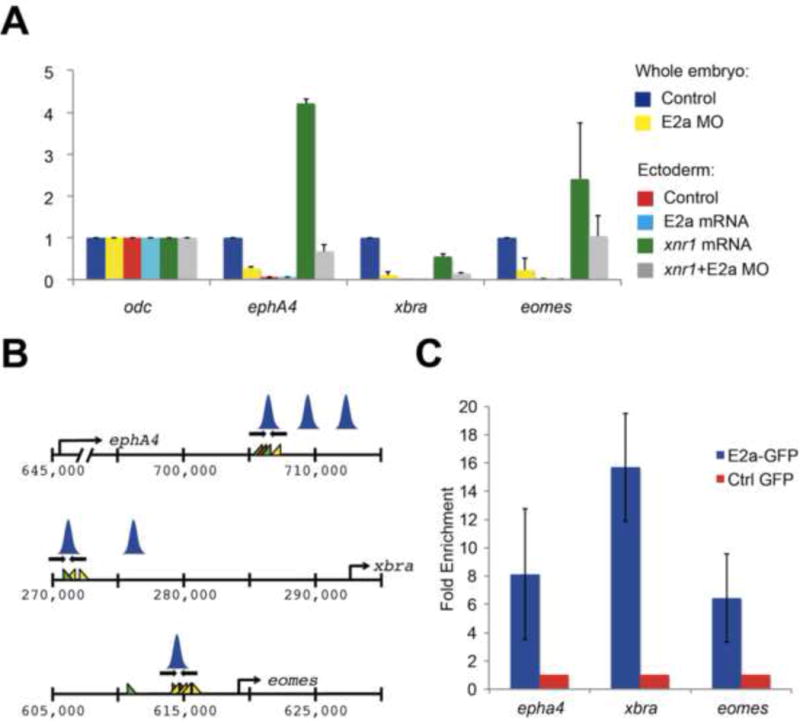
A) qPCRs comparing expression of xbra, eomes, and epha4 in whole stage 10.5 control or e2a-depleted embryos, and in ectoderm explants. Expression levels are normalized to odc and to control ectoderm explants. Error bars represent the standard deviation for three biological replicates, with two technical replicates per biological replicate. Note: although error bars for eomes expression are large, reflecting biological variation between groups of explants, eomes expression was lower in E2a MO+Xnr1 ectoderm than Xnr1 ectoderm in every biological replicate. B) Regulatory regions for eomes, epha4, and xbra were identified from ChIP-Seq analysis in Figure 1, and regulatory regions that are occupied by Smad2/3 in both control and e2a-depleted embryos were analyzed for E2a occupancy. Primers, Smad, FoxH1, and E2a (SCA) binding sites as well as the positions of Smad2/3 peaks are shown as in Figure 2. C) ChIP-qPCR for mE2a-GFP shows significant enrichment at regulatory regions for eomes, xbra, and epha4. qPCRs are normalized to expression in embryos injected with GFP alone, and to expression of an off-peak primer. See also Figure S3.
E2a directly targets regulatory regions of dorsal mesoderm and morphogenesis genes
We next tested explicitly whether dorsal mesoderm genes and morphogenesis genes are targeted directly by E2a. To this end, we first identified putative regulatory regions in genes that require E2a for their expression, but that have intact Smad2/3 binding in the absence of E2a (Table 1). We examined the sequence of regions that are bound by Smad2/3 in both the presence and absence of E2a to identify putative E2a binding sites in these regions (SCA motifs, Yoon et al., 2011). We find that SCA motifs frequently occur within 5kb of these regions. To test whether E2a binds these regions, we performed ChIP-qPCR using mE2a-GFP at putative regulatory regions for three genes that require E2a for transcription: eomes, xbra, and epha4 (Figure 4B). We find that mE2a-GFP is significantly enriched at these regulatory regions relative to ChIP performed with GFP alone (Figure 4C). This suggests that these three genes, and likely others from Table 1, are direct targets of E2a for transcriptional activation.
E2a-EnR fusion proteins repress axial mesoderm development
We next asked whether E2a’s function as a transcriptional activator is required for normal development. We have shown that E2a directly targets the dorsal mesoderm genes xbra and eomes, and the morphogenesis gene epha4, and that E2a is required for Nodal-dependent activation of these genes, but it was not yet clear which early developmental functions were driven by this role of E2a, as opposed to its role in Lefty repression. We were specifically interested in whether E2a acts as an activator in gastrulation morphogenesis, as this process is poorly rescued by inhibition of Lefty in E2a depleted embryos. Notably, several of the 46 Smad2/3 target genes downregulated in E2a depleted embryos (Table 1) are regulators of cytoskeletal changes and cell movements, suggesting these genes might mediate bottle cell formation and gastrulation movements as targets of E2a activation (epha4, as well as cdc42ep4, lpar2, efna1, lpar2, map2k6, pcdh8.2). Overexpression of e2a mRNA has minimal effects on gastrulation, apart from local inhibition of lefty expression (Figure 2 and data not shown). Therefore, to separate E2a’s activator and repressor functions, we made an obligate repressor by fusing E2a to the repressor domain of Engrailed. We then injected 250pg of E2a-EnR or 20ng of E2A MO in both blastomeres at the two-cell stage. We harvested these embryos and stage matched controls at stage 11 and examined their morphology and gene expression. We find that E2a-EnR injected embryos have delayed blastopore closure (Figure 5A) and like E2a depleted embryos, have reduced expression of xbra in injected cells. In contrast to E2a depleted embryos, E2a-EnR injected cells have reduced lefty expression, demonstrating that E2a-EnR represses lefty. Later in development, embryos injected with E2a-EnR have defects in dorsal and axial mesoderm development, with truncated tails and spina bifida. E2a-EnR injected embryos also showed reduced expression of several genes we had found to be downregulated in E2a morphants by RNA-Seq, including the muscle markers myoD and myf5 and the notochord marker foxA1 (Figure 5B). Ventral mesoderm development appeared to be unaffected, as E2a-EnR injected embryos have normal expression of the heart marker nkx2.5. These phenotypic effects are less dramatic than the failure of blastopore formation and loss of mesendoderm specification seen in E2a morphants. We have shown that proper gastrulation morphogenesis and specification of axial mesoderm are inhibited by E2a when acting as a repressor, and reveal that these functions rely instead on E2a acting as an activator. Interestingly, the E2a-EnR repressor fusion closely phenocopies the effects of an Xbra-EnR repressor fusion protein, which also causes a shortened axis and reduced axial mesoderm (Conlon et al., 1996; Kitaguchi et al., 2002). These effects are consistent with our observation that E2a directly targets both early axial mesoderm genes such as eomes and xbra, and early morphogenesis genes such as epha4 (Evren et al., 2014; Park et al., 2011). It also supports observations from our RNA-Seq analysis that E2a is normally required for expression of many axial mesoderm genes, including xbra, myod, myf5, and foxa1, and that E2a-EnR conversely targets these genes for repression. We therefore conclude E2a must usually act to activate expression of genes associated with gastrulation morphogenesis and axial mesoderm development, as well as to repress lefty expression.
Figure 5. E2a fused to the Engrailed repressor domain causes mild blastopore formation defects and loss of axial mesoderm, and synergizes with low doses of lefty mRNA.
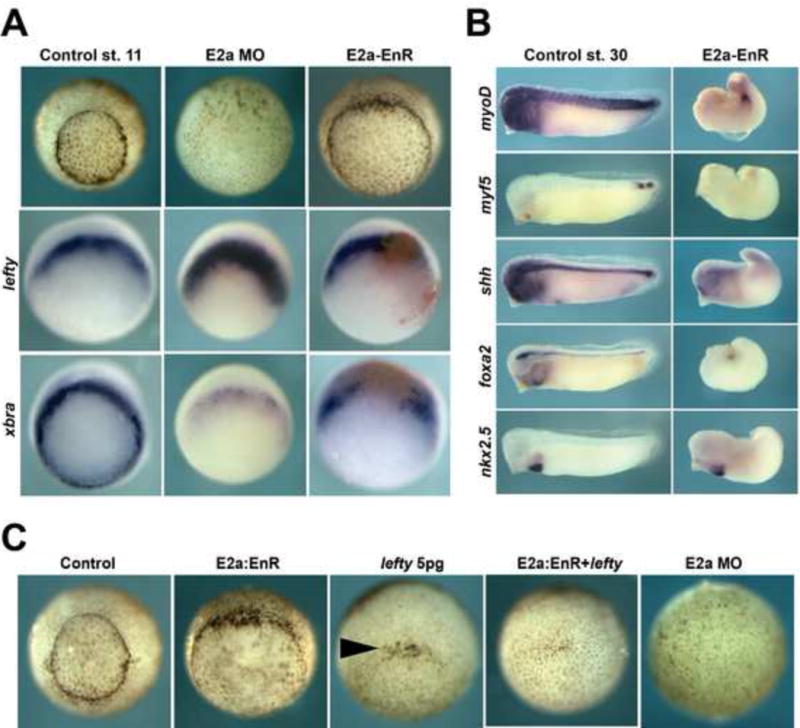
A) Effects of E2a-EnR injection (both cells at the 2-cell stage) at stage 11. Embryos injected with E2a-EnR have delayed blastopore closure relative to uninjected control embryos, but far better blastopore formation than E2a morphants. Stage 10.5 E2a-EnR embryos express more xbra than E2a morphants, but less than control embryos. E2a-EnR injected embryos express less lefty than E2a morphants. B) Effects of E2a-EnR injection (both cells at the 2-cell stage) at stage 30. E2a-EnR injected embryos show reduced expression of muscle markers (myoD, myf5), and notochord markers (shh, foxA2), but normal expression of the heart marker nkx2.5. C) E2a-Enr and 5pg of lefty mRNA each cause only modest effects on blastopore closure, but embryos injected with both mRNAs show a failure of bottle cell formation similar to E2a morphants. Arrowhead indicates bottle cells.
E2a functions as both an activator and a repressor to drive gastrulation
As we have revealed that E2A functions as both an activator and repressor, we next sought to examine whether these two activities could combine to phenocopy the severe phenotype of E2a depleted embryos, in which the blastopore fails to form. To this end, we injected lefty mRNA alone, e2a-enR alone, lefty and e2a-enR together or E2a MO into X. tropicalis embryos and examined the resulting effects on bottle cell formation. We chose 5pg of lefty mRNA as equivalent to the level of lefty expression in E2a depleted embryos (Supplemental Figure S2), and 250pg of e2a-enR mRNA as a dose that robustly represses mesoderm development. We find that this dose of lefty alone causes incomplete inhibition of bottle cell formation and that E2a-EnR, while causing a delay in gastrulation, does not affect bottle cell formation (Figure 5C). Surprisingly, we find lefty and e2a-enR synergize to repress all bottle cell formation, strongly suggesting that each of these contributes partially to E2a’s role in bottle cell formation (27/29 embryos with no bottle cells)(Figure 5C). We conclude that E2a has two functions: lefty repression and dorsal gene activation, and that inhibiting both functions is necessary for full recapitulation of the E2a loss-of-function phenotype, as inhibition of either function alone leads to more modest effects on gastrulation.
Discussion
Multiprotein transcriptional complexes can have diverse functions depending on their conformation and genomic position. The Smad2/3 complex mediates a wide variety of cellular and developmental events, including mesendoderm development, left/right axis specification, cell division, metastasis, and pluripotency (Schier, 2009; Shen, 2007). Smad2/3 associates with many potential cofactors, but the effect that these cofactors have on diversifying the roles of Smad2/3 are largely unknown, especially in the context of the in vivo embryo. Here we find evidence that a Smad2/3 transcriptional cofactor, E2a, regulates early development in two ways: through transcriptional repression of the Nodal inhibitor lefty and through transcriptional activation of dorsal mesoderm and morphogenesis genes.
E2a plays an essential role in mesendoderm development by repressing transcription of the Nodal inhibitor Lefty. As a consequence, in E2a depleted embryos, Lefty is dramatically upregulated, leading to a failure of mesendoderm cell fate specification. We find that E2a achieves this role by directing the position of Smad2/3 at the lefty genomic locus. We find that Smad2/3 can occupy one of two positions at the lefty locus, and in the absence of E2a, Smad2/3 is displaced from its usual position to one nearer the lefty transcriptional start site. As a result, lefty transcription is dramatically upregulated (Figure 6A). This upregulation has strongest effect in dorsal vegetal cells, and requires endogenous Nodal signals, demonstrating that the effect of E2a is specifically through the Nodal signaling pathway, through the positioning of Smad2/3. We favor a mechanism by which E2a excludes Smad2/3 from associating with region 2 of lefty, such that it fails to associate with transcriptional coactivators. This may be a more general mechanism at other sites where Smad2/3 positioning is gained or shifted in the absence of E2a. In hESCs, pluripotency is maintained by the repressive activity of a complex made up of Taz/Yap/Tead4, Oct4, and Smad2/3 (TSO) at mesendoderm gene enhancers, which is relieved and switched to activation in the presence of Activin (Beyer et al., 2013). Our data demonstrate that E2a plays a similar role in governing the switch between repression and activation at some enhancers in the early embryo. In the absence of E2a, many genes (3000+) are upregulated, suggesting that E2a acts broadly as a transcriptional repressor, and many genes gain Smad2/3 binding (571) or have shifted Smad2/3 binding (192). At some loci, E2a may also act more directly as a repressor by recruiting corepressors, as does in association with ETO/MTG corepressors in hematopoiesis (Gow et al., 2014; Guo et al., 2009; Zhang et al., 2004). However, Lefty is by far the most critical target of E2a repression for early cell fate specification, as downregulation of Lefty is sufficient to rescue expression of mesoderm genes in E2a depleted embryos. An intriguing remaining question is why E2a competes with Smad2/3 binding at this locus, given that E2a and Smad2/3 are able to directly associate, and co-occupy transcriptional regulatory regions at other genes (Figure 4 and Yoon et al., 2011). One potential explanation is that E2a and Smad may have context-specific associations with other bHLH proteins, such as HEB, which may influence either Smad binding choice or coactivator recruitment. While our earlier work suggests that HEB is not required for early embryonic development, it is expressed throughout the mesoderm at gastrulation (Yoon et al., 2011), and may play a role redundantly with E2a.
Figure 6. Model of Smad2/3 and E2a interactions in transcriptional regulation.
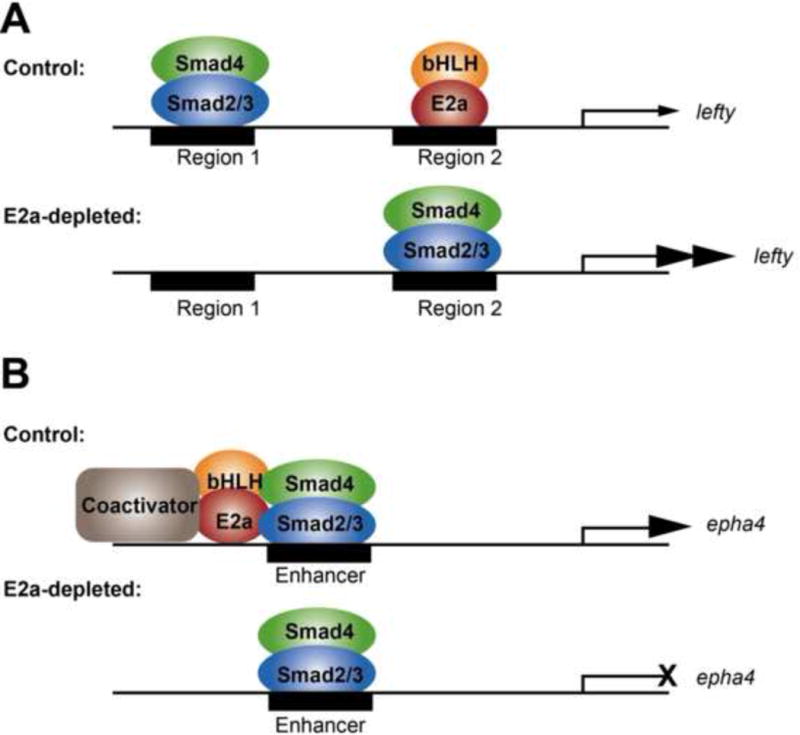
A) At the lefty locus. In control embryos, Smad2/3 normally occupies region 1, while E2a normally occupies region 2. Transcription of lefty is weak. In the absence of E2a, Smad2/3 moves to preferentially occupy region 2, and transcription of lefty is upregulated. B) At dorsal mesoderm loci. In control embryos, Smad2/3 occupancy at enhancers is coupled by E2a to coactivators, and transcription of the target gene (epha4 is shown as an example) is robust. In the absence of E2a, the coupling of Smad2/3 to transcriptional coactivators is lost, and transcription of the target gene is reduced.
E2a is also required at 46 critical early developmental Smad2/3 target genes, not for Smad2/3 positioning, but for their transcriptional activation (Table 1). This role is distinct from repression of Lefty. These genes include many well-known transcription factors regulating early mesendoderm development, as well as several regulators of cell shape changes and movements that act in gastrulation morphogenesis. This is substantiated by ChIP of E2a, which demonstrates that E2a binds directly to the regulatory regions of several of these genes including eomes, xbra, and epha4. At these loci, Smad2/3 is positioned normally, but transcription is inhibited, demonstrating that E2a is needed more directly for their activation. In hESCs, the localization of E2a to mesendoderm genes is strongly dependent on Nodal signaling, suggesting that in this activator capacity, E2a requires Smad2/3 to properly target mesendoderm genes (Yoon et al., 2011). The activity of E2a as a transcriptional activator has dramatic consequences for gastrulation morphogenesis. Notably, epha4 has recently been shown to regulate dorsal mesoderm involution through xbra (Evren et al., 2014), and so the regulation of these two genes by E2a explains why the loss of E2a has dramatic consequences on gastrulation movements. When E2a is forced to lose its activator role by becoming a repressor, gastrulation movements and axial mesoderm formation are perturbed. For these genes, we hypothesize that E2a is critical for coupling Smad2/3 binding to transcriptional activation (Figure 6B). In other cell types, E2a is essential for recruitment of transcriptional coactivators like CPB and p300 (Bayly et al., 2004; Bradney et al., 2003), and we consider it likely that in the embryo, E2a also acts as a bridge between Smad2/3 and these coactivators. Some genes, like xbra, may be regulated by E2a both through its repression of Lefty and through direct transcriptional activation. Thus, E2a represents another layer in the finely balanced regulation of Nodal signaling and its targets in the early embryo.
Overall, we demonstrate that E2a has critical functions, both as a repressor and activator, in the localization and function of the Smad2/3 transcriptional complex. These findings highlight that perturbation of a single component of a transcriptional complex can have dramatic consequences for behavior of the complex in vivo. The Smad2/3 complex differentially regulates many genes at gastrulation, with different effects in distinct cell types. This study reveals that E2a strongly influences both binding site choice of the Smad2/3 complex throughout the genome, and the consequences of Smad2/3 binding on gene expression. By preventing strong transcription of lefty and by allowing transcriptional activation of other Nodal target genes, E2a has a profound effect on many aspects of early development regulated by Nodal signaling.
Experimental Procedures
GEO accession number for all deep sequencing data: GSE56169.
Xenopus tropicalis embryo culture
Xenopus tropicalis husbandry procedures were performed according to NIH and IACUC approved protocols. Embryos were generated by natural mating as described (Yoon et al., 2011; Khokha et al., 2002). Staging was assessed according to (Niewkoop and Faber, 1967). For microsurgery details, see Supplemental experimental procedures.
RNA-Seq library preparation
Total RNA was isolated by collecting duplicate cohorts of 100 embryos for control and E2a-depleted embryos. 20ug of starting total RNA was used for each library. The Illumina HiSeq platform was used for sequencing. Reads were aligned to the X. tropicalis 4.1 genome version and corresponding annotation. FPKM normalization and downstream differential expression analysis was carried out as described (Tan et al., 2013). See supplemental experimental procedures for additional details.
ChIP-Seq library preparation
Duplicate cohorts of 200 control or E2a-depleted embryos were fixed for ChIP as described (Blythe et al., 2009; Wills et al., 2014). Immunoprecipitation was carried out using anti-Smad2/3 (Santa Cruz Biotechnology sc-8332) or anti-GFP antibodies (life technologies #A11122, lot 1296649). Sequencing and analysis was carried out on the Illumina hi-Seq platform as described (Wills et al., 2014). In brief, libraries were aligned to the X. tropicalis 4.1 genome version using bwa. Peaks were called using MACS2. Peaks were assigned to nearest neighboring genes using HOMER, using the xentro2 annotation tool. Comparisons of peak positions were carried out using BedTools. Analyses comparing genes implicated in ChIP-Seq and genes implicated in RNA-Seq were done using in house Perl scripts or Unix tools.
See supplemental experimental procedures for ChIP-qPCR primers.
Microinjection
Morpholino oligonucleotide design and microinjection
E2A morpholino oligonucleotide sequence:
TGTCATCCTCTGCTGTTGATTCATT (Yoon, Wills et al., 2011)
Lefty morpholino oligonucleotide sequence:
CATGTGCTAGTGACACCCATCTTGC
mRNA synthesis and injection
Linearized plasmid DNA was transcribed with Sp6 polymerase, using NTP mix containing 5′ methyl-G cap (mmessage mmachine, Ambion). mRNA was precipitated first with LiCl, then re-precipitated using ammonium acetate, quantitated, resuspended and stored at −80°C. Injections were performed at stages and with amounts specified in each experiment.
mRNA plasmids and amounts used:
X. tropicalis E2a: IMAGE 7660124, 250pg per blastomere
Mouse E2a: IMAGE clone 2631291, 500pg per blastomere
E2a:GFP (see below for clone details), 250pg per blastomere
E2a:EnR (see below for clone details), 250pg per blastomere
X. laevis Xnr1: 100pg per embryo
Whole mount in situ hybridization
Embryos were developed to the desired stage and then fixed in MEMFA for 2–6 hours at room temperature or overnight at 4°C. X. tropicalis multi-basket in situ hybridization protocols were followed as described in (Khokha et al., 2002). For plasmids used for in situ hybridization probes see Supplemental experimental procedures.
Quantitative RT-PCR
Total RNA was isolated from injected or uninjected embryos (3 per experiment) or animal caps (10 per experiment). RNA was treated with DNAse 1 (Invitrogen catalog # 18068-015), and cDNA synthesized using SuperScript III reverse transcriptase (Invitrogen). Quantitative RT-PCR analysis was performed using BioRad SYBR green labeling system and the BioRad iCycler PCR machine, Sybr Green mix, and analysis software. For PCR details and primers, see Supplemental experimental procedures.
E2a Fusion protein constructs
Mouse E2a-GFP fusion was made by fusing the carboxy terminus of mouse E2a to the amino terminus of eGFP on a CS2 backbone. X. tropicalis E2a-EnR fusion was made by fusing the Engrailed repressor domain to the carboxy terminus of X. tropicalis E2a, on a pCMV Sport6 backbone. For details see Supplemental experimental procedures.
Supplementary Material
Acknowledgments
We thank Kin-Fai Au (Stanford), Rakhi Gupta (Stanford), Duygu Ucar (Stanford and Jackson Laboratory), and Taejoon Kwon (UT Austin) for assistance with bioinformatics software and analysis. We thank Eduardo Gonzalez-Maldonado and Rakhi Gupta for assistance with preparation of Smad2/3 ChIP-SEQ libraries, and for ChIP-PCR primers for eomes. This project was supported in part by NRSA fellowship 1F32 DK089643-01 to AW and by grants NIH R01GM103787, NIH R01HD076839, and NIH R01GM095346 to JB. Sequencing data associated with this project are deposited to NCBI under Geo accession number GSE56169.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing interests.
References
- Bayly R, Chuen L, Currie RA, Hyndman BD, Casselman R, Blobel GA, LeBrun DP. E2A-PBX1 interacts directly with the KIX domain of CBP/p300 in the induction of proliferation in primary hematopoietic cells. J Biol Chem. 2004;279:55362–55371. doi: 10.1074/jbc.M408654200. [DOI] [PubMed] [Google Scholar]
- Beyer TA, Weiss A, Khomchuk Y, Huang K, Ogunjimi AA, Varelas X, Wrana JL. Switch enhancers interpret TGF-beta and Hippo signaling to control cell fate in human embryonic stem cells. Cell Rep. 2013;5:1611–1624. doi: 10.1016/j.celrep.2013.11.021. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradney C, Hjelmeland M, Komatsu Y, Yoshida M, Yao TP, Zhuang Y. Regulation of E2A activities by histone acetyltransferases in B lymphocyte development. J Biol Chem. 2003;278:2370–2376. doi: 10.1074/jbc.M211464200. [DOI] [PubMed] [Google Scholar]
- Branford WW, Yost HJ. Lefty-dependent inhibition of Nodal- and Wnt-responsive organizer gene expression is essential for normal gastrulation. Curr Biol. 2002;12:2136–2141. doi: 10.1016/s0960-9822(02)01360-x. [DOI] [PubMed] [Google Scholar]
- Chen C, Shen MM. Two modes by which Lefty proteins inhibit nodal signaling. Curr Biol. 2004;14:618–624. doi: 10.1016/j.cub.2004.02.042. [DOI] [PubMed] [Google Scholar]
- Chen WY, Zhang J, Geng H, Du Z, Nakadai T, Roeder RG. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev. 2013;27:1596–1609. doi: 10.1101/gad.216192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AM, Thisse B, Thisse C, Wright CV. The lefty-related factor Xatv acts as a feedback inhibitor of nodal signaling in mesoderm induction and L-R axis development in xenopus. Development. 2000;127:1049–1061. doi: 10.1242/dev.127.5.1049. [DOI] [PubMed] [Google Scholar]
- Cheng SK, Olale F, Brivanlou AH, Schier AF. Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2004;2:E30. doi: 10.1371/journal.pbio.0020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Evren S, Wen JW, Luu O, Damm EW, Nagel M, Winklbauer R. EphA4-dependent Brachyury expression is required for dorsal mesoderm involution in the Xenopus gastrula. Development. 2014 doi: 10.1242/dev.111880. [DOI] [PubMed] [Google Scholar]
- Germain S, Howell M, Esslemont GM, Hill CS. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 2000;14:435–451. [PMC free article] [PubMed] [Google Scholar]
- Gow CH, Guo C, Wang D, Hu Q, Zhang J. Differential involvement of E2A-corepressor interactions in distinct leukemogenic pathways. Nucleic Acids Res. 2014;42:137–152. doi: 10.1093/nar/gkt855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Hu Q, Yan C, Zhang J. Multivalent binding of the ETO corepressor to E proteins facilitates dual repression controls targeting chromatin and the basal transcription machinery. Mol Cell Biol. 2009;29:2644–2657. doi: 10.1128/MCB.00073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Wills A, Ucar D, Baker J. Developmental enhancers are marked independently of zygotic Nodal signals in Xenopus. Dev Biol. 2014;395:38–49. doi: 10.1016/j.ydbio.2014.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Kitaguchi T, Mizugishi K, Hatayama M, Aruga J, Mikoshiba K. Xenopus Brachyury regulates mesodermal expression of Zic3, a gene controlling left-right asymmetry. Dev Growth Differ. 2002;44:55–61. doi: 10.1046/j.1440-169x.2002.00624.x. [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, et al. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meno C, Gritsman K, Ohishi S, Ohfuji Y, Heckscher E, Mochida K, Shimono A, Kondoh H, Talbot WS, Robertson EJ, et al. Mouse Lefty2 and zebrafish antivin are feedback inhibitors of nodal signaling during vertebrate gastrulation. Mol Cell. 1999;4:287–298. doi: 10.1016/s1097-2765(00)80331-7. [DOI] [PubMed] [Google Scholar]
- Niewkoop PD, Faber J. Normal Table of Xenopus laevis. Amsterdam: North Holland Publishing Company; 1967. reprinted 1994 Garland Publishing, New York. [Google Scholar]
- Park EC, Cho GS, Kim GH, Choi SC, Han JK. The involvement of Eph-Ephrin signaling in tissue separation and convergence during Xenopus gastrulation movements. Dev Biol. 2011;350:441–450. doi: 10.1016/j.ydbio.2010.12.012. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal morphogens. Cold Spring Harb Perspect Biol. 2009;1:a003459. doi: 10.1101/cshperspect.a003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MM. Nodal signaling: developmental roles and regulation. Development. 2007;134:1023–1034. doi: 10.1242/dev.000166. [DOI] [PubMed] [Google Scholar]
- Slagle CE, Aoki T, Burdine RD. Nodal-dependent mesendoderm specification requires the combinatorial activities of FoxH1 and Eomesodermin. PLoS Genet. 2011;7:e1002072. doi: 10.1371/journal.pgen.1002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–216. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AK, Arnold SJ, Trotter MW, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AE, Gupta R, Chuong E, Baker JC. Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis. Methods. 2013 doi: 10.1016/j.ymeth.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills AE, Gupta R, Chuong E, Baker JC. Chromatin immunoprecipitation and deep sequencing in Xenopus tropicalis and Xenopus laevis. Methods. 2014;66:410–421. doi: 10.1016/j.ymeth.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Crabtree GR. ATP-dependent chromatin remodeling in neural development. Curr Opin Neurobiol. 2009;19:120–126. doi: 10.1016/j.conb.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SJ, Wills AE, Chuong E, Gupta R, Baker JC. HEB and E2A function as SMAD/FOXH1 cofactors. Genes Dev. 2011;25:1654–1661. doi: 10.1101/gad.16800511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, Roeder RG. E protein silencing by the leukemogenic AML1-ETO fusion protein. Science. 2004;305:1286–1289. doi: 10.1126/science.1097937. [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


