Abstract
Although collagenolytic matrix metalloproteinases (MMPs) possess common domain organizations, there are subtle differences in their processing of collagenous triplehelical substrates. In this study, we have incorporated peptoid residues into collagen model triple-helical peptides and examined MMP activities toward these peptomeric chimeras. Several different peptoid residues were incorporated into triplehelical substrates at subsites P3, P1, P1′, and P10′ individually or in combination, and the effects of the peptoid residues were evaluated on the activities of full-length MMP-1, MMP-8, MMP-13, and MMP-14/MT1-MMP. Most peptomers showed little discrimination between MMPs. However, a peptomer containing N-methyl Gly (sarcosine) in the P1′ subsite and N-isobutyl Gly (NLeu) in the P10′ subsite was hydrolyzed efficiently only by MMP-13 [nomenclature relative to the α1(I)772–786 sequence]. Cleavage site analysis showed hydrolysis at the Gly-Gln bond, indicating a shifted binding of the triple helix compared to the parent sequence. Favorable hydrolysis by MMP-13 was not due to sequence specificity or instability of the substrate triple helix but rather was based on the specific interactions of the P7′ peptoid residue with the MMP-13 hemopexin-like domain. A fluorescence resonance energy transfer triple-helical peptomer was constructed and found to be readily processed by MMP-13, not cleaved by MMP-1 and MMP-8, and weakly hydrolyzed by MT1-MMP. The influence of the triple-helical structure containing peptoid residues on the interaction between MMP subsites and individual substrate residues may provide additional information about the mechanism of collagenolysis, the understanding of collagen specificity, and the design of selective MMP probes.
Graphical abstract
Matrix metalloproteinases (MMPs) make up a family of zinc-dependent proteolytic enzymes with the ability to catalyze the degradation of extracellular matrix components, including collagens. Some MMPs are able to catalyze hydrolysis of one or more of the interstitial (types I-III) collagens within their triple-helical domains. This group of MMPs consists of MMP-1, MMP-2, MMP-8, MMP-9, MMP-13, MMP-14/MT1-MMP, and MMP-15/MT2-MMP.1–4 The aforementioned MMPs, except MMP-2 and MMP-9, possess the same domain organization and, for efficient collagenolytic activity, require both the catalytic (CAT) and hemopexin-like (HPX) domains.5–10 The linker region between these domains also plays a role in collagenolysis, either by directing binding of the substrate11 or by allowing for proper orientation of the CAT and HPX domains.12,13 MMP-2 and MMP-9 belong to the gelatinase MMP subfamily and possess three fibronectin type II (FN II) inserts within their CAT domains. The FN II inserts offer similar type I collagen binding sites.14–16
The sequence specificities of collagenolytic MMPs have been extensively examined utilizing single-stranded peptides, peptide libraries, and phage-display libraries. These include studies of MMP-1, MMP-2, and MMP-8,17,18 MMP-13,19,20 and MT1-MMP.18,21–23 However, there are only a few studies that described the effects of amino acid substitutions within a triplehelical context on MMP activity.16,24–28
Peptoids (oligomers of N-substituted glycine residues) represent a class of polymers that are not found in nature. They differ from peptides in the manner of side-chain attachment and can be considered as peptide mimics in which the side chain has been shifted from the chiral α-carbon atom in a peptide to the achiral nitrogen. In polypeptoids, N-substituted glycine residues are connected through polyimide bonds. Peptoids lack secondary amide hydrogen atoms and thus are incapable of forming hydrogen bonds characteristic of the secondary structures found in peptides and proteins. Polypeptoids are also resistant to proteolysis.29 The modular structure of peptoids, ease of synthesis, and high compatibility with existing peptide chemistry synthetic protocols make peptoids an ideal tool for structure–activity and drug discovery-related studies.
Peptide–peptoid hybrids (peptomers) have proved to be valuable tools for analyzing proteolysis. Peptoid residues have been successfully incorporated into synthetic30 and endogenous peptide inhibitors, which allowed for the formation of potent and protease resistant peptide-based inhibitors.31 Systematic substitution of natural amino acids using peptoid residues can be considered as “peptoid scanning” that in principle is similar to the Ala scan commonly employed and provides valuable structural insight into protein–protein interactions.32
Goodman and co-workers reported the first examples of triple-helical peptide (THP)–peptoid hybrids.33 In a series of studies, the influence of NLeu on triplehelical structure was investigated.34–38 Results showed that peptoid-containing (Gly-Pro-NLeu)9 structures formed more stable triple helices than (Gly-Pro-Pro)10 and clearly demonstrated that the peptoid residue NLeu is an excellent proline surrogate.
THPs have been powerful tools for elucidating MMP kinetic behavior and substrate cleavage selectivity. Taking into account the previous introduction of peptoid residues into a serine protease inhibitor structure31 that also revealed interesting structural information,39 we examined the effect of peptoid residues on triple-helical stability and on the activity of collagenolytic MMPs, namely MMP-1, MMP-8, MMP-13, and MT1-MMP, toward triple-helical peptomers. We hypothesized that that probing the CAT and HPX domains of collagenolytic MMPs via THPs with varying contents of peptoid residues would reveal different binding site interactions. To test this hypothesis, we assembled 10 different THP-based peptomers incorporating peptoid residues at subsites P3, P1, P1′, and P10′ individually or in combination and evaluated their activities toward collagenases. Our starting point was the native collagen-like THP based on the α1(I)772–786 sequence.
EXPERIMENTAL PROCEDURES
All chemicals were molecular biology or peptide synthesis grade and purchased from ThermoFisher Scientific (Waltham, MA) and Sigma-Aldrich (St. Louis, MO). Fmoc-N-Me-Ile-OH was purchased from Chemimpex International (Wood Dale, IL). Enzymes were purchased from R&D Systems and EMD Biochemicals. The full-length MMP-14/MT1-MMP used in this study is the enzyme ectodomain also termed soluble MT1-MMP (sMT1-MMP).
Peptomer and Peptide Synthesis
The parent peptide sequence was based on type I collagen α1-chain residues 772–786, which contains the MMP cleavable Gly775–Ile776 bond (Table 1). Peptides and peptomers were synthesized by Fmoc solid-phase chemistry using TentaGel S Ram resin (Advanced ChemTech, Louisville, KY), with a substitution level of 0.26 mmol/g. Peptide synthesis was conducted on the Liberty (CEM, Matthews, NC) automated microwave-assisted peptide synthesizer equipped with a Discover microwave module. Fmoc-labeled amino acids were coupled using 5 equiv of each amino acid, 4.9 equiv of N, N, N′,N′-tetramethyl-O-(6-chloro-1H–benzotriazol-1-yl)uronium hexafluorophosphate, and 8 equiv of N-methylmorpholine (microwave power of 25 W at 50 °C, for 300 s). Peptoid residue N-Me-glycine (sarcosine, Sar) was introduced as a standard amino acid (Fmoc-Sar-OH) on the peptide synthesizer. The NIle and NLeu peptoid residues were introduced manually by the two-step submonomeric method involving coupling bromoacetic acid and the corresponding amine (S)-(+)-2-butylamine and isobutylamine, respectively, according to the standard protocol.40 Fluorogenic analogues were synthesized as described above using Fmoc-Lys(Mca) and Fmoc-Lys(Dnp) to incorporate the fluorophore/quencher pair (see further discussion below) (Table 1).
Table 1.
Triple-Helical Peptides and Peptomersa
| compound | sequence | modification type | MW observed [M + H]+ (theoretical) |
melting point Tm (°C) |
|---|---|---|---|---|
| α1(I)772–786 THP |
(GPO)4-GPQG↓IAGQRGVVGLO(GPO)4-NH2 | parent sequence | 3558.8 (3557.8) | 38 |
| MS-1 | (GPO)4-GPQG-NIle-AGQRGVVGLO(GPO)4-NH2 | P1′ = NIle | 3558.8 (3557.8) | 36 |
| MS-2 | (GPO)4-GPQG-Sar-AGQRGVVGLO(GPO)4-NH2 | P1′ = Sar | 3517.8 (3516.8) | 40 |
| MS-3 | (GPO)4GPQ-Sar-IAGQRGVVGLO(GPO)4-NH2 | P1 = Sar | 3572.8 (3571.8) | 18 |
| MS-4 | (GPO)4G-Sar-QGIAGQRGVVGLO(GPO)4-NH2 | P3 = Sar | 3532.8 (3531.8) | 34 |
| MS-7 | (GPO)4GPQGIAGQRGVVG-NLeu-O(GPO)4-NH2 | P10′ = NLeu | 3558.8 (3557.8) | 42.5 |
| MS-8 | (GPO)4GPQG-Sar-AGQRGVVG-NLeu-O(GPO)4- NH2 |
P1′ = Sar, P10′ = NLeu | 3517.8 (3516.8) | 43.5 |
| MS-9 | (GPO)4GPQ-Sar-IAGQRGVVG-NLeu-GLO(GPO)4- NH2 |
P1 = Sar, P10′ = NLeu | 3572.8 (3571.8) | 24.5 |
| MS-10 | (GPO)4GPQGWAGQRGVVG-NLeu-O(GPO)4-NH2 | P1′ = Trp, P10′ = NLeu | 3631.8 (3630.8) | 34 |
| pfTHP-1 | (GPO)4GP-Lys(Mca)-G-Sar-AGQRGV-Lys(Dnp)-G- NLeu-O(GPO)4-NH2 |
P1′ = Sar, P10′ = NLeu, fluorogenic substrate |
3927.8 (3926.8) | 36 |
| Ac-pfTHP-1 | Ac-(GPO)4-GP-Lys(Mca)-G-Sar-AGQRGV-Lys(Dnp) G-NLeu-O(GPO)4-NH2 |
P1′ = Sar, P10′ = NLeu, acetylated fluorogenic substrate |
3969.8 (3968.8) | 40 |
O is 4-hydroxy-l-proline. The cleavage site of the parent compound is indicated with a down arrow.
Crude peptides and peptomers were purified using RP-HPLC on an Agilent 1260 Infinity series preparative HPLC instrument equipped with a Vydac C18 column (15–20 µm, 300 Å, 250 mm × 22 mm). The elution gradient was from 5 to 50% B in 60 min (where A was 0.1% TFA in H2O and B was 0.1% TFA in acetonitrile), with a flow rate of 10 mL/min, and detection at λ = 220 and 280 nm. The HPLC fractions were combined, frozen, and lyophilized. Peptide and peptomer purity was evaluated on an Agilent 1260 Infinity analytical HPLC instrument using a Vydac C18 column (5 µm, 300 Å, 150 mm × 4.6 mm), with an analytical gradient from 2 to 98% B over 20 min, a flow rate of 1 mL/min, and detection at λ = 220 and 280 nm. MALDI-TOF MS analysis was performed using an Applied Biosystems (Carlsbad, CA) Voyager DE-PRO Biospectrometry workstation with a α-cyano-4-hydroxycinnamic acid/2,5-dihydroxybenzoic acid matrix.
Circular Dichroism (CD) Spectroscopy
Peptides and peptomers were dissolved in 0.5% acetic acid to a concentration of 25 µM and equilibrated at 4 °C (>24 h) to facilitate triplehelix formation. Peptide concentrations were determined by HPLC analysis using a calibration curve. Triple-helical structure was evaluated by near-UV CD spectroscopy using a Jasco (Easton, MD) J-810 spectropolarimeter with a path length of 1 mm. Thermal transition curves were obtained by recording the molar ellipticity ([θ]) at λ = 225 nm with an increase in temperature of 20 °C/h over the range of 5–80 °C. The temperature was controlled by a JASCO PTC-348WI temperature control unit. The THP melting temperature (Tm) was defined as the inflection point in the transition region (first derivative). The spectra were normalized by designating the highest [θ]225 as 100% folded and the lowest [θ]225 as 0% folded.
Metalloproteinase Activation
All MMPs (except MT1-MMP) were activated41 by mixing equal volumes of stock and activator to a final concentration of 1 mM p-aminomercuric acetate, incubated for 60 min in a 37 °C water bath, diluted to 100 nM in ice-cold TSB*Zn buffer [50 mM Tris, 150 mM NaCl, 0.02% NaN3, 0.01% Brij-35, 10 mM CaCl2, and 1 µM ZnCl2 (pH 7.5)], and kept on ice to prevent autoproteolysis. MT1-MMP was activated by incubation of proMT1-MMP with 0.1 equiv of rhTrypsin-3 for 1 h at 37 °C. After MT1-MMP activation, remaining trypsin-3 activity was quenched by addition of 100 equiv of AEBSF (R&D Systems) and incubation for 15 min at room temperature. Immediately after activation, the enzyme was diluted in cold TSB*Zn buffer. Enzyme aliquots were kept on wet ice and used the same day. MMP activity was initially evaluated using the Knight single-stranded peptide and compared with prior data. 42,43 In this way, Knight single-stranded peptide activity was used as an indicator of enzyme integrity rather than tissue inhibitor of metalloproteinase titration as performed previously.25
HPLC-Monitored Hydrolysis of Peptomeric THPs by MMPs
All peptomers were dissolved in TSB and equilibrated at 4 °C for 24 h prior to experiments to facilitate formation of the triple helix. Triple-helical structure was additionally evaluated using CD spectroscopy. Peptomers were incubated with active MMP-1, MMP-8, MMP-13, or MT1-MMP in TSB*Zn buffer, using 20 nM enzyme at a 1:1000 (enzyme:peptomer) molar ratio for 24 h at room temperature. In the case of peptomers MS-2, MS-7, and MS-8, a separate experiment was also conducted at 37 °C. The enzymatic hydrolysis reaction was quenched using 50 mM EDTA and the mixture subjected to RP-HPLC analysis. The analysis was performed in triplicate. The reaction products were evaluated on an Agilent 1260 Infinity analytical HPLC instrument using a Vydac C18 column (5 µm, 300 Å, 150 mm × 4.6 mm), with an analytical gradient from 2 to 98% B over 20 min, a flow rate of 1 mL/min, and detection at λ = 220 nm. MALDI-TOF MS analysis was also performed to determine cleavage products. The peaks on HPLC chromatograms were integrated and used for the determination of the extent of hydrolysis.
Kinetic Assays
All proteinases were activated as described above. FRET peptomers were dissolved in TSB and equilibrated at 4 °C for 24 h to facilitate triple-helix formation. Peptomer concentrations were determined using a Thermo Scientific (Waltham, MA) NanoDrop 1000 instrument via a wavelength scan at λ = 363 nm and εDnp = 15900 M−1 cm−1. Enzyme kinetic assays were performed in triplicate and evaluated with a BioTek (Winooski, VT) Synergy 4 plate reader running Gen5 software as described previously.43 In brief, a range of peptide concentrations was created by diluting in a 1:1 ratio a 100 µM stock solution of peptide 12 times. A 76 µL volume of sample was loaded; the plate was read, and 4 µL of a 20× enzyme stock solution (100 nM) was added. The kinetic protocol at 30 °C included shaking for 30 s followed by reading each well every 8 s (for 600 s) to determine initial reaction rates. Plates were stored at 30 °C for 24 h before a final reading. The peptide concentration of 25 µM was analyzed by HPLC to determine the percentage of reaction completeness with 100% cleavage RFU = [(24 h RFU) – (0 h RFU)] × 100/ (percentage of peptide cleaved), where RFU is relative fluorescence units. Enzyme activity was determined as VCorr = [peptide](initial rate)/(100% cleavage RFU). Analysis of substrate concentration versus reaction velocity was achieved by Lineweaver–Burke, Hanes–Woolf, and Eadie–Hofstee plots as well as Michaelis–Menten regression with GraphPad Prism. When the four modes of analysis correlated well, the experiment was considered a success.
Molecular Modeling
The starting structures for the THP and the MMP·THP complex were obtained from our previously published NMR-derived complex of MMP-1 with α1(I)772–786 THP.13 Molecular dynamics simulations were performed using the Desmond software package44 with the OPLS-AA 2005 force field.45 The α1(I)772–786 THP was used as a reference structure for MS-8. The peptoid residues at positions 17 and 26 were modified manually to Sar and NLeu, respectively. N- and C-termini were capped with acetyl and N-methyl groups, respectively. In each case, the peptide/peptomer was placed in an orthorhombic TIP3P water box with 10 Å of buffer in each direction and neutralized with Cl− counterions, followed by the addition of 0.15 M NaCl buffer. The model systems were relaxed prior to the simulations using a default relaxation protocol in Maestro46 that includes both restrained and unrestrained minimizations followed by four short MD simulations in which restraints were gradually removed. A NPT simulation of 20 ns was then performed at 300 K with a recording interval of 2 ps for the trajectory and energy. All simulations were conducted under periodic boundary conditions. The Nose-Hoover thermostat method was used to maintain a constant temperature of 300 K with a relaxation time of 1 ps. Nonbonded interactions were modeled using a short-range cutoff method, with a cutoff radius of 9 Å, and a long-range smooth particle mesh Ewald method, with an Ewald tolerance of 1 × 10−9. All other parameters were at their default settings. The output MD trajectories were analyzed using VMD.47 Hydrogen bonds were analyzed using the VMD HBonds plugin. A hydrogen bond was defined by a donor– acceptor distance of <3.4 Å and a donor–H⋯acceptor angle of <35°.
Modeling of MMP-13 interactions was performed as follows. The MS-8 structure was aligned with the THP·MMP-1 complex structure13 using heavy atoms of residues P1′ and P10′ so that Gln779 occupied the S1′ subsite. This MMP-1· MS-8 complex model was used for initial qualitative structure analysis. Also, the MMP-1·MS-8 complex served as a template for the MMP-13·MS-8 complex that was obtained by homology modeling using Modeler software48 and the MMP-13 sequence (UniProt49 accession number P45452). Next, the MMP-13· MS-8 complex was prepared for MD simulation using the protein preparation module of the Desmond package, as described above. The enzyme·peptomer complex was placed in an orthorhombic TIP3P water box with 15 Å of buffer in each direction and neutralized with Na+ counterions, followed by the addition of 0.15 M NaCl buffer. The model systems were relaxed prior to the simulations. An NPT simulation of 5 ns was then performed at 300 K with a recording interval of 4 ps for the trajectory and 2 ps for energy. The output MD trajectories were analyzed using Maestro and VMD.47
RESULTS
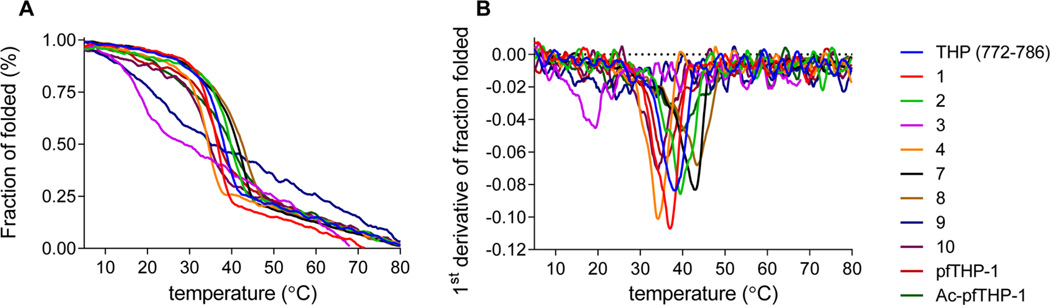
Triple-helical peptomer analogues based on the type I collagen α1-chain 772–786 sequence were synthesized and characterized (Table 1). The α1(I)772–786 sequence contains the Gly775–Ile776 MMP cleavable site 50 The rationale for each peptoid substitution is described below. After synthesis and purification, peptomers were analyzed by circular dichroism (CD) spectroscopy. CD melting curves were subsequently used to evaluate the melting temperatures (Tm values) of the peptomers (Figure 1). Each peptomer was tested for proteolytic susceptibility at room temperature using full-length human MMP-1, MMP-8, MMP-13, and MT1-MMP. The native α1(I)772–786 THP served as a control.
Figure 1.
CD properties of triple-helical peptides and peptomers. (A) Thermal transition curves of triple-helical peptides and peptomers. Readings were taken at λ = 225 nm and normalized to fraction folded. (B) Melting point evaluation of triple-helical peptides and peptomers based on thermal transition curves.
Primary MMP specificity is determined by the enzyme S1′ subsite (nomenclature of Schechter and Berger51). Thus, the first analogue contained NIle substituted for Ile in the P1′ subsite (Table 1, analogue MS-1). None of the tested collagenases was able to hydrolyze the peptomer. NIle in the P1′ subsite did not greatly affect the stability of the triple helix, as the peptomer Tm was lower than the reference THP Tm by only 2 °C (Table 1). Because MS-1 was not a substrate, its possible inhibitory properties were tested. No inhibition for any of the MMPs was observed up to an MS-1 concentration of 200 µM (data not shown). An analogue containing N-methyl Gly (sarcosine, Sar) in the P1′ subsite (MS-2) was slightly more stable (by 2 °C) than the parent THP (Table 1) and was not hydrolyzed by any of the MMPs after incubation for 24 h (at room temperature). This peptomer did not have any inhibitory properties (data not shown).
In collagenous MMP substrates, the P1 subsite is typically occupied by a Gly residue. The position of this amino acid is common among many MMP substrates.52,53 The simplest peptoid residue mimicking Gly is its N-methylated analogue, Sar (see above). MS-3, which contained Sar at subsite P1, was dramatically less stable than the parent THP (ΔTm = 20 °C) (Table 1 and Figure 1). MS-3 was readily hydrolyzed by all MMPs without any particular selectivity. Hydrolysis occurred between Sar775 and Ile776 as determined by MALDI-TOF MS analysis.
Peptomer MS-4, containing Sar at subsite P3, was prepared next. This subsite is typically occupied by a Pro residue that is favored by all collagenases.16 In theory, replacement of a secondary amino acid with a peptoid residue should not cause a substantial change in THP structure or stability. Indeed, MS-4 had a Tm value 4 °C lower than that of the parent peptide (Table 1 and Figure 1). When tested for proteolytic susceptibility, MS-4 was not hydrolyzed by any of the tested MMPs.
Having tested three crucial residues that interact with the CAT domain, we next sought to map interactions between peptomer THPs and the HPX domain. Prior studies indicated that the P10′ subsite is important for collagenous substrate interaction with MMP-1, and this interaction is via the HPX domain.13,54 Thus, MS-7 contained NLeu at subsite P10′. Surprisingly, the substitution at this position resulted in stabilization of the triple helix by 4.5 °C (Table 1 and Figure 1). Incubation of MS-7 with the collagenolytic MMPs showed the peptomer to be a better substrate than the parent THP (data not shown). MS-7 was hydrolyzed at the Gly775–Ile776 bond by all enzymes.
The influence of substitution at subsite P10′ in combination with the P1′ or P1 subsites was next pursued. MS-8 containing Sar and NLeu at subsites P1′ and P10′, respectively, was the most thermally stable peptomer with a Tm of 43.5 °C (Table 1 and Figure 1). Conversely, MS-9, which contained Sar and NLeu at subsites P1 and P10′, respectively, has a Tm value 13.5 °C below that of the parent THP (Table 1 and Figure 1). Overall, substitution of the P10′ subsite Leu residue with NLeu stabilized all THP constructs examined. MS-8 was not hydrolyzed by either MMP-1 or MMP-8. MT1-MMP exhibited low levels of activity toward MS-8 (<2%) and, on the basis of HPLC analysis, hydrolyzed the peptomer at a site different than Gly-Sar (data not shown). MMP-13 had the highest activity toward MS-8 (~10%), and hydrolysis occurred between Gly778 and Gln779 only. In contrast, MS-9 was readily hydrolyzed by all MMPs without any significant selectivity, and the cleavage, as expected from results with MS-3, was observed between Sar775 and Ile776. In an attempt to improve the selectivity MS-7, the Ile in the P1′ subsite was replaced with a bulky Trp residue (MS-10). MS-10 was resistant to MMP-1 hydrolysis, while the other collagenolytic MMPs cleaved it at the Gly775-Trp776 bond. MT1-MMP hydrolysis was the least selective, occurring at several bonds.
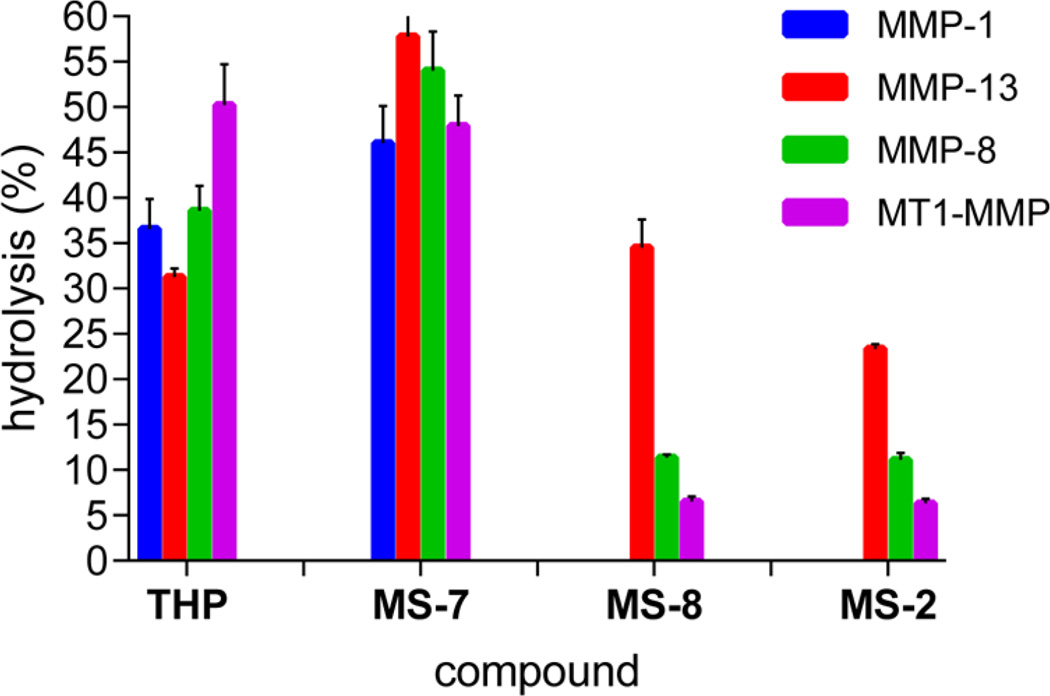
Taking into account the improved stability of peptomers containing NLeu at subsite P10′, we examined their proteolytic susceptibility at 37 °C (Figure 2). MS-7 was a much better substrate than the parent THP peptide. The greatest (2-fold) difference was observed with MMP-13. MT1-MMP was the only collagenase that had no preference for P10′ subsite substitution with a peptoid residue. At 37 °C, MS-2 (P1′ Sar residue) became a weak substrate for all MMPs except MMP-1, which did not hydrolyze it. MMP-13 hydrolyzed MS-2 4 times faster than MT1-MMP and 3 times faster than MMP-8. The best selectivity was obtained for MS-8 (P1′ Sar residue, P10′ NLeu residue), in that it was favored by MMP-13. MMP-1 was unable to process MS-8, while MMP-8 and MT1-MMP hydrolyzed it 3 and 5 times slower, respectively, than MMP-13. Interestingly, the cleavage pattern was different for each MMP. MMP-13 had a distinct cleavage site, the Gly778– Gln779 bond, while MT1-MMP cleaved the Gly781–Val782 and Arg780–Gly781 bonds, corresponding to the P6′–P7′ and P5′–P6′ subsites of the parent THP, respectively. The MMP-8 cleavage site was found to be the Gly781–Val782 bond.
Figure 2.
Screening of the susceptibility of triple-helical peptides and peptomers toward MMP hydrolysis at 37 °C after 24 h as measured by RP-HPLC analysis.
A FRET analogue of MS-8, in which subsites P5 and P5′ contained the fluorophore/quencher pair of Mca/Dnp each attached via the ε-amino group of Lys, was synthesized (Table 1). The peptomeric fluorogenic THP (pfTHP-1) formed a stable triplehelical structure with a Tm of 36 °C (Figure 1 and Table 1). To further improve the stability of the pfTHP, the N-terminus was acetylated (Ac-pfTHP-1). N-Terminal acetylation is useful for stabilizing collagen mimetic structures by removing the repulsive N-terminal charges.55 Ac-pfTHP-1 was more stable than pfTHP-1 (Figure 1), exhibiting a melting point of 40 °C (Table 1).
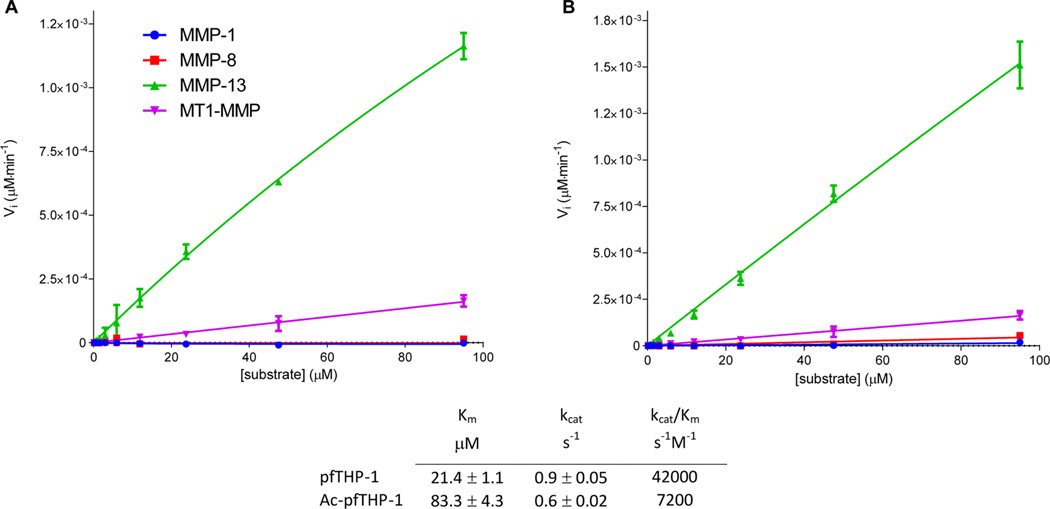
MMP-1 and MMP-8 were unable to hydrolyze pfTHP-1 and Ac-pfTHP-1. In a fashion similar to that of the unlabeled peptomers, pfTHP-1 and Ac-pfTHP-1 were cleaved by MMP-13 and MT1-MMP at different sites. MMP-13 had a distinct cleavage site between Gly778 and Gln779, while MT1-MMP had two cleavage sites, the Gly781–Val782 and Arg780–Gly781 bonds. RP-HPLC analysis of 25 µM Ac-pfTHP-1 incubated at 30 °C for 24 h indicated <1% hydrolysis by MMP-1, <2% hydrolysis by MMP-8, 22% hydrolysis by MT1-MMP, and 65% hydrolysis by MMP-13 (data not shown). The determination of kinetic parameters for hydrolysis of pfTHP-1 and Ac-pfTHP-1 was unsuccessful in the cases of MMP-1 and MMP-8 because of their minimal activity (Figure 3). The obtained KM, kcat, and kcat/KM values for MMP-13 hydrolysis (Figure 3) were in the range of typical triple-helical fluorogenic peptide substrates reported previously.16,28 In the case of MT1-MMP, analysis using the Michaelis–Menten kinetic model was difficult to interpret because of the low activity and multiple cleavage sites. The noncollagenolytic MMP-3 was not able to hydrolyze Ac-pfTHP-1 (data not shown).
Figure 3.
Initial velocities (Vi) for MMP hydrolysis of (A) pfTHP-1 and (B) Ac-pfTHP-1 at 30 °C. Kinetic parameters were then determined for MMP-13 (table).
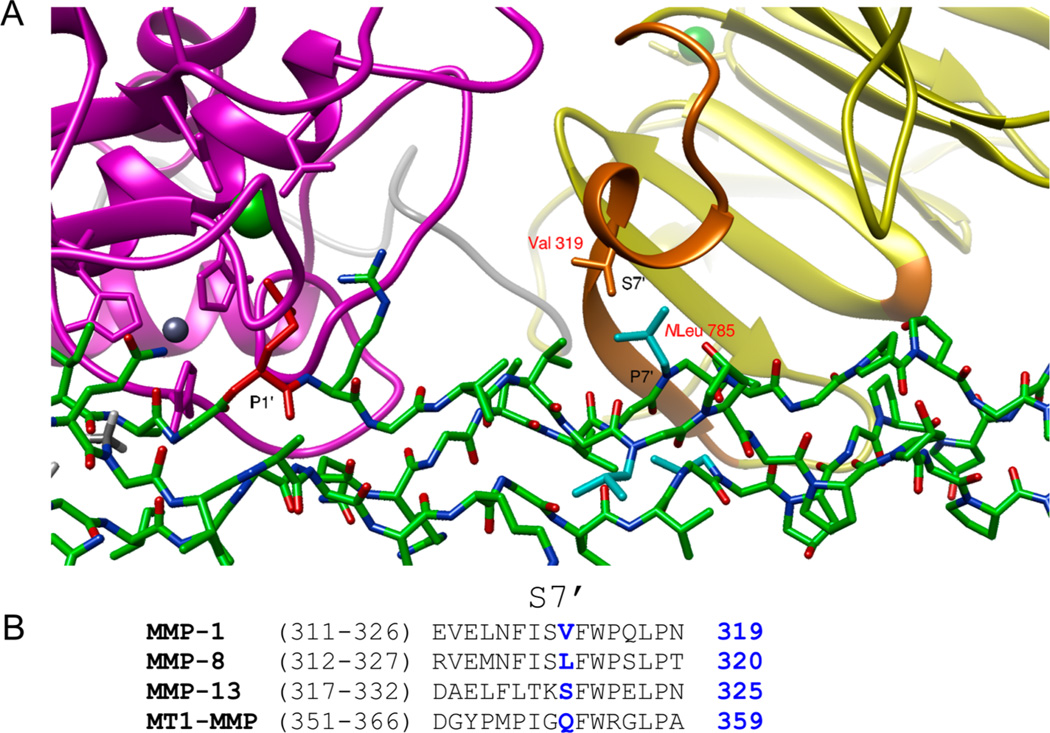
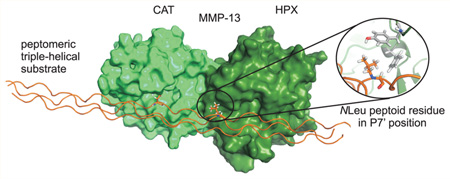
To examine the selectivity of MMP-13 toward MS-8, a molecular modeling study was undertaken. Briefly, a starting structure of a complex of MMP-1 with the α1(I)772–786 THP determined by NMR and published previously was used.13 The native THP molecule was modified manually to include peptoid residues Sar776 and NLeu785. The alignment of the MS-8 P1 subsite with the THP P1 subsite provided the relative orientation of the peptomer in relation to the enzyme (Figure 4). This model showed that the position of NLeu785 changes from the P10′ subsite to the P7′ subsite because of the new Gly–Gln cleavage site. In this case, the middle-chain NLeu785 may incur a steric clash with the S7′ residue located at helix α1 between blades I and II of the HPX domain (Figure 4). The different composition of this region among collagenases together with slightly different orientations of the HPX domains may explain the selectivity of MMP-13 toward hydrolysis of MS-2 and pfTHP-1.
Figure 4.
(A) Molecular modeling of MS-8 interacting with MMP-1. The P7′ residue (middle strand) is sterically hindered during interaction with the S7′ pocket (THP, green; CAT domain, magenta; HPX domain, yellow; HPX residues interacting with the THP, orange; P1′ residue, red; P7′ residue, cyan). (B) Sequence alignment of the α1 helix region of the HPX domain containing the S7′ pocket. The possible S7′ residue causing hindrance is colored blue and numbered in blue.
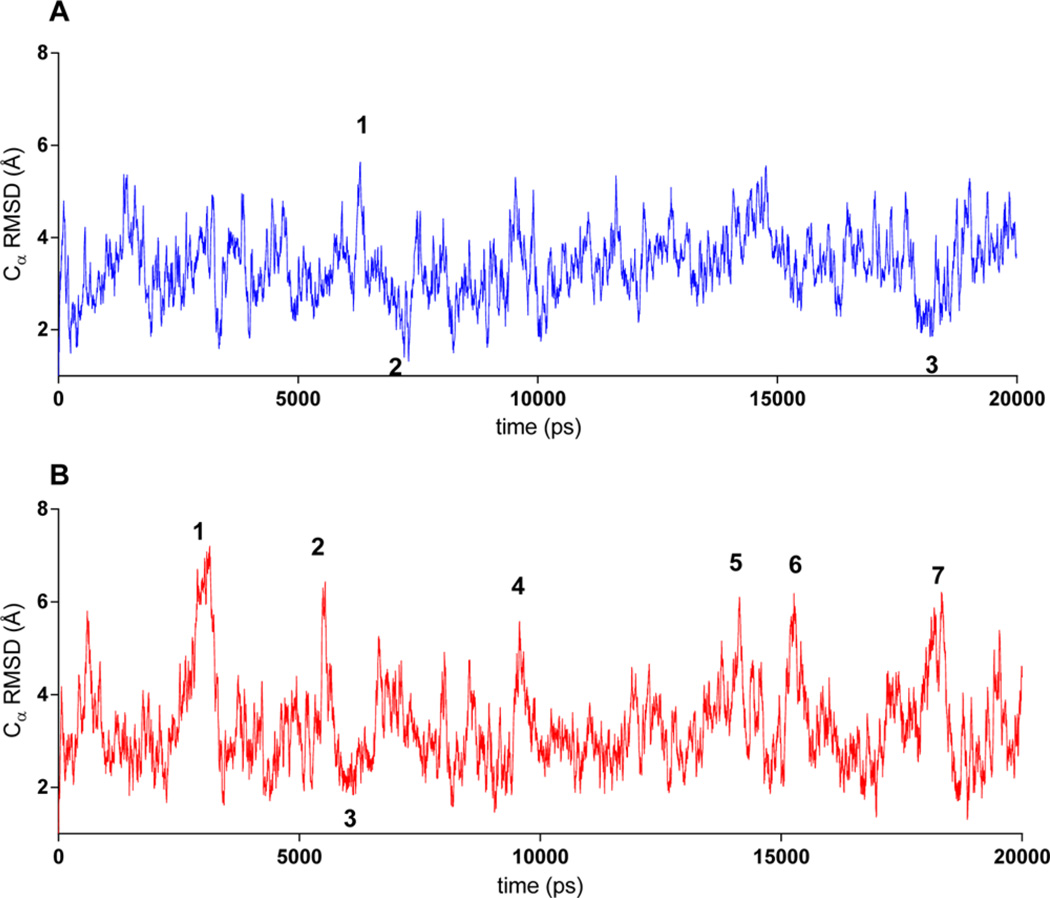
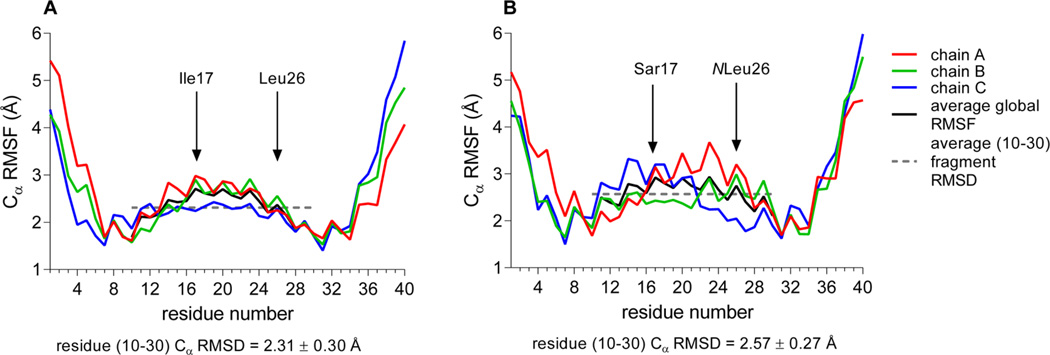
To evaluate the influence of peptoid residues on collagen-like structural stability, two independent 20 ns long molecular dynamics (MD) simulations were run for the α1(I)772–786 THP and MS-8. The simulations were run using a TIP3P water model and OPLS_2005 force field.45 This system has been used previously to study collagen-like peptides.56,57 The triplehelical structures of THP and MS-8 were stable during the entire simulation time. The simulation trajectory was analyzed to determine the possible change in the stability and/or flexibility of the triple-helical structure. This was performed using root-mean-square deviation (rmsd) and root-mean-square fluctuation (rmsf) analysis, where rmsd is the deviation from the starting structure and rmsf shows which residues are moving the most over the course of the simulation. The rmsd was calculated with respect to the initial conformation as a function of time. The average Cα rmsd values for trajectories (Figure 5) showed that MS-8 was slightly more flexible than the parent THP. The high- and low-Cα rmsd value snapshot structures (Figure 5, designated with numbers) were identified and compared (Figure 6). There was no significant difference between the MS-8 and THP structures as shown by the analysis of the rmsf of all residues (Figure 7). In both cases, the N- and C-terminal residues (residues 1–9 and 31–40, respectively) were more flexible than the central residues (10–30). The analysis showed that both compounds behave similarly, with MS-8 being slightly more flexible in the central region because of the presence of peptoid residues at positions 17 and 26.
Figure 5.
rmsd calculated for Cα atoms in (A) THP and (B) MS-8 during a 20 ns simulation time. Numbers correspond to snapshot structures depicted in Figure 6.
Figure 6.
Structure snapshots of (A) THP and (B) MS-8 with high and low Cα rmsd values identified in MD simulations, numbered accordingly from a trajectory (panels A and B of Figure 5, respectively).
Figure 7.
rmsf calculated for Cα atoms in (A) THP and (B) MS-8 during a 20 ns simulation time. The average fragment rmsf (dashed line) was calculated for residues 10–30.
Because the stability of a triple-helical structure is partially attributed to a main-chain hydrogen bond network,58 the occupancies of main-chain hydrogen bonds during the simulation time were investigated. The results show no significant influence of peptoid residues on the inter-main-chain hydrogen bond network (Table S1 of the Supporting Information). This is due to the fact that the peptoid residues Sar17 and NLeu26 are located at position X of the Gly-X-Y triplets, pointing outside the triple-helical main chain. Residues 17 and 26 act as hydrogen bond acceptors through carbonyl groups. Residue 17 (in both models) accepts hydrogen bonds from Gly16 and Gly19 during almost the whole simulation time (occupancy of >90%). The Leu26 residue in the THP accepts hydrogen bonds from Gly28 and Gly25 (occupancies of 99 and 92%, respectively). The presence of NLeu26 introduces more disturbances into the hydrogen bond network around this residue. The Gly28⋯NLeu26 H-bond reduces its presence by nearly 30% of the time, while the Gly25⋯NLeu26 H-bond is present 17% less often (Table S1 of the Supporting Information).
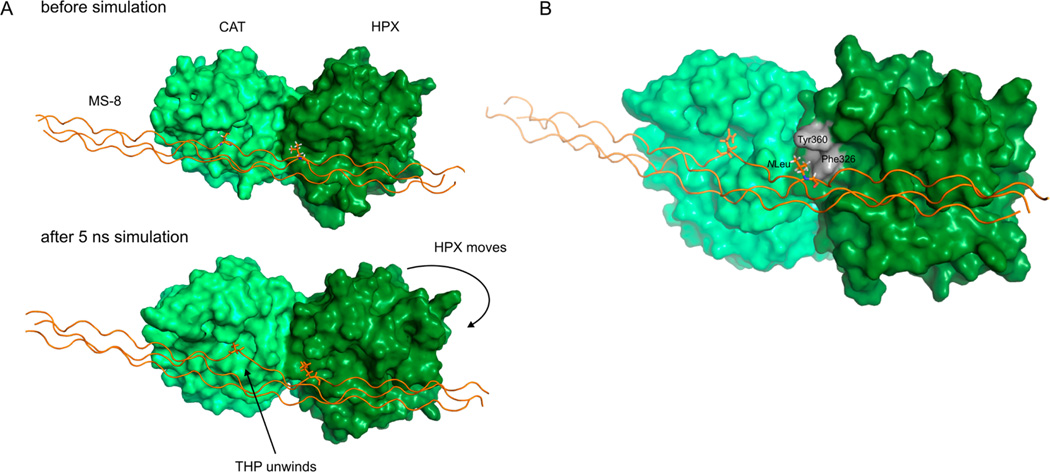
A short MD simulation of MMP-13 in complex with MS-8 was run to investigate the possible role of the P7′ NLeu in binding of MS-8 with the enzyme. Because no experimentally determined MMP-13·collagen-like peptide complex exists and the relative orientation of the CAT and HPX domains with ligand present can differ significantly from that without ligand,59,60 we utilized the experimentally determined (NMR) model of MMP-1 with the α1(I)772–786 THP published previously.13 The sequences of MMP-1 and MMP-13 are 50% identical, and they have the same length of linker domain between the CAT and HPX domains. The crude MMP-13·MS-8 model was subjected to a 5 ns MD simulation, which was conducted using the Desmond/Maestro package. The simulation showed that MS-8 interacts with both the CAT and HPX domains in an expected manner, where the P1′ Gln779 occupies the S1′ pocket, Sar773 (of the leading chain) interacts with the S3 subsite, while NLeu782 (of the middle chain) occupies the S7′ subsite. During the simulation, the HPX domain moves and maximizes contact with the triple-helical substrate. As the HPX domain moves, the triple-helical region close to the P1′ subsite relaxes its structure (Figure 8A and movie in the Supporting Information). The P7′ subsite occupied by NLeu782 forms hydrophobic interactions with the S7′ subsite that includes Phe326 and Tyr360. Remarkably, the peptoid residue in the P7′ position allows for easy accommodation of the side chain that would not be possible in the case of a natural amino acid (Figure 8B).
Figure 8.
Analysis of the MD simulation of the MMP-13·MS-8 complex. Panel A shows the relative HPX domain movement conjugated with simultaneous destabilization and/or unwinding of MS-8. Panel B shows the interaction of the P7′ NLeu782 residue with the S7′ subsite containing hydrophobic residues Phe326 and Tyr360. A video showing domain movement (morphing) between the beginning and end frames of a 5 ns trajectory is available as Supporting Information.
DISCUSSION
Peptoid residues influence the backbone geometry of the resulting peptomer because of the greater flexibility of peptoid monomers. Peptoids alter the pattern of backbone hydrogen bonding and interactions of side chains of the substituted residue (due to the Cα to N side-chain shift and side-chain reorientation) with the interacting partner. These unique features of peptoids may aid in revealing salient features of protein–protein interactions.32 For example, the X-ray crystallographic structure of a peptomeric analogue of SFTI-1 containing NLys in the P1 subsite revealed an interesting mechanism of proteolytic resistance.39 The peptomer also provided insights into the mechanism of action of the serine protease family.
To understand the molecular basis for collagen stability and metabolism, a number of approaches have been used, including the application of synthetic mimics of collagen. Since 1994, after the high-resolution structure of a collagen model triplehelical peptide became available,61 considerable research has focused on synthetic mimics of collagen that contain guest residues, tethers, peptoid residues, or analogues of the prevalent 4(R)-hydroxyproline residues.62,63
The study presented here has utilized peptoid residue probes to examine triple-helical peptide stability and processing. Studies by the Goodman group showed that (Gly-NLeu-Pro)n constructs form stable triple-helical structures and (Gly-NLeu-Pro)n constructs are more stable that (Gly-Pro-NLeu)n analogues.35 Interestingly, an Ac-(Gly-Pro-Sar)10-Gly-OH analogue did not exhibit triple-helical structure.64 At present, a single substitution by NLeu within a Gly784-NLeu785-Hyp786 triplet (MS-7) improved the melting point compared with that of the Gly784-Leu785-Hyp786 parent (ΔTm = +4.5 °C). In a similar fashion, the Gly775-Sar776-Ala777 construct (MS-2) provided a triple helix more stable than that of the parent Gly775-Ile776-Ala777 construct (ΔTm = +2 °C). Interestingly, MS-8, which contained both substitutions, had a cooperative stability effect (ΔTm = +5.5 °C). The single substitution of the Ile in a peptomer containing the Gly775-NIle776-Ala777 construct (MS-1) resulted in a slightly lower triple-helix stability (ΔTm = −2 °C). The difference between the stabilities of MS-1 and MS-2 may be explained by the different local geometry of a peptoid residue in the Gly-Nxx-Ala triplet and a sum of steric and hydrophobic effects that are important for inter- and intrachain interactions in aqueous solvents. This observation correlates with a study in which the influence of a single Nxx peptoid residue within Ac-(Gly-NLeu-Pro)3-Gly-Nxx-Pro-(Gly-NLeu-Pro)3-NH2 was tested.37 The effect of the solvent on the main-chain geometry of peptoids/peptomers has been studied experimentally and theoretically.65–68 Results show that water strengthens the tendency of the methylated peptide bond in peptoids to adopt the cis configuration. According to quantum chemical calculations, in vacuo for both studied compounds, the lowest-energy conformer is in the trans configuration, with the energy of the cis N-terminal amide bond conformer being ~2 kcal/mol higher. However, in water, the energies of these two conformers are almost equal.66
Triple-helical peptides have been powerful tools for elucidating MMP kinetic behavior and substrate cleavage selectivity. Here, amino acids at subsites P3, P1, P1′, and P10′ were replaced with peptoid residues mimicking natural amino acids. The different peptoid substitutions had different impacts on activities of MMPs. The changes introduced into subsites P3, P1, and P10′ of the α1(I)772–786 THP did not allow for refinement of selectivity among the collagenolytic MMPs. Upon substitution of P1′ subsite Ile776 with Sar (MS-2), a selective proteolytic susceptibility by MMP-13 was observed. MS-2 was hydrolyzed efficiently (>20%) only by MMP-13. MALDI-TOF MS analysis showed cleavage products corresponding to hydrolysis of the Gly778–Gln779 bond.
Aligning the sequences of MS-2, MS-7, MS-8, and the parent THP may explain the selectivity of MMP-13 (Figure 9). MS-2 and MS-8, containing Sar773 or the combination of Sar773 and NLeu782, promote cleavage of the Gly778–Gln779 bond, where Gln becomes the new P1′ subsite residue. The hydrolysis site of this Gly–Gln bond is found in native type II collagen by MMP-13.69 Upon examination of the alignment of sequences (Figure 9), it is apparent that when Gln779 becomes the P1′ subsite residue, another peptoid residue (NLeu) becomes the new P7′ subsite residue. Also, the Arg780 residue becomes the new P2′ subsite residue, and Gly is found at subsites P4 and P3′, all of which are favored by MMP-13.53
Figure 9.

Comparison of MS-2, MS-7, MS-8, Ac-pfTHP-1, and THP sequences. Red is for the peptoid residue, magenta for the fluorophore/ quencher attached through the ε-amino group of Lys, and green for terminal fragments.
The selectivity observed for MMP-13 could be the result of (a) preferred sequence specificity, (b) instability of the triple helix upon peptoid incorporation, and/or (c) altered interactions of the triple helix with MMP CAT and/or HPX domains. The Gly–Gln bond is hydrolyzed at 28, 10, and 34% of the rate of the Gly–Ile bond for MMP-1, MMP-8, and MMP-2, respectively.17 Thus, sequence specificity alone cannot explain the observed selectivity, as MMP-1 has a reasonable activity toward Gly–Gln bonds. Other, subtle changes in triplehelical sequence affect both MMP-1 and MMP-1325,70–72 but do not explain the lack of MMP-1 hydrolysis observed here.
One could argue that the observed MMP-13 selectivity toward the MS-8 peptomer is a result of the change in triplehelical structure of the peptomeric THP compared to reference THP. Our CD data (Figure 1) together with MD simulation results (Figures 5–7) do not support this hypothesis. Another possible explanation of MMP-13 selectivity could be a result of a different HPX domain orientation toward the THP substrate. Although the cleavage site is shifted, MS-2 and MS-8 still extend to the P20′ subsite of MMP-13. Prior studies have shown that the MMP-13 HPX domain has significant interactions with the triple helix until at least the P17′ subsite.16 Farndale and co-workers recently confirmed the importance of the MMP-13 HPX domain for collagen recognition and cleavage.73 MMP-13 residues of the HPX domain interacting with the triple-helical substrate P13′ and P16′ subsites were main determinants for MMP-13 collagen recognition and/or processing.73 In NMR-based investigations of full-length MMP-1-catalyzed collagenolysis, residues 291, 292, and 311–326 belonging to the HPX domain were found to be interacting with the THP.13 Similar sites of interaction between MMP-1 and the α1(I)772–786 THP were observed by hydrogen–deuterium exchange mass spectrometry.70 These sites may differ between MMP-1 and MMP-13 on the basis of different conformations of the CAT and HPX domains observed in the X-ray crystallographic structures of the two enzymes.74,75 Interestingly, we found that the S7′ subsite did not have a substantial influence on MMP-13 HPX domain recognition of peptomers, yet an Ala scan found this subsite to affect MMP-13 binding to THPs.73 A comparison of HPX residues composing the S7′ subsite pocket reveals differences between MMPs (Figure 4B). The incorporation of the NLeu peptoid residue in subsite P7′ allowed precise mapping of this interaction, which would be rather difficult to obtain using amino acids. The NLeu residue seems to be in the right orientation toward the loop/turn region of the HPX domain S7′ subsite (Figure 4A).
As MS-8 allowed for differentiation of hydrolysis rates between collagenolytic MMPs, a FRET analogue of the peptomer was synthesized to create a potential discriminatory probe. The FRET peptomer Ac-pfTHP-1 was thermally stable (Tm = 40 °C). When tested at 30 °C, it showed excellent selectivity toward MMP-13 compared with MMP-1, MMP-8, and MT1-MMP (Figure 3).
Incorporation of peptoid residues into collagen-like peptides may find several interesting applications. Such peptomeric hybrids can be used to search for novel MMP exosites and in some cases may serve as proteolytically resistant scaffolds carrying collagen-specific recognition sites. For example, the use of peptoid residues may inhibit MMP processing of the α1(I)772–786 sequence while maintaining the cell adhesion properties of this site. Along these lines, the (Gly-Pro-NLeu)10-Gly-Pro-NH2 collagen mimetic has been shown to stimulate the attachment and growth of corneal epithelial cells and fibroblasts. Application of triple-helical peptomers could lead to the development of new, collagen-based biomaterials, resistant to MMP hydrolysis.
In conclusion, we report here the use of peptoids incorporated into a native collagen peptide sequence that allowed for mapping essential MMP–collagen interactions. Peptomers containing peptoid residues in the P3 and P7′ subsites are good substrates of MMP-13 and may be applied for the design of selective probes of this collagenase.
Supplementary Material
Acknowledgments
Funding
This research was supported by the National Cancer Institute of the National Institutes of Health via Grant CA098799 (to G.B.F.).
ABBREVIATIONS
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- CAT
catalytic
- Dnp
2,4-dinitrophenyl residue
- FN II
fibronectin type II domain
- FRET
fluorescence resonance energy transfer
- HCTU
N, N, N′, N′-tetramethyl-O-(6-chloro-1H–benzotriazol-1-yl)uranium hexafluorophosphate
- HPX
hemopexin domain
- Mca
7-methoxycoumarin-4-acetyl residue
- MD
molecular dynamics
- MMP-1
matrix metalloproteinase 1/interstitial collagenase
- MMP-13
matrix metalloproteinase 13/collagenase 3
- MMP-14/MT1-MMP
matrix metalloproteinase 14/membrane type 1 matrix metalloproteinase 1
- MMP-8
matrix metalloproteinase 8/neutrophil collagenase
- NIle
(S)-(+)-2-butylamine
- NLeu
N-isobutyl glycine
- NPT
isothermal–isobaric ensemble
- RFU
relative fluorescence unit
- rmsd
root-mean-square deviation
- rmsf
root-mean-square fluctuation
- RP-HPLC
reverse-phase high-performance liquid chromatography
- Sar
sarcosine (N-methyl glycine)
- THP
triple-helical peptide
- TIP3P
transferable intermolecular potential three-point water model
Footnotes
ASSOCIATED CONTENT
A table containing occupancies of the main-chain hydrogen bonds as a function of time and a video showing domain movement between beginning and end frames (morphing) of a 5 ns long MMP-13·MS-8 complex simulation trajectory. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00110.
The authors declare no competing financial interest.
REFERENCES
- 1.McCawley LJ, Matrisian LM. Matrix metalloproteinases: They’re not just for matrix anymore. Curr. Opin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 2.Overall CM. Molecular determinants of metalloproteinase substrate specificity. Mol. Biotechnol. 2002;22:51–86. doi: 10.1385/MB:22:1:051. [DOI] [PubMed] [Google Scholar]
- 3.Morrison CJ, Overall CM. TIMP-independence of MMP-2 activation by MT2-MMP is determined by contributions of both the MT2-MMP catalytic and hemopexin domains. J. Biol. Chem. 2006;281:26528–26539. doi: 10.1074/jbc.M603331200. [DOI] [PubMed] [Google Scholar]
- 4.Bigg HF, Rowan AD, Barker MD, Cawston TE. Activity of matrix metalloproteinase-9 against native collagen types I and III. FEBS J. 2007;274:1246–1255. doi: 10.1111/j.1742-4658.2007.05669.x. [DOI] [PubMed] [Google Scholar]
- 5.Clark IM, Cawston TE. Fragments of human fibroblast collagenase. Purification and characterization. Biochem. J. 1989;263:201–206. doi: 10.1042/bj2630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knäuper V, Osthues A, DeClerk YA, Langley KE, Bläser J, Tschesche H. Fragmentation of human polymorphonuclear-leukocyte collagenase. Biochem. J. 1993;291:847–854. doi: 10.1042/bj2910847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knauper V, Cowell S, Smith B, Lopez-Otin C, O’Shea M, Morris H, Zardi L, Murphy G. The role of the C-terminal domain of human collagenase-3 (MMP-13) in the activation of procollagenase-3, substrate specificity, and tissue inhibitor of metalloproteinase interaction. J. Biol. Chem. 1997;272:7608–7616. doi: 10.1074/jbc.272.12.7608. [DOI] [PubMed] [Google Scholar]
- 8.Murphy G, Allan JA, Willenbrock F, Cockett MI, O’Connell JP, Docherty AJ. The role of the C-terminal domain in collagenase and stromelysin specificity. J. Biol. Chem. 1992;267:9612–9618. [PubMed] [Google Scholar]
- 9.Ohuchi E, Imai K, Fujii Y, Sato H, Seiki M, Okada Y. Membrane type I matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 1997;272:2446–2451. doi: 10.1074/jbc.272.4.2446. [DOI] [PubMed] [Google Scholar]
- 10.Hurst DR, Schwartz MA, Ghaffari MA, Jin Y, Tschesche H, Fields GB, Sang QX. Catalytic- and ecto-domains of membrane type 1-matrix metalloproteinase have similar inhibition profiles but distinct endopeptidase activities. Biochem. J. 2004;377:775–779. doi: 10.1042/BJ20031067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Souza SJ, Pereira HM, Jacchieri S, Brentani RR. Collagen/collagenase interaction: Does the enzyme mimic the conformation of its own substrate? FASEB J. 1996;10:927–930. doi: 10.1096/fasebj.10.8.8666171. [DOI] [PubMed] [Google Scholar]
- 12.Iyer S, Visse R, Nagase H, Acharya KR. Crystal structure of an active form of human MMP-1. J. Mol. Biol. 2006;362:78–88. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertini I, Fragai F, Luchinat C, Melikian M, Toccafondi M, Lauer JL, Fields GB. Structural Basis for Matrix Metalloproteinase 1 Catalyzed Collagenolysis. J. Am. Chem. Soc. 2012;134:2100–2110. doi: 10.1021/ja208338j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steffensen B, Wallon UM, Overall CM. Extracellular matrix binding properties of recombinant fibronectin type II-like modules of human 72-kDa gelatinase/type IV collagenase. High affinity binding to native type I collagen but not native type IV collagen. J. Biol. Chem. 1995;270:11555–11566. doi: 10.1074/jbc.270.19.11555. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Chen Z, Wang Y, Yamada Y, Steffensen B. Functional basis for the overlap in ligand interactions and substrate specificities of matrix metalloproteinases-9 and −2. Biochem. J. 2005;392:127–134. doi: 10.1042/BJ20050650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robichaud TK, Steffensen B, Fields GB. Exosite interactions impact matrix metalloproteinase collagen specificities. J. Biol. Chem. 2011;286:37535–37542. doi: 10.1074/jbc.M111.273391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagase H, Fields GB. Human matrix metalloproteinase specificity studies using collagen sequence-based synthetic peptides. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 18.Turk BE, Huang LL, Piro ET, Cantley LC. Determination of protease cleavage site motifs using mixture-based oriented peptide libraries. Nat. Biotechnol. 2001;19:661–667. doi: 10.1038/90273. [DOI] [PubMed] [Google Scholar]
- 19.Deng S-J, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, Carter HL, III, Neugebauer J, Pahel G, Weiner MP, Moss ML. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J. Biol. Chem. 2000;275:31422–31427. doi: 10.1074/jbc.M004538200. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen FH, Yeung N, Kiefer L, Murphy G, Lopez-Otin C, Vitek MP, Moss ML. Use of a multipleenzyme/multiple reagent assay system to quantify activity levels in samples containing mixtures of matrix metalloproteinases. Biochemistry. 2004;43:2987–2995. doi: 10.1021/bi036063m. [DOI] [PubMed] [Google Scholar]
- 21.Mucha A, Cuniasse P, Kannan R, Beau F, Yiotakis A, Basset P, Dive V. Membrane type-1 matrix metalloproteinase and stromelysin-3 cleave more efficiently synthetic substrates containing unusual amino acids in their P1′ positions. J. Biol. Chem. 1998;273:2763–2768. doi: 10.1074/jbc.273.5.2763. [DOI] [PubMed] [Google Scholar]
- 22.Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K, Wierzba K, Yamada Y. Identification of substrate sequences for membrane type-1 matrix metalloproteinase using bacteriophage peptide display library. Biochem. Biophys. Res. Commun. 1999;266:308–313. doi: 10.1006/bbrc.1999.1816. [DOI] [PubMed] [Google Scholar]
- 23.Kridel SJ, Sawai H, Ratnikov BI, Chen EI, Li W, Godzik A, Strongin AY, Smith JW. A unique substrate binding mode discriminates membrane type 1-matrix metalloproteinase (MT1-MMP) from other matrix metalloproteinases. J. Biol. Chem. 2002;277:23788–23793. doi: 10.1074/jbc.M111574200. [DOI] [PubMed] [Google Scholar]
- 24.Minond D, Lauer-Fields JL, Nagase H, Fields GB. Matrix metalloproteinase triple-helical peptidase activities are differentially regulated by substrate stability. Biochemistry. 2004;43:11474–11481. doi: 10.1021/bi048938i. [DOI] [PubMed] [Google Scholar]
- 25.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Visse R, Nagase H, Fields GB. The roles of substrate thermal stability and P2 and P1′ subsite identity on matrix metalloproteinase triple-helical peptidase activity and collagen specificity. J. Biol. Chem. 2006;281:38302–38313. doi: 10.1074/jbc.M606004200. [DOI] [PubMed] [Google Scholar]
- 26.Wu H, Byrne MH, Stacey A, Goldring MB, Birkhead JR, Jaenisch R, Krane SM. Generation of collagenase-resistant collagen by site-directed mutagenesis of murine pro α1(I) collagen gene. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5888–5892. doi: 10.1073/pnas.87.15.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasty KA, Wu H, Byrne M, Goldring MB, Seyer JM, Jaenisch R, Krane SM, Mainardi CL. Susceptibility of type I collagen containing mutated α1(I) chains to cleavage by human neutrophil collagenase. Matrix. 1993;13:181–186. doi: 10.1016/s0934-8832(11)80001-6. [DOI] [PubMed] [Google Scholar]
- 28.Minond D, Lauer-Fields JL, Cudic M, Overall CM, Pei D, Brew K, Moss ML, Fields GB. Differentiation of secreted and membrane-type matrix metalloproteinase activities based on substitutions and interruptions of triple-helical sequences. Biochemistry. 2007;46:3724–3733. doi: 10.1021/bi062199j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller SM, Simon RJ, Ng S, Zuckermann RN, Kerr JM, Moos WH. Proteolytic studies of homologous peptide and N-substituted glycine peptoid oligomers. Bioorg. Med. Chem. Lett. 1994;4:2657. [Google Scholar]
- 30.Lee S-G, Chmielewski J. Cross-linked Peptoid-Based Dimerization Inhibitors of HIV-1 Protease. ChemBioChem. 2010;11:1513–1516. doi: 10.1002/cbic.201000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stawikowski M, Stawikowska R, Jaskiewicz A, Zablotna E, Rolka K. Examples of peptide-peptoid hybrid serine protease inhibitors based on the trypsin inhibitor SFTI-1 with complete protease resistance at the P1-P1′ reactive site. ChemBioChem. 2005;6:1057–1061. doi: 10.1002/cbic.200400412. [DOI] [PubMed] [Google Scholar]
- 32.Park M, Wetzler M, Jardetzky TS, Barron AE. A Readily Applicable Strategy to Convert Peptides to Peptoid-based Therapeutics. PLoS One. 2013;8:e58874. doi: 10.1371/journal.pone.0058874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goodman M, Wang LL, Feng Y. Synthesis and characterization of sequential peptide-peptoid copolymers. Polym. Prepr. Am. Chem. Soc., Div. Polym. Chem.) 1994;35:767–768. [Google Scholar]
- 34.Feng Y, Melacini G, Taulane JP, Goodman M. Collagen-based structures containing the peptoid residue N-isobutylglycine (Nleu): Synthesis and biophysical studies of Gly-Pro-Nleu sequences by circular dichroism, ultraviolet absorbance, and optical rotation. Biopolymers. 1996;39:859–872. doi: 10.1002/(SICI)1097-0282(199612)39:6%3C859::AID-BIP10%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 35.Feng Y, Melacini G, Goodman M. Collagen-based structures containing the peptoid residue N-isobutylglycine (Nleu): Synthesis and biophysical studies of Gly-Nleu-Pro sequences by circular dichroism and optical rotation. Biochemistry. 1997;36:8716–8724. doi: 10.1021/bi962980z. [DOI] [PubMed] [Google Scholar]
- 36.Melacini G, Feng Y, Goodman M. Collagen-based structures containing the peptoid residue N-isobutylglycine (Nleu): Conformational analysis of Gly-Nleu-Pro sequences by 1H-NMR and molecular modeling. Biochemistry. 1997;36:8725–8732. doi: 10.1021/bi962981r. [DOI] [PubMed] [Google Scholar]
- 37.Kwak J, Jefferson EA, Bhumralkar M, Goodman M. Triple helical stabilities of guest-host collagen mimetic structures. Bioorg. Med. Chem. 1999;7:153–160. doi: 10.1016/s0968-0896(98)00230-2. [DOI] [PubMed] [Google Scholar]
- 38.Johnson G, Jenkins M, McLean KM, Griesser HJ, Kwak J, Goodman M, Steele JG. Peptoid-containing collagen mimetics with cell binding activity. J. Biomed. Mater. Res. 2000;51:612–624. doi: 10.1002/1097-4636(20000915)51:4<612::aid-jbm9>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 39.Krzywda S, Jaskolski M, Rolka K, Stawikowski MJ. Structure of a proteolytically resistant analogue of (NLys)-5SFTI-1 in complex with trypsin: Evidence for the direct participation of the Ser214 carbonyl group in serine protease-mediated proteolysis. Acta Crystallogr. 2014;D70:668–675. doi: 10.1107/S1399004713032252. [DOI] [PubMed] [Google Scholar]
- 40.Stawikowski MJ. Peptoids and peptide-peptoid hybrid biopolymers as peptidomimetics. Methods Mol. Biol. 2013;1081:47–60. doi: 10.1007/978-1-62703-652-8_4. [DOI] [PubMed] [Google Scholar]
- 41.Itoh Y, Binner S, Nagase H. Steps involved in activation of the complex of pro-matrix metalloproteinase 2 (progelatinase A) and tissue inhibitor of metalloproteinases (TIMP)-2 by 4-aminophenylmercuric acetate. Biochem. J. 1995;308(Part 2):645–651. doi: 10.1042/bj3080645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lauer-Fields JL, Sritharan T, Stack MS, Nagase H, Fields GB. Selective hydrolysis of triple-helical substrates by matrix metalloproteinase-2 and −9. J. Biol. Chem. 2003;278:18140–18145. doi: 10.1074/jbc.M211330200. [DOI] [PubMed] [Google Scholar]
- 43.Lauer-Fields JL, Whitehead JK, Li S, Hammer RP, Brew K, Fields GB. Selective modulation of matrix metalloproteinase 9 (MMP-9) functions via exosite inhibition. J. Biol. Chem. 2008;283:20087–20095. doi: 10.1074/jbc.M801438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowers KJ, Chow E, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE. Scalable algorithms for molecular dynamics simulations on commodity clusters. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing; ACM, Tampa, FL. 2006. p. 84. [Google Scholar]
- 45.Banks JL, Beard HS, Cao Y, Cho AE, Damm W, Farid R, Felts AK, Halgren TA, Mainz DT, Maple JR, Murphy R, Philipp DM, Repasky MP, Zhang LY, Berne BJ, Friesner RA, Gallicchio E, Levy RM. Integrated Modeling Program, Applied Chemical Theory (IMPACT) J. Comput. Chem. 2005;26:1752–1780. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maestro, release 2014–3. New York: Schrödinger, LLC; 2014. [Google Scholar]
- 47.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J. Mol. Graphics. 1996;14:27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 48.Fiser A, Sali A. Modeller: Generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 49.Apweiler R, Bairoch A, Wu CH. Protein sequence databases. Curr. Opin. Chem. Biol. 2004;8:76–80. doi: 10.1016/j.cbpa.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Fields GB. Interstitial collagen catabolism. J. Biol. Chem. 2013;288:8785–8793. doi: 10.1074/jbc.R113.451211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem. Biophys. Res. Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 52.Nagase H. Substrate Specificity of MMPs. In: Clendeninn N, Appelt K, editors. Matrix Metalloproteinase Inhibitors in Cancer Therapy. Totowa, NJ: Humana Press; 2001. pp. 39–66. [Google Scholar]
- 53.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, Murphy G, Enghild JJ, Hohenester E, Nagase H. Structural insights into triple-helical collagen cleavage by matrix metalloproteinase 1. Proc. Natl. Acad. Sci. U.S.A. 2012;109:12461–12466. doi: 10.1073/pnas.1204991109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodman M, Bhumralkar Jefferson EA, Kwak J, Locardi E. Collagen mimetics. Biopolymers. 1998;47:127–142. doi: 10.1002/(SICI)1097-0282(1998)47:2<127::AID-BIP2>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 56.Andersson IE, Andersson CD, Batsalova T, Dzhambazov B, Holmdahl R, Kihlberg J, Linusson A. Design of glycopeptides used to investigate class II MHC binding and T-cell responses associated with autoimmune arthritis. PLoS One. 2011;6:e17881. doi: 10.1371/journal.pone.0017881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson IE, Batsalova T, Haag S, Dzhambazov B, Holmdahl R, Kihlberg J, Linusson A. (E)-Alkene and ethylene isosteres substantially alter the hydrogen-bonding network in class II MHC A(q)/glycopeptide complexes and affect T-cell recognition. J. Am. Chem. Soc. 2011;133:14368–14378. doi: 10.1021/ja2038722. [DOI] [PubMed] [Google Scholar]
- 58.Shoulders MD, Raines RT. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bertini I, Calderone V, Fragai M, Jaiswal R, Luchinat C, Melikian M, Mylonas E, Svergun DI. Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12. J. Am. Chem. Soc. 2008;130:7011–7021. doi: 10.1021/ja710491y. [DOI] [PubMed] [Google Scholar]
- 60.Cerofolini L, Fields GB, Fragai M, Geraldes CF, Luchinat C, Parigi G, Ravera E, Svergun DI, Teixeira JM. Examination of matrix metalloproteinase-1 in solution: A preference for the pre-collagenolysis state. J. Biol. Chem. 2013;288:30659–30671. doi: 10.1074/jbc.M113.477240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 Å resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 62.Jenkins CL, Raines RT. Insights on the conformational stability of collagen. Nat. Prod. Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 63.Fields GB. Synthesis and biological applications of collagen-model triple-helical peptides. Org. Biomol. Chem. 2010;8:1237–1258. doi: 10.1039/b920670a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Y, Melacini G, Taulane JP, Goodman M. Collagen-based structures containing the peptoid residue N-isobutylglycine (Nleu): Synthesis and biophysical studies of Gly-Pro-Nleu sequences by circular dichroism, ultraviolet absorbance, and optical rotation. Biopolymers. 1996;39:859–872. doi: 10.1002/(SICI)1097-0282(199612)39:6%3C859::AID-BIP10%3E3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 65.Uejio JS, Schwartz CP, Duffin AM, England A, Prendergast D, Saykally RJ. Monopeptide versus monopeptoid: Insights on structure and hydration of aqueous alanine and sarcosine via X-ray absorption spectroscopy. J. Phys. Chem. B. 2010;114:4702–4709. doi: 10.1021/jp911007k. [DOI] [PubMed] [Google Scholar]
- 66.Walesa R, Broda MA. Solvent effects on the conformational preferences of model peptoids. MP2 study. J. Pept. Sci. 2014;20:203–211. doi: 10.1002/psc.2601. [DOI] [PubMed] [Google Scholar]
- 67.Kee Kang Y, Sook Park H. Ab initio conformational study of N-acetyl-l-proline-N′,N′-dimethylamide: A model for polyproline. Biophys. Chem. 2005;113:93–101. doi: 10.1016/j.bpc.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Laursen JS, Engel-Andreasen J, Fristrup P, Harris P, Olsen CA. Cis-trans amide bond rotamers in β-peptoids and peptoids: Evaluation of stereoelectronic effects in backbone and side chains. J. Am. Chem. Soc. 2013;135:2835–2844. doi: 10.1021/ja312532x. [DOI] [PubMed] [Google Scholar]
- 69.Lauer-Fields JL, Nagase H, Fields GB. Use of Edman degradation sequence analysis and matrix-assisted laser desorption/ionization mass spectrometry in designing substrates for matrix metalloproteinases. J. Chromatogr. A. 2000;890:117–125. doi: 10.1016/s0021-9673(00)00396-4. [DOI] [PubMed] [Google Scholar]
- 70.Lauer-Fields JL, Chalmers MJ, Busby SA, Minond D, Griffin PR, Fields GB. Identification of specific hemopexin-like domain residues that facilitate matrix metalloproteinase collagenolytic activity. J. Biol. Chem. 2009;284:24017–24024. doi: 10.1074/jbc.M109.016873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lauer JL, Bhowmick M, Tokmina-Roszyk D, Lin Y, Van Doren SR, Fields GB. The role of collagen charge clusters in the modulation of matrix metalloproteinase activity. J. Biol. Chem. 2014;289:1981–1992. doi: 10.1074/jbc.M113.513408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu Z, Visse R, Inouye M, Nagase H, Brodsky B. Defining requirements for collagenase cleavage in collagen type III using a bacterial collagen system. J. Biol. Chem. 2012;287:22988–22997. doi: 10.1074/jbc.M112.348979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Howes JM, Bihan D, Slatter DA, Hamaia SW, Packman LC, Knauper V, Visse R, Farndale RW. The recognition of collagen and triple-helical toolkit peptides by MMP-13: Sequence specificity for binding and cleavage. J. Biol. Chem. 2014;289:24091–24101. doi: 10.1074/jbc.M114.583443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iyer S, Visse R, Nagase H, Acharya KR. Crystal structure of an active form of human MMP-1. J. Mol. Biol. 2006;362:78–88. doi: 10.1016/j.jmb.2006.06.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stura EA, Visse R, Cuniasse P, Dive V, Nagase H. Crystal structure of full-length human collagenase 3 (MMP-13) with peptides in the active site defines exosites in the catalytic domain. FASEB J. 2013;27:4395–4405. doi: 10.1096/fj.13-233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grab B, Miles AJ, Furcht LT, Fields GB. Promotion of fibroblast adhesion by triple-helical peptide models of type I collagen-derived sequences. J. Biol. Chem. 1996;271:12234–12240. doi: 10.1074/jbc.271.21.12234. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.