Induced pluripotent stem cells (iPSCs) derived from patients with types 1, 2, and 3 Gaucher disease (GD) were differentiated to CD34+/CD45+/CD43+/CD143+ hematopoietic progenitor cells and their developmental potential was examined. The results suggest that GD hematopoietic progenitors have decreased erythroid potential as well as increased and aberrant myelopoiesis. Abnormal myelopoiesis might contribute to the pathological properties of Gaucher macrophages, central to GD manifestations. The hematopoietic developmental defects observed reflect hematologic abnormalities in GD patients, demonstrating the utility of GD-iPSCs for modeling this disease.
Keywords: Induced pluripotent stem cells, Hematopoiesis, Myeloid cells, Monocyte, Stem/progenitor cell, Hematopoietic stem cells, Erythropoiesis
Abstract
Gaucher disease (GD) is the most common lysosomal storage disease resulting from mutations in the lysosomal enzyme glucocerebrosidase (GCase). The hematopoietic abnormalities in GD include the presence of characteristic Gaucher macrophages that infiltrate patient tissues and cytopenias. At present, it is not clear whether these cytopenias are secondary to the pathological activity of Gaucher cells or a direct effect of GCase deficiency on hematopoietic development. To address this question, we differentiated induced pluripotent stem cells (iPSCs) derived from patients with types 1, 2, and 3 GD to CD34+/CD45+/CD43+/CD143+ hematopoietic progenitor cells (HPCs) and examined their developmental potential. The formation of GD-HPCs was unaffected. However, these progenitors demonstrated a skewed lineage commitment, with increased myeloid differentiation and decreased erythroid differentiation and maturation. Interestingly, myeloid colony-formation assays revealed that GD-HPCs, but not control-HPCs, gave rise to adherent, macrophage-like cells, another indication of abnormal myelopoiesis. The extent of these hematologic abnormalities correlated with the severity of the GCase mutations. All the phenotypic abnormalities of GD-HPCs observed were reversed by incubation with recombinant GCase, indicating that these developmental defects were caused by the mutated GCase. Our results show that GCase deficiency directly impairs hematopoietic development. Additionally, our results suggest that aberrant myelopoiesis might contribute to the pathological properties of Gaucher macrophages, which are central to GD manifestations. The hematopoietic developmental defects we observed reflect hematologic abnormalities in patients with GD, demonstrating the utility of GD-iPSCs for modeling this disease.
Significance
This study showed that hematopoietic progenitors from patients with Gaucher disease (GD) have intrinsic developmental abnormalities that reflect characteristic clinical manifestations. These abnormalities include decreased erythroid potential and abnormal myelopoiesis. GD hematopoietic progenitors gave rise to aberrant macrophage-like cells, suggesting that abnormal myelopoiesis may contribute to the pathological properties of Gaucher macrophages. All the hematopoietic abnormalities observed were reversed by incubation with recombinant glucocerebrosidase, which is used to treat patients with type 1 GD. The results suggest that enzyme replacement therapy could help normalize clinical parameters in these patients, not only through recombinant glucocerebrosidase uptake by Gaucher macrophages, which are the intended target, but also potentially by acting directly on hematopoietic progenitors. The results shown here provide new insights into the etiology of GD hematopoietic abnormalities, and highlight the utility of GD iPSC for modeling the disease and therapeutic development.
Introduction
Gaucher disease (GD) is the most common of the lysosomal storage disorders, which collectively account for ∼1:5,000 live births [1]. The causative gene in GD encodes the lysosomal membrane-associated enzyme glucocerebrosidase (GCase). Mutations in GCase lead to enzyme deficiency and the accumulation of its glucosyl sphingolipid substrates in lysosomes [1–3]. GD is classified into three distinct clinical subtypes based on the presence and severity of neurological involvement. Type 1 GD is the most common and mildest form of the disease, with patients presenting with hepatosplenomegaly, bone disease, and hematologic abnormalities. In addition to these visceral symptoms, patients with types 2 and 3 GD also present with neurological manifestations. Type 2 is the most severe, acute infantile neuronopathic form, and type 3 is a subacute, chronic neuronopathic form [4].
The most distinct hallmark of GD is the presence of Gaucher cells [5]. These cells are lipid-engorged macrophages [6–8] that infiltrate patient tissues, including the bone marrow, spleen, and liver. Gaucher cells are erythrophagocytic and often have remnants of red blood cells (RBCs) owing to their inability to digest cell membrane lipids [8–10]. The infiltration of these cells into patient tissues is believed to be central to the visceral and hematopoietic defects in GD [4, 11]. Other hematopoietic involvement in GD includes thrombocytopenia, which affects 60% of patients, and anemia, affecting 36% of patients [11, 12]. In addition to anemia, abnormal RBC morphology [13, 14] and aggregation [14, 15] have been described. GD patients also exhibit an increased risk of developing multiple myeloma [11, 16], and it has been proposed that this is due to increased production of cytokines by Gaucher cells [11, 17, 18]. The molecular mechanisms leading to these hematopoietic manifestations are currently poorly understood. In the present study, we sought to examine whether GCase deficiency causes intrinsic developmental defects that might contribute to the hematopoietic abnormalities seen in patients with GD.
The difficulty in obtaining hematopoietic progenitors cells (HPCs) from GD patients, in particular from pediatric populations with types 2 and 3 GD, has limited investigations into whether GCase enzyme deficiency directly impairs the developmental potential of hematopoietic progenitors. To overcome these limitations, we used induced pluripotent stem cells (iPSCs) derived from GD patients (GD-iPSCs) as an in vitro disease model. iPSCs generated through the introduction of transcription factors identified as master regulators of pluripotency [19, 20] offer a unique opportunity for in vitro disease modeling (reviewed in [21–24]). Using GD-iPSCs from patients with types 1, 2, and 3 GD, we recently showed that GD-iPSC-derived macrophages exhibit a striking delay in their ability to clear phagocytosed RBCs, a characteristic hallmark of GD. We also found that the extent of this defect correlated with the severity of the mutations and that this phenotype was completely reversed by incubation with recombinant GCase (rGCase) [10].
In the present study, we found that iPSC-derived HPCs from patients representing all three clinical subtypes of GD (GD-HPCs) have intrinsic developmental abnormalities. GD-HPCs exhibited enhanced myeloid differentiation and gave rise to abnormal macrophage-like cells, strongly suggesting that GD myelopoiesis is compromised. GD-HPCs also exhibited deficient erythroid differentiation and maturation, which may contribute to the cytopenias observed in patients. Restoration of GCase activity in GD-HPCs through incubation with rGCase corrected the erythroid and myeloid differentiation abnormalities, including abrogating the appearance of abnormal macrophage-like cells. These findings offer unique insights into the etiology of the hematopoietic abnormalities caused by GCase deficiency and highlight the utility of iPSC-derived HPCs for modeling GD and their potential for drug discovery.
Materials and Methods
Cells
The human control and type 1, 2, and 3 GD-iPSC lines used in the present study are listed in supplemental online Table 1. All these iPSC lines have been previously described [10, 18]. They include fibroblast-derived iPSCs from a patient with type 1 GD (N370S/N370S), a patient with type 2 GD (W184R/D409H), another patient with type 2 GD (L444P/RecNci1), and a patient with type 3 GD (L444P/L444P). Control iPSC lines included iPS-DF4-7T.A cells purchased from WiCell Repository (Madison, WI, http://www.wicell.org), and MJ, an iPSC line derived from foreskin fibroblasts. DR4 mouse embryonic fibroblasts (MEFs) were obtained from 13.5E embryos of DR4 male mice (The Jackson Laboratory, Bar Harbor, ME, http://www.jax.org) [25] and CF1 female mice (Charles River Laboratories, Frederick, MD, http://www.criver.com), and maintained in fibroblast culture medium. The iPSC lines were grown on irradiated DR4 MEFs in standard iPSC medium (Dulbecco’s modified Eagle’s medium-F12 [Invitrogen, Carlsbad, CA, http://www.lifetechnologies.com], 20% [vol/vol] Knockout Serum Replacement [Invitrogen], 1% [vol/vol] nonessential amino acids, 0.5% [vol/vol] l-glutamine, 0.5% [vol/vol] penicillin/streptomycin, 0.1 mM β-mercaptoethanol, and 16–30 ng/ml basic fibroblast growth factor [bFGF; Stemgent, Cambridge, MA, http://www.stemgent.com). The iPSC lines were passaged every 5–7 days by treatment with 2 mg/ml collagenase type IV (Invitrogen) and cell scraping. All the work with the human iPSC lines described in the present study was performed with approval from the institutional review board and embryonic stem cell research oversight committees.
Hematopoietic Differentiation
Control and GD-iPSCs were differentiated into hematopoietic cells, as previously described [26] and diagrammed in supplemental online Figure 1. For embryoid body (EB) formation, the iPSC lines were placed on adaptation medium for 24 hours. The adaptation medium composition was StemSpanSFEM (serum-free expansion medium; StemCell Technologies, Vancouver, BC, Canada, http://www.stemcell.com), 15% (vol/vol) ES-Cult fetal bovine serum (FBS; StemCell Technologies), 50 μg/ml ascorbic acid, 1% (vol/vol) ExCyte (EMD Millipore, Billerica, MA, http://www.emdmillipore.com), 1% (vol/vol) penicillin/streptomycin (Invitrogen), 0.5% (vol/vol) insulin-transferrin-selenium-ethanolamine (Life Technologies, Carlsbad, CA, http://www.lifetechnologies.com). After 24 hours, the iPSCs were detached from the plates by treatment with 2 mg/ml Dispase (Invitrogen). The detached cells were resuspended in adaptation medium and added to semisolid methylcellulose medium in ultra-low attachment plates (Corning, Corning, NY, http://www.corning.com). The methylcellulose medium composition was MethoCult H4236 (StemCell Technologies), 15% (vol/vol) ES-Cult FBS (StemCell Technologies), 3.5% (vol/vol) PFHM II (Life Technologies), 50 μg/ml ascorbic acid, and 0.5% (vol/vol) ExCyte (EMD Millipore). EBs were collected on day 3 and resuspended in liquid differentiation medium (StemPro34 [Life Technologies], 1% [vol/vol] l-glutamine, and 0.4 mM 1-Thioglycerol [Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). The medium was supplemented with the following cytokines: 50 ng/ml bone morphogenetic protein 4 (Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com), 50 ng/ml vascular endothelial growth factor A (VEGFa; Miltenyi Biotec), 50 ng/ml basic fibroblast growth factor (bFGF; Stemgent), and 5 μg/ml heparan sulfate (Sigma-Aldrich). The culture medium was replaced on day 6. On day 8, the EBs were collected and disaggregated using 1 mg/ml collagenase type IV (Invitrogen), and 0.5–1.0 × 105 EB cells per well in 6-well plates or 5.0–9.0 × 105 EB cells per T75 culture flask were plated. Before plating the EB cells, wells and flasks were coated with 10 μg/ml fibronectin (Life Technologies) overnight at 4°C. The plated cells were cultured in complete endothelial growth medium-2 (EGM-2 BulletKit; Lonza, Walkersville, MD, http://www.lonza.com) supplemented with 25 ng/ml VEGFa (Miltenyi Biotec). EGM-2 and VEGFa supplement were replaced the next day and every 2 days thereafter. After 3 days in EGM-2/VEGFa culture, nonadherent HPCs arose from clusters of cobblestone-like adherent cells. These nonadherent cells were collected on days 3, 5, and 7 of EGM-2/VEGFa culture and analyzed by flow cytometry, or were collected on day 3 and plated for methylcellulose hematopoietic colony formation assays, as described in the next section.
Hematopoietic Colony-Forming Assays
Methylcellulose colony formation assays were performed to determine hematopoietic progenitor frequency, as previously described [26]. Suspension HPCs collected on day 3 of EGM-2/VEGFa culture were resuspended in StemSpanSFEM medium (StemCell Technologies), and added to MethoCult serum-free methylcellulose medium (StemCell Technologies). Next, 4 × 104 cells per 35-mm gridded culture dish were seeded in 1.5 ml of methylcellulose. Granulocyte/macrophage progenitors (colony-forming unit granulocyte macrophage [CFU-GM], CFU-G, and CFU-M) were assayed in MethoCult SF H4536, and erythroid progenitors (CFU-E) were assayed in MethoCult SF H4236 supplemented with 3 U/ml erythropoietin (EPO; R&D Systems, Minneapolis, MN, http://www.rndsystems.com) and 10 ng/ml each of interleukin-6 (IL-6) and IL-3 (R&D Systems). After 8–10 days, the cultures received an additional 1 ml of the corresponding methylcellulose. Hematopoietic colony formation was scored at 16–20 days of culture, and cells were collected for flow cytometric analysis at this time. The colony counts were scored and averaged between duplicates, and the experiments were repeated independently at least three times.
Flow Cytometry
For flow cytometric analysis, HPCs from EGM-2/VEGFa culture were collected on days 3, 5, and 7, and hematopoietic colonies grown in methylcellulose were washed with phosphate-buffered saline (PBS) and collected between days 16 and 20. Next, 0.5–1.0 × 105 cells were resuspended in 100 μl of 5% FBS/PBS and incubated for 20 minutes on ice in the dark with the indicated fluorochrome-conjugated antibodies listed, singly or in dual combination. For hemoglobin expression analysis, a cell permeabilization kit (catalog no. GAS004; Invitrogen) was used per the manufacturer’s instructions. Isotype-matched control antibodies were used to control for nonspecific binding. Spectral compensation was achieved using CaliBRITE Beads (eBioscience, Inc., San Diego, CA, http://www.ebioscience.com), singly stained for each fluorochrome according to the manufacturer’s instructions. The cells were washed with PBS, centrifuged, and resuspended in PBS for data acquisition using a BD LSRII flow cytometer (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com). At least 10,000 events were acquired for each tube and analyzed using FlowJo software (Tree Star, Inc., Ashland, OR, http://www.treestar.com).
Antibodies for Flow Cytometry
The following antibodies were used for flow cytometric analysis: CD34-PE (catalog no. 555822), CD45-APC (catalog no. 555485), CD43-fluorescein isothiocyanate (FITC; catalog no. 555475), CD143/ACE/BB9 (catalog no. 557929), CD14-APC (catalog no. 555399), CD71-APC (catalog no. 551374), CD235a-PE (catalog no. 555570), and hemoglobin-ε-FITC (catalog no. 552829). All the antibodies were purchased from BD Biosciences.
May-Grünwald-Giemsa Stain
The methylcellulose was washed out with PBS, and the adherent cell clusters from the myeloid colony formation assays were stained with May-Grünwald (MG 500)-Giemsa (G-9641; Sigma-Aldrich) according to the manufacturer’s instructions.
Immunofluorescence Analysis
The methylcellulose was washed out with PBS. Adherent cell clusters from myeloid colony formation assays were fixed with 4% (vol/vol) paraformaldehyde for 15 minutes, and blocked in PBS containing 8% FBS/0.05% sodium azide. The cells were stained with FITC-CD14 (catalog no. sc-1182; Santa Cruz Biotechnology Inc., Santa Cruz, CA, http://www.scbt.com) at a concentration of 1:50 in PBS/8% FBS/0.05% sodium azide with 2 mg/ml saponin for 1 hour. The cell nuclei were stained with 4′6-diamidino-2-phenylindole-containing mounting medium (Vectashield; Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com). FITC-isotype (catalog no. 555748; BD Biosciences) was used as a negative control.
Imaging
Nonfluorescent images were captured using an inverted Nikon Eclipse TE-2000 microscope with Nikon Imaging Systems (NIS)-Elements AR 3.0 collection software (Nikon, Tokyo, Japan, http://www.nikon.com). Fluorescent images were captured using an inverted Nikon Eclipse TE-2000S microscope with NIS-Elements BR 3.1 collection software. ImageJ64 software (NIH, Bethesda, MD) was used to compile the fluorescent overlay images.
Treatment With recombinant GCase
For treatment with recombinant human GCase, EGM-2/VEGFa cultures were supplemented with 0.24 U/ml Cerezyme (Genzyme, Cambridge, MA, http://www.genzyme.com), and rGCase was replenished every other day. The same concentration of rGCase was also added to methylcellulose colony formation assays and replenished after 8–10 days. Cerezyme was obtained from patient infusion remnants.
Statistical Analysis
The colony formation data were analyzed using the nonparametric Kruskal-Wallis test comparing the pooled controls against each of the patient cases for all colony counts, using the SPSS statistical software package, release 22.0.0 (IBM Corp., Armonk, NY, http://www-01.ibm.com/software/analytics/spss/). The data for marker expression were analyzed using Prism software, version 4.0c (GraphPad Software, Inc., San Diego, CA, http://www.graphpad.com) and two-tailed unpaired Student’s t tests with a confidence level for significance of 95%.
Results
GCase Deficiency Does Not Impair HPC Generation
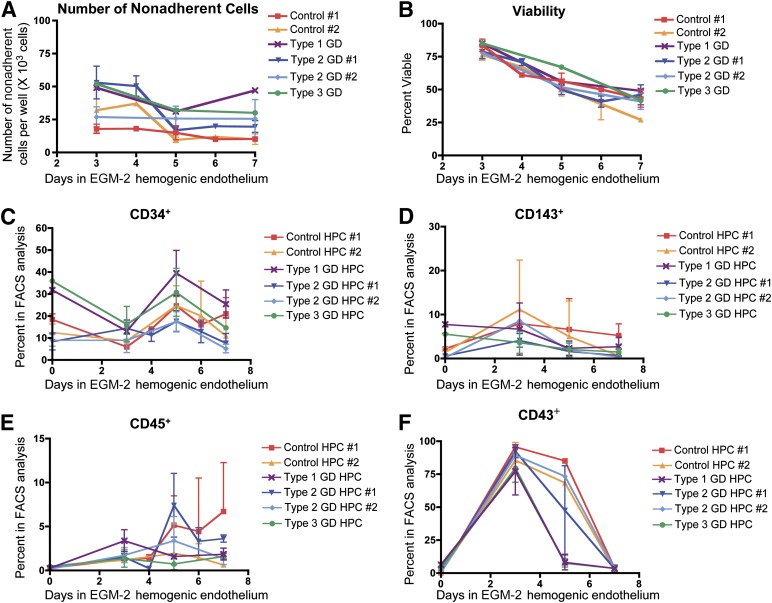
In order to investigate the effects of GCase enzyme deficiency on the hematopoietic system, control iPSCs and GD-iPSCs representing all three clinical subtypes of GD (supplemental online Table 1) were differentiated to HPCs using a previously established protocol [26] (supplemental online Fig. 1). As shown in Figure 1A and 1B, all GD- and control iPSC lines gave rise to HPCs with comparable efficiency and viability, as assessed by trypan blue exclusion. We then characterized the iPSC-derived HPCs according to the expression of known HPC markers, namely CD34 [26], CD45 [26], CD143/angiotensin converting enzyme (ACE/BB9) [27], and CD43 (leukosialin) [28, 29]. The expression kinetics of these markers was comparable between the control and types 1, 2, and 3 GD-HPCs (Fig. 1C–1F; supplemental online Fig. 2). These results suggest that GCase enzyme deficiency does not compromise HPC generation.
Figure 1.
Generation and characterization of HPC lines. The indicated control and type 1, 2, and 3 GD-HPCs were collected from EGM-2 hemogenic endothelium culture at the indicated times. (A): Ordinates indicate the number of viable HPCs collected from hemogenic endothelium per well. No difference was found in the generation efficiency for this population between the GD and control lines (mean ± SEM). (B): The percentage of viable HPCs collected from the hemogenic endothelium culture was determined by trypan blue exclusion. No difference was found in viability between the GD-HPCs and control HPCs (mean ± SEM). (C–F): Control and type 1, 2, and 3 GD-HPCs were collected from EGM-2 hemogenic endothelium culture at the indicated time points and analyzed for hematopoietic progenitor markers using flow cytometry: CD34 (C), CD143/ACE/BB9 (D), CD45 (E), and CD43 (F). The expression levels and kinetics of marker expression were comparable between the GD-HPCs and control-HPCs (mean ± SEM). Abbreviations: EGM-2, endothelial growth medium-2; FACS, fluorescence-activated cell sorting; GD, Gaucher disease; HPC, hematopoietic progenitor cell.
GD-HPCs Have Altered Myeloid and Erythroid Differentiation Potential
In order to assess the multilineage potential of GD-HPCs, we performed colony formation assays specific for either the myeloid or erythroid lineages. Both control and GD-HPCs gave rise to myeloid and erythroid colonies with typical morphologies (supplemental online Fig. 3).
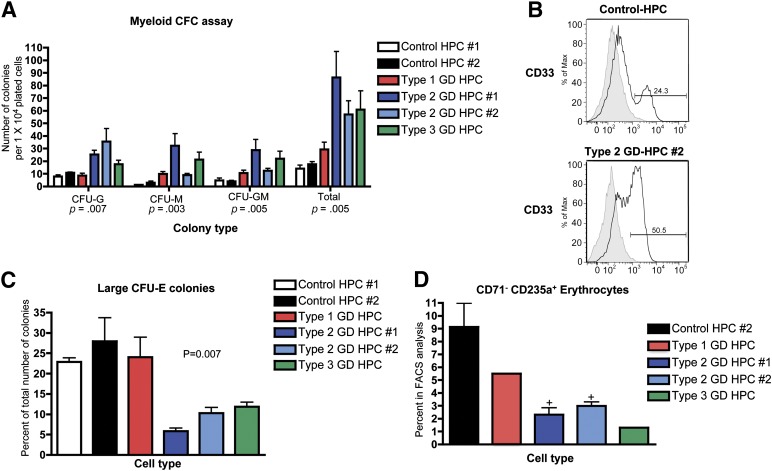
To perform myeloid colony formation assays, control and GD-HPCs were plated in methylcellulose medium containing G-CSF and GM-CSF. GD-HPCs exhibited increased differentiation potential toward the myeloid lineage, as assessed by the increased formation of myeloid colonies (Fig. 2A). Although patient-specific differences were observed, this increased propensity for myeloid differentiation correlated with disease severity. The mutant HPCs from mild type 1 GD showed only a slight increase in myeloid differentiation, while HPCs from the more severe types 2 and 3 GD had the largest increases (Fig. 2A). The increase in myeloid differentiation potential in GD-HPCs compared with control HPCs was confirmed by flow cytometric analysis of myeloid colonies, using antibodies to the myeloid progenitor marker CD33. The myeloid colonies derived from the most severe type 2 GD-HPCs exhibited increased expression of CD33 compared with those derived from the control HPCs (Fig. 2B; supplemental online Fig. 4A).
Figure 2.
Skewed myeloid and erythroid potential of GD-HPCs. (A): The indicated control and GD-HPCs were assayed for myeloid-specific colony formation. The data demonstrate an increased differentiation potential of GD-HPCs toward the myeloid lineage. The p values from the nonparametric Kruskal-Wallis test are indicated (mean ± SEM). (B): Myeloid colonies collected from methylcellulose were analyzed for expression of the myeloid progenitor marker CD33 by flow cytometry. Representative histograms show that type 2 GD-HPC-derived myeloid colonies have increased CD33 expression. Gray shaded line represents isotype control. (C): Control and GD-HPCs were assayed for erythroid-specific colony formation. The results show a decreased percentage of large CFU-E colonies (those consisting of ≥50 cells) from GD-HPCs. Shown is the p value from the nonparametric Kruskal-Wallis test (mean ± SEM). (D): The indicated control HPC- and GD-HPC-derived erythroid colonies were collected from methylcellulose assays and analyzed for the erythroid markers CD71 and CD235a using flow cytometry. The data show that the percentage of mature erythrocytes with the expression profile of CD71− CD235a+ was reduced in GD-HPC-derived compared with control HPC-derived erythroid colonies. +, p < .05, Student’s t test (mean ± SEM). Abbreviations: CFC, colony forming cell; CFU, colony forming unit; E, erythroid; G, granulocyte; GD, Gaucher disease; HPC, hematopoietic progenitor cell; M, macrophage.
To perform erythroid colony formation assays, control and GD-HPCs were plated in methylcellulose medium containing EPO. The appearance of CFU-E colonies in methylcellulose is shown in supplemental online Figure 4B. Differentiation of types 2 and 3 GD-HPCs resulted in decreased frequencies of the larger, more mature erythroid colonies consisting of more than 50 cells compared with those in the controls (Fig. 2C). We then performed flow cytometric analysis for the erythroid markers CD71 (transferrin receptor) and CD235a (glycophorin A). CD71 is highly expressed on early erythroid precursor cells, and its expression decreases as the cells mature to CD235a+ erythroblasts [30–32]. Flow cytometric analysis revealed a decrease in the CD71− CD235a+ erythrocyte population within erythroid colonies derived from GD-HPCs compared with controls (Fig. 2D). This deficiency in erythroid differentiation and maturation also correlated with disease severity, with HPCs from types 2 and 3 GD exhibiting more impaired erythroid differentiation than type 1 GD-HPCs (Fig. 2C). No difference was found between the expression of the different hemoglobin (Hb) forms between the control HPC- and GD-HPC-derived erythroid colonies, with Hb-ε being the primary form (supplemental online Fig. 4C; data not shown).
Together, our colony formation assays and flow cytometric data showed that GD-HPCs display an increased propensity for differentiation along the myeloid lineage and decreased differentiation and maturation toward the erythroid lineage. These abnormalities may contribute to the anemia and the presence of large numbers of pathological macrophages in patients with GD [11].
Reversal of Hematologic Phenotype by Treatment With Recombinant GCase
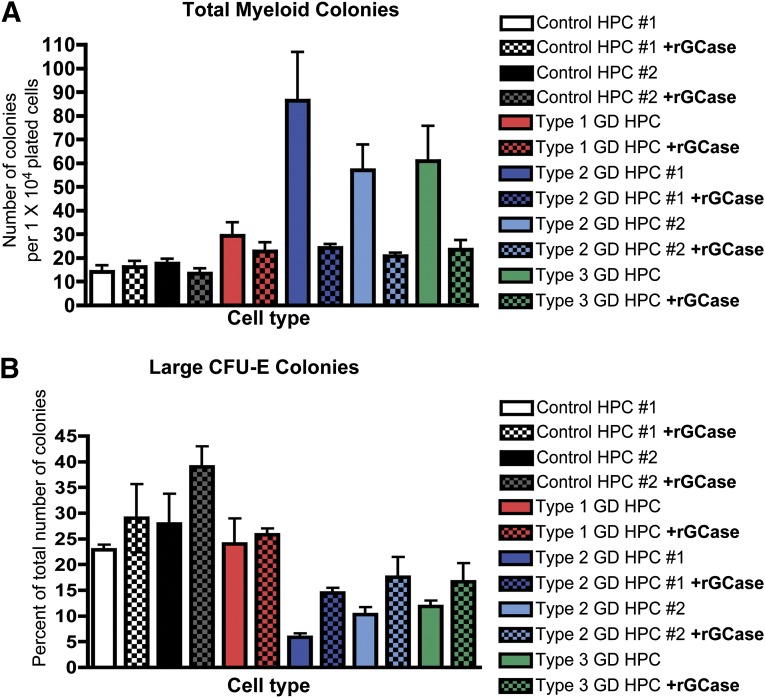
To determine whether GCase enzyme deficiency was directly responsible for altering the GD-HPC differentiation potential, we generated HPCs in the presence of rGCase and performed myeloid and erythroid colony formation assays in methylcellulose supplemented with rGCase. rGCase treatment reversed the increased myeloid differentiation potential of types 1, 2, and 3 GD-HPCs, decreasing the total number of myeloid colonies (Fig. 3A) and the number of CFU-G, CFU-M, and CFU-GM colonies (supplemental online Fig. 5A) to levels similar to those of the controls. rGCase treatment had no significant effect on control HPC myeloid differentiation. Similarly, flow cytometric analysis showed that treatment with rGCase reversed the increase in the percentage of CD33+ myeloid progenitors derived from type 2 GD-HPCs to control levels (supplemental online Fig. 5B).
Figure 3.
Recombinant GCase reverses the myeloid and erythroid abnormalities of GD-HPCs. Recombinant GCase was added to EGM-2 hemogenic endothelium cultures to correct the enzyme deficiency in GD-HPCs, and colony formation was assayed. (A): Treatment with rGCase reversed the increased myeloid differentiation within all GD-HPC lines, bringing the total number of myeloid colonies scored to levels similar to that of the controls. rGCase treatment had no significant effect on controls (mean ± SEM). (B): Treatment with rGCase increased the percentage of large CFU-E colonies from GD-HPCs. rGCase treatment had no significant effect on the controls (mean ± SEM). Abbreviations: CFU-E, colony-forming unit-erythroid; GCase, glucocerebrosidase; GD, Gaucher disease; HPC, hematopoietic progenitor cell; rGCase, recombinant glucocerebrosidase.
Incubation of GD-HPCs with rGCase also increased erythroid differentiation, as evidenced by the increased formation of larger CFU-E colonies after enzyme treatment (Fig. 3B). rGCase treatment had no significant effect on control HPC erythroid differentiation. Flow cytometric analysis showed that rGCase treatment also restored erythroid maturation, increasing the percentage of CD71− CD235a+ mature erythrocytes derived from type 2 GD-HPCs to control levels (supplemental online Fig. 5C).
The reversal of the skewed myeloid and erythroid differentiation potential of GD-HPCs by rGCase treatment strongly suggests that the hematopoietic abnormalities we observed were caused by GCase deficiency.
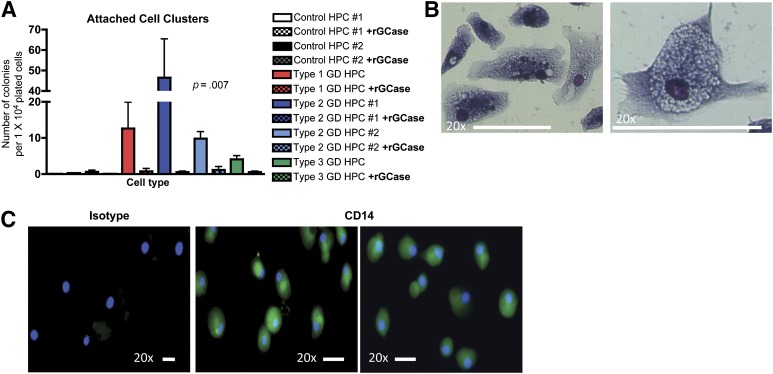
GD-HPCs Give Rise to Clusters of Abnormal Macrophage-Like Cells
While performing myeloid colony formation assays from GD-HPCs, we observed the persistent appearance of adherent cell clusters. GD-HPCs gave rise to numerous adherent cell clusters containing many cells, while control HPCs gave rise to very few of these clusters, which contained only a few cells (Fig. 4A). The frequency of adherent cell clusters varied among the different GD genotypes, with the highest number corresponding to type 2 GD patient 1. Treatment of GD-HPCs with rGCase during their generation in EGM-2 culture and during methylcellulose colony formation assays completely abrogated the appearance of these adherent cells (Fig. 4A), indicating that their abnormal presence was caused by GCase deficiency. To characterize these adherent cells, we performed May-Grünwald-Giemsa staining. The adherent cells had a macrophage-like morphology (Fig. 4B). To determine whether these cells, which appeared in myeloid colony assays only, expressed macrophage markers, we performed immunofluorescence staining for the macrophage marker CD14. These adherent cells expressed CD14, consistent with their macrophage-like morphological appearance (Fig. 4C). These results suggest that aberrant myelopoiesis might contribute to the pathological properties of Gaucher macrophages, which are central players in GD.
Figure 4.
GD-HPCs give rise to adherent macrophage-like cells. (A): The indicated control and GD-HPCs were assayed for myeloid-specific colony formation in methylcellulose. GD-HPCs gave rise to adherent cell clusters that were absent from control HPCs, and treatment with rGCase abrogated the appearance of adherent cell clusters from GD-HPC cultures. Shown are p values from the nonparametric Kruskal-Wallis test (mean ± SEM). (B): Adherent cell clusters were stained with May-Grünwald-Giemsa. Representative image of adherent cell clusters derived from type 2 GD-HPCs showing macrophage-like morphology. Scale bars = 50 μm; magnification ×20. (C): Adherent cell clusters were stained with antibodies to the macrophage marker CD14. Representative fluorescent images show the expression of CD14 within adherent cells derived from type 2 GD-HPCs. Left: Staining with isotype control. Blue, 4′,6-diamidino-2-phenylindole; green, CD14. Scale bars = 50 μm; magnification ×20. Abbreviations: GD, Gaucher disease; HPC, hematopoietic progenitor cell; rGCase, recombinant glucocerebrosidase.
Discussion
In the present study, we used GD-iPSCs to examine whether GCase deficiency has a direct effect on the developmental potential of the hematopoietic lineage. We found that GCase deficiency did not affect the formation of HPCs. However, their multipotency was skewed toward increased myeloid differentiation and decreased erythroid differentiation. Additionally, GD-HPCs, but not control HPCs, gave rise to a population of abnormal macrophage-like cells, another indication of abnormalities in the lineage that gives rise to hallmark Gaucher macrophages. All these abnormalities were reversed by incubation with rGCase, which is used to successfully treat type 1 GD [33, 34]. The hematologic abnormalities of mutant HPCs we observed in vitro recapitulate clinical manifestations in patients with GD, suggesting that GD-iPSCs are a valuable tool to model GD.
It has been suggested that the infiltration of patient tissues by pathological Gaucher cells is primarily responsible for the hematopoietic and visceral abnormalities present in GD [4]. In this view, overly active phagocytic macrophages may cause a reduction in circulating RBCs and other hematopoietic cells, leading to cytopenias and to the formation of lipid-engorged, pathological Gaucher cells [7–9]. However, it is also possible that developmental abnormalities within the mutant HPC population are responsible for, or contribute to, the hematologic manifestations in these patients. Earlier studies have reported impaired erythroid and myeloid differentiation after treatment of CD34+ HPCs from normal subjects with conduritol B epoxide (CBE), which caused 99% inhibition of the wild-type enzyme [35]. In our GD-iPSC-HPC model, we observed an increase in myeloid differentiation, in contrast to the findings reported in those pharmacological inhibition studies. Although we do not know the reasons for this discrepancy, the two studies are not directly comparable. In our GD-iPSC model, the mutant GCase proteins are expressed, which is closer to physiological conditions than inhibition of wild-type GCase enzyme. In addition, any gain-of-function activity of the mutant proteins [36], which may play a role in the pathology of GD, is preserved in our system. Finally, the possibility that CBE has off-target effects that contribute to the observed phenotype cannot be ruled out. Animal models with a targeted deletion in the GCase gene within the hematopoietic and mesenchymal stem cell compartment [37] have also been used to study the pathology of GD. While these animal models recapitulate the hallmarks of GD, including the presence of Gaucher cells in tissues, cytopenias, and an increased incidence of myeloma [37–39], these studies did not clarify whether the hematologic abnormalities observed were secondary to Gaucher cell infiltration, or due to intrinsic defects in the hematopoietic lineage. To directly address these questions, we used disease-specific iPSCs as an in vitro model for GD hematopoiesis. As some of the pathological abnormalities in types 2 and 3 GD may start in utero [40], our GD-iPSC model provides an opportunity to investigate the effects of GCase deficiency through the developmental steps from pluripotency to lineage-restricted hematopoietic cells.
GCase mutations did not affect the generation or viability of HPCs derived from types 1, 2, and 3 GD, suggesting that the effects of GCase deficiency on hematopoiesis lie downstream of the generation of the HPC pool. Our results showed that GD-HPCs had an increased differentiation potential toward the myeloid lineage. This finding correlated with the severity of the GCase mutations, with the mildest type 1 GD-HPCs only showing a modest increase in myeloid differentiation, while the most severe types 2 and 3 GD-HPCs exhibited the largest increase. One of the most interesting findings of the present study was the identification of clusters of abnormal adherent cells, which arose in myeloid colony assays from GD-HPCs but not control HPCs. These conspicuous cell clusters failed to appear when the assays were performed in the presence of rGCase, indicating that their formation was a result of GCase deficiency. The appearance of adherent, CD14+ macrophage-like cells in our colony assays suggests that the myeloid lineage abnormalities of GD-HPCs might not be confined to increased production of myeloid progenitors, but also aberrant maturation, which might contribute to the pathological properties of Gaucher macrophages. Gaucher cells are pathological lipid-engorged macrophages, which infiltrate patient tissues, including the spleen, liver, and bone marrow [4]. Together, the tissue infiltration of Gaucher cells and their altered inflammatory profile [18] are believed to be central to GD pathology and the increased risk of developing multiple myeloma [4, 11, 17]. Although the results of colony-formation assays in vitro cannot be extrapolated to in vivo myelopoiesis, our results suggest the possibility that the presence of pathological macrophages in the bone marrow might not be solely due to their infiltration of this tissue, but perhaps also due to their origination there.
Our results showed that erythroid differentiation of GD-HPCs was also compromised. Although control-HPCs produced many large, robust erythroid colonies, GD-HPC-derived erythroid colonies remained small. This suggests a defect in the more primitive erythroid progenitors, which are responsible for generating larger colonies owing to an increased proliferative capacity [41]. There was also decreased maturation toward CD71− CD235a+ erythroid progenitors within GD-HPC-derived erythroid colonies. This deficiency in erythroid differentiation and maturation correlated with disease severity. It should be noted that iPSC-derived erythroid progenitors are not indicative of definitive hematopoiesis, as these cells express primarily Hb-ε [26, 42]. Our results suggest that GCase deficiency skews the balance between the myeloid and erythroid lineages, potentially by altering the fate decisions within the primitive common progenitor that gives rise to both these lineages [43–45]. The diminished erythroid potential of GD-HPCs might contribute to the anemia present in GD patients [11]. Taken together, our results indicate that GCase deficiency affects early differentiation and maturation in the myeloid and erythroid lineages, and that the increased differentiation toward the myeloid lineage might be coupled to a decrease in erythroid differentiation. The in vitro phenotypes we observed reflect known clinical abnormalities in the erythroid lineage in GD patients. Importantly, all the hematopoietic abnormalities of GD-HPCs we observed were almost completely reversed by incubation with rGCase, demonstrating that these developmental defects were caused by GCase deficiency. Enzyme replacement therapy (ERT) using rGCase is a very effective treatment for type 1 GD. It reduces the burden of Gaucher macrophages and alleviates the cytopenias in these patients. The phenotypic reversal by rGCase we observed suggests that ERT could help normalize the clinical parameters in GD patients, not only through rGCase uptake by Gaucher macrophages [46], but also by acting directly on HPCs. We should note that although the GD-iPSCs we used in these studies were derived from 4 GD patients carrying the most frequent GCase mutations in types 1, 2, and 3 GD [47–49], additional studies with more patients representative of each genotype are required to validate our conclusions.
Conclusion
Our results demonstrate an impaired developmental potential of GD-HPCs due to GCase enzyme deficiency that reflects clinical manifestations. We showed that GCase deficiency results in altered hematopoietic differentiation such that myeloid differentiation is enhanced and erythroid differentiation and maturation are deficient. Our results also suggest that aberrant myelopoiesis might contribute to the generation of pathological macrophages, which are central to the manifestations of GD. GD-iPSCs will be an invaluable tool to identify the mechanisms that underlie the pathophysiology of GD.
Supplementary Material
Acknowledgments
This work was supported by the Maryland Stem Cell Research Fund (MSCRF) (Grants 2009-MSCRFII-0082-00 and 2007-MSCRFE-0110-00 to R.A.F. and Grants 2011-MSCRF II-0008-00 and 2007-MSCRF II-0379-00 to E.T.Z.), the March of Dimes (Grant 6-FY10-334 to R.A.F.), and NIH/National Heart, Lung, and Blood Institute Grant U01HL099775 (to E.T.Z.). T.S.P. and O.A. are recipients of MSCRF postdoctoral fellowship grants.
Author Contributions
J.A.S.: project direction, experiment planning and performance, data analysis and interpretation, manuscript writing; T.S.P.: provision of study materials, experiment performance, data analysis and interpretation; D.M., L.M.P., Y.L., and O.A.: experiment performance, data analysis and interpretation; E.S.: provision of study materials and patient data; S.M.B.: data analysis and interpretation; E.T.Z.: provision of study materials, data analysis and interpretation; R.A.F.: project direction; data analysis and interpretation, manuscript writing.
Disclosure of Potential Conflicts of Interest
S.M.B. is a compensated consultant on EMD Serono Advisory Board. The other authors indicated no potential conflicts of interest.
References
- 1.Vitner EB, Platt FM, Futerman AH. Common and uncommon pathogenic cascades in lysosomal storage diseases. J Biol Chem. 2010;285:20423–20427. doi: 10.1074/jbc.R110.134452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenbloom BE, Weinreb NJ. Gaucher disease: A comprehensive review. Crit Rev Oncog. 2013;18:163–175. doi: 10.1615/critrevoncog.2013006060. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski GA. Gaucher disease and other storage disorders. Hematology Am Soc Hematol Educ Program. 2012;2012:13–18. doi: 10.1182/asheducation-2012.1.13. [DOI] [PubMed] [Google Scholar]
- 4.Jmoudiak M, Futerman AH. Gaucher disease: Pathological mechanisms and modern management. Br J Haematol. 2005;129:178–188. doi: 10.1111/j.1365-2141.2004.05351.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhao H, Grabowski GA. Gaucher disease: Perspectives on a prototype lysosomal disease. Cell Mol Life Sci. 2002;59:694–707. doi: 10.1007/s00018-002-8458-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parkin JL, Brunning RD. Pathology of the Gaucher cell. Prog Clin Biol Res. 1982;95:151–175. [PubMed] [Google Scholar]
- 7.Boven LA, van Meurs M, Boot RG, et al. Gaucher cells demonstrate a distinct macrophage phenotype and resemble alternatively activated macrophages. Am J Clin Pathol. 2004;122:359–369. doi: 10.1309/BG5V-A8JR-DQH1-M7HN. [DOI] [PubMed] [Google Scholar]
- 8.Machaczka M, Klimkowska M, Regenthal S, et al. Gaucher disease with foamy transformed macrophages and erythrophagocytic activity. J Inherit Metab Dis. 2011;34:233–235. doi: 10.1007/s10545-010-9241-0. [DOI] [PubMed] [Google Scholar]
- 9.Bitton A, Etzell J, Grenert JP, et al. Erythrophagocytosis in Gaucher cells. Arch Pathol Lab Med. 2004;128:1191–1192. doi: 10.5858/2004-128-1191-EIGC. [DOI] [PubMed] [Google Scholar]
- 10.Panicker LM, Miller D, Park TS, et al. Induced pluripotent stem cell model recapitulates pathologic hallmarks of Gaucher disease. Proc Natl Acad Sci USA. 2012;109:18054–18059. doi: 10.1073/pnas.1207889109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hughes D, Cappellini MD, Berger M, et al. Recommendations for the management of the haematological and onco-haematological aspects of Gaucher disease. Br J Haematol. 2007;138:676–686. doi: 10.1111/j.1365-2141.2007.06701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas AS, Mehta A, Hughes DA. Gaucher disease: Haematological presentations and complications. Br J Haematol. 2014;165:427–440. doi: 10.1111/bjh.12804. [DOI] [PubMed] [Google Scholar]
- 13.Bratosin D, Tissier JP, Lapillonne H, et al. A cytometric study of the red blood cells in Gaucher disease reveals their abnormal shape that may be involved in increased erythrophagocytosis. Cytometry B Clin Cytom. 2011;80:28–37. doi: 10.1002/cyto.b.20539. [DOI] [PubMed] [Google Scholar]
- 14.Franco M, Collec E, Connes P, et al. Abnormal properties of red blood cells suggest a role in the pathophysiology of Gaucher disease. Blood. 2013;121:546–555. doi: 10.1182/blood-2012-07-442467. [DOI] [PubMed] [Google Scholar]
- 15.Adar T, Ben-Ami R, Elstein D, et al. Aggregation of red blood cells in patients with Gaucher disease. Br J Haematol. 2006;134:432–437. doi: 10.1111/j.1365-2141.2006.06199.x. [DOI] [PubMed] [Google Scholar]
- 16.Ayto R, Hughes DA. Gaucher disease and myeloma. Crit Rev Oncog. 2013;18:247–268. doi: 10.1615/critrevoncog.2013006061. [DOI] [PubMed] [Google Scholar]
- 17.Barak V, Acker M, Nisman B, et al. Cytokines in Gaucher’s disease. Eur Cytokine Netw. 1999;10:205–210. [PubMed] [Google Scholar]
- 18.Panicker LM, Miller D, Awad O, et al. Gaucher iPSC-derived macrophages produce elevated levels of inflammatory mediators and serve as a new platform for therapeutic development. Stem Cells. 2014;32:2338–2349. doi: 10.1002/stem.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Cherry AB, Daley GQ. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med. 2013;64:277–290. doi: 10.1146/annurev-med-050311-163324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cherry AB, Daley GQ. Reprogramming cellular identity for regenerative medicine. Cell. 2012;148:1110–1122. doi: 10.1016/j.cell.2012.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azevedo JL, Feldman RA. Tinkering with transcription factors uncovers plasticity of somatic cells. Genes Cancer. 2010;1:1089–1099. doi: 10.1177/1947601911401908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goodridge HS. Induced pluripotent stem cell-derived myeloid phagocytes: Disease modeling and therapeutic applications. Drug Discov Today. 2014;19:774–780. doi: 10.1016/j.drudis.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 25.Tucker KL, Wang Y, Dausman J, et al. A transgenic mouse strain expressing four drug-selectable marker genes. Nucleic Acids Res. 1997;25:3745–3746. doi: 10.1093/nar/25.18.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park TS, Zimmerlin L, Zambidis ET. Efficient and simultaneous generation of hematopoietic and vascular progenitors from human induced pluripotent stem cells. Cytometry A. 2013;83:114–126. doi: 10.1002/cyto.a.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zambidis ET, Park TS, Yu W, et al. Expression of angiotensin-converting enzyme (CD143) identifies and regulates primitive hemangioblasts derived from human pluripotent stem cells. Blood. 2008;112:3601–3614. doi: 10.1182/blood-2008-03-144766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou HY, Yang XY, Tao LJ, et al. Characterization of human embryonic stem cell-derived hematopoietic progenitor phenotype. In Vitro Cell Dev Biol Anim. 2010;46:733–737. doi: 10.1007/s11626-010-9337-8. [DOI] [PubMed] [Google Scholar]
- 29.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsee DK, Pinkus GS, Yu H. CD71 (transferrin receptor): An effective marker for erythroid precursors in bone marrow biopsy specimens. Am J Clin Pathol. 2010;134:429–435. doi: 10.1309/AJCPCRK3MOAOJ6AT. [DOI] [PubMed] [Google Scholar]
- 31.van den Akker E, Satchwell TJ, Pellegrin S, et al. The majority of the in vitro erythroid expansion potential resides in CD34(−) cells, outweighing the contribution of CD34(+) cells and significantly increasing the erythroblast yield from peripheral blood samples. Haematologica. 2010;95:1594–1598. doi: 10.3324/haematol.2009.019828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen K, Liu J, Heck S, et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cox TM. Gaucher disease: Clinical profile and therapeutic developments. Biologics. 2010;4:299–313. doi: 10.2147/BTT.S7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidransky E, LaMarca ME, Ginns EI. Therapy for Gaucher disease: Don’t stop thinking about tomorrow. Mol Genet Metab. 2007;90:122–125. doi: 10.1016/j.ymgme.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 35.Berger J, Lecourt S, Vanneaux V, et al. Glucocerebrosidase deficiency dramatically impairs human bone marrow haematopoiesis in an in vitro model of Gaucher disease. Br J Haematol. 2010;150:93–101. doi: 10.1111/j.1365-2141.2010.08214.x. [DOI] [PubMed] [Google Scholar]
- 36.Hughes DA, Pastores GM. The pathophysiology of GD—Current understanding and rationale for existing and emerging therapeutic approaches. Wien Med Wochenschr. 2010;160:594–599. doi: 10.1007/s10354-010-0864-4. [DOI] [PubMed] [Google Scholar]
- 37.Enquist IB, Nilsson E, Ooka A, et al. Effective cell and gene therapy in a murine model of Gaucher disease. Proc Natl Acad Sci USA. 2006;103:13819–13824. doi: 10.1073/pnas.0606016103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mistry PK, Liu J, Yang M, et al. Glucocerebrosidase gene-deficient mouse recapitulates Gaucher disease displaying cellular and molecular dysregulation beyond the macrophage. Proc Natl Acad Sci USA. 2010;107:19473–19478. doi: 10.1073/pnas.1003308107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavlova EV, Wang SZ, Archer J, et al. B cell lymphoma and myeloma in murine Gaucher’s disease. J Pathol. 2013;231:88–97. doi: 10.1002/path.4227. [DOI] [PubMed] [Google Scholar]
- 40.Orvisky E, Sidransky E, McKinney CE, et al. Glucosylsphingosine accumulation in mice and patients with type 2 Gaucher disease begins early in gestation. Pediatr Res. 2000;48:233–237. doi: 10.1203/00006450-200008000-00018. [DOI] [PubMed] [Google Scholar]
- 41.Eaves C, Lambie K. Atlas of Human Hematopoietic Colonies. Vancouver, BC, Canada: Stem Cell Technologies; 1995. [Google Scholar]
- 42.Slukvin II. Hematopoietic specification from human pluripotent stem cells: Current advances and challenges toward de novo generation of hematopoietic stem cells. Blood. 2013;122:4035–4046. doi: 10.1182/blood-2013-07-474825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 44.Nakajima H. Role of transcription factors in differentiation and reprogramming of hematopoietic cells. Keio J Med. 2011;60:47–55. doi: 10.2302/kjm.60.47. [DOI] [PubMed] [Google Scholar]
- 45.Chotinantakul K, Leeanansaksiri W. Hematopoietic stem cell development, niches, and signaling pathways. Bone Marrow Res. 2012;2012:270425. doi: 10.1155/2012/270425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brady RO. Enzyme replacement for lysosomal diseases. Annu Rev Med. 2006;57:283–296. doi: 10.1146/annurev.med.57.110104.115650. [DOI] [PubMed] [Google Scholar]
- 47.Hruska KS, LaMarca ME, Scott CR, et al. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29:567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 48.Chen M, Wang J. Gaucher disease: Review of the literature. Arch Pathol Lab Med. 2008;132:851–853. doi: 10.5858/2008-132-851-GDROTL. [DOI] [PubMed] [Google Scholar]
- 49.Koprivica V, Stone DL, Park JK, et al. Analysis and classification of 304 mutant alleles in patients with type 1 and type 3 Gaucher disease. Am J Hum Genet. 2000;66:1777–1786. doi: 10.1086/302925. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.