Sen et al. find that lack of sustained histone H3K36 methylation is commensurate with increased cryptic transcription in a subset of genes in old cells and with shorter life span. In contrast, deletion of the K36me2/3 demethylase Rph1 increases H3K36me3 within these genes, suppresses cryptic transcript initiation, and extends life span.
Keywords: aging, epigenetics, H3K36 methylation, cryptic transcription
Abstract
Epigenetic mechanisms, including histone post-translational modifications, control longevity in diverse organisms. Relatedly, loss of proper transcriptional regulation on a global scale is an emerging phenomenon of shortened life span, but the specific mechanisms linking these observations remain to be uncovered. Here, we describe a life span screen in Saccharomyces cerevisiae that is designed to identify amino acid residues of histones that regulate yeast replicative aging. Our results reveal that lack of sustained histone H3K36 methylation is commensurate with increased cryptic transcription in a subset of genes in old cells and with shorter life span. In contrast, deletion of the K36me2/3 demethylase Rph1 increases H3K36me3 within these genes, suppresses cryptic transcript initiation, and extends life span. We show that this aging phenomenon is conserved, as cryptic transcription also increases in old worms. We propose that epigenetic misregulation in aging cells leads to loss of transcriptional precision that is detrimental to life span, and, importantly, this acceleration in aging can be reversed by restoring transcriptional fidelity.
The use of yeast as an aging model has become more popular because of the conservation of several fundamental aging pathways also found in higher eukaryotes and its readily available genetic tools (Wasko and Kaeberlein 2013). Aging yeast cells have features that are distinct from young cells, including a fragmented nucleolus, slow growth, sterility, accumulation of extrachromosomal rDNA circles (ERCs), and loss of heterozygosity (Sinclair et al. 1998). Such outcomes are reflective of epigenetic changes driving chromatin reorganization and reprogramming of gene expression.
Several distinct mechanisms underlying aging-associated epigenetic alterations have been reported. In yeast, the NAD+-dependent deacetylases Sir2 and Hst2 maintain genome stability by suppressing the formation of ERCs (Blander et al. 2004). However, during replicative aging, Sir2 levels decline, and the remaining protein relocalizes to the nucleolus, leading to elevated H4K16 acetylation in subtelomeric loci, causing gene derepression (Kennedy et al. 1995; Dang et al. 2009). Furthermore, Sir2 relocalization to the nucleolus is accompanied by a loss of silencing at the MAT (mating type) locus, resulting in sterility (Smeal et al. 1996). Sir2 orthologs in worms, flies, and mammals (sirtuins) are also reported to promote longevity and/or delay age-related pathologies (Giblin et al. 2014). Additionally, the appropriate dynamics of H3K56 acetylation via the action of Asf1/Rtt109 and deacetylases Hst3/Hst4 are important for maintaining proper histone protein expression. Histone loss is a hallmark of old yeast cells, and deletion of Asf1/Rtt019 or Hst3/4 shortens life span (Dang et al. 2009; Feser et al. 2010). Also, aging yeast cells show a global loss of histones from the genome, and inactivation of the Hir complex or overexpression of histones extends life span (Feser et al. 2010; Hu et al. 2014). Similar depletion of histone levels has been reported in aged human cells (O'Sullivan et al. 2010; Adams et al. 2013). Other factors include chromatin remodelers that function in calorie restriction pathways to promote life span. In yeast, nutrient depletion deactivates the ISW2 complex conferring life span extension by up-regulating the stress response pathway (Dang et al. 2014). In worms, nutrient depletion reduces insulin-like signaling allowing for nuclear localization of DAF16/FOXO transcription factors that cooperate with SWI/SNF to promote stress response and longevity (Riedel et al. 2013). Finally, observations in worms reveal that mutations in the ASH2 complex that decrease H3K4me3 levels extend life span through three generations (Greer et al. 2011). Conversely, mutations in the H3K4me3 demethylase RBR2 shorten life span (Han and Brunet 2012). There is significant evidence of epigenetic changes with mammalian aging as well (Das and Tyler 2012; Adams et al. 2013; Shah et al. 2013; Wood and Helfand 2013).
Despite the body of evidence in support of the epigenetic model of aging, there has been no systematic screen for other modifications or chromatin pathways in the process. In this study, we report the results of a population-based high-throughput life span screen using a large-scale histone H3/H4 mutant library and identify H3K36 as an important residue modulating life span.
H3K36 is methylated by Set2 in yeast, which can add up to three methyl groups on the lysine side chain (Strahl et al. 2002). K36 dimethylation and trimethylation states are recognized by the Rpd3S complex that deacetylates H3/H4 N-terminal tails and thereby suppresses initiation of transcripts from intragenic cryptic promoters (Smolle and Workman 2013). Trimethylated K36 residues also interact with chromatin remodelers ISW1b and Chd1 while inhibiting the binding of Asf1 that exchanges histones over gene bodies (Smolle et al. 2012; Venkatesh et al. 2012). Collectively, H3K36 methylation is critical to restoring chromatin structure after RNA polymerase II (Pol II) passage and prevents spurious cryptic transcription (Butler and Dent 2012). Furthermore, there are two lysine demethylases in the JmjC-domain family that act on H3K36. These are the monomethyl and dimethyl demethylase Jhd1 and the dimethyl and trimethyl demethylase Rph1 (Kim and Buratowski 2007; Klose et al. 2007; Kwon and Ahn 2011).
In this study, we show that the loss of H3K36me3 in aged yeast cells results in the production of intragenic short transcripts and a shorter life span. Deletion of the Rph1 demethylase can reverse the aging phenotype by preserving H3K36me3 levels and suppressing the production of these spurious transcripts. We detected these shorter cryptic transcripts in the aged nematode Caenorhabditis elegans, suggesting a conserved role for this pathway in aging. Overall, we present a model in which H3K36me3 promotes longevity by enhancing transcription fidelity.
Results
A high-throughput screen for histone mutations altering life span
Several histone post-translational modifications have been shown to change with age in model systems from yeast to mammalian cells and tissues (Feser and Tyler 2011). More importantly, several of these marks have been shown to play causal roles in life span modulation. However, these studies were based on a few well-characterized marks monitored by specific antibodies. We reasoned that a systematic longevity screen assessing a large set of histone marks might provide more insight into the causative relationship between histone modifications and aging.
Thanks to the tractability of yeast genetics, systematic histone mutant libraries have been made available (Dai et al. 2008; Nakanishi et al. 2008). We used the histone H3/H4 systematic mutant library developed in the Boeke laboratory (Dai et al. 2008) to carry out a longevity screen for the following reasons. First, all mutations in this library are integrated into the yeast genome; therefore, complications caused by variations in natural loss rates of the histone plasmid during aging are avoided. Second, each mutant carries two unique barcodes that can be used for identification and quantification; hence, the library is compatible with high-throughput screening. Third, all modifiable residues are mutated into both similarly and oppositely charged residues, thereby providing greater insight into potential roles of corresponding modifications.
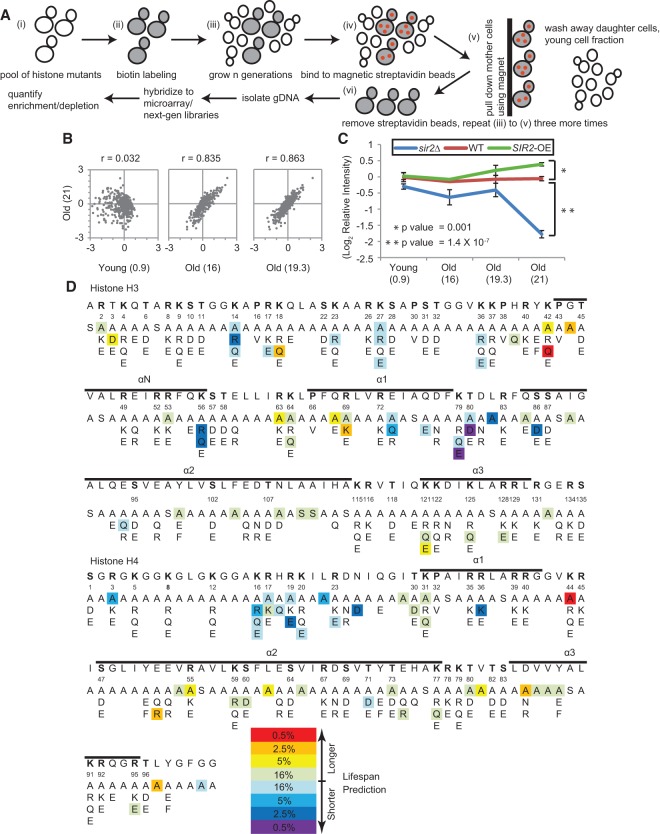
The yeast replicative life span assay is time-consuming and labor-intensive and is not well-suited for screening approaches. Based on our experience with the old mother cell sorting assay (Dang et al. 2009), we developed a novel high-throughput method for life span screening (Fig. 1A). Briefly, a pool of all mutants from the library was labeled with biotin, aged, and sorted multiple times to purify young and old fractions. Mutants with a longer life span are expected to be enriched in old fractions, while mutants with shorter life spans will be depleted. To quantify the relative enrichment for each mutant in young and old fractions, genomic DNA was extracted from each fraction and subjected to either barcode microarray or barcode sequencing analysis. As a control for the life span screen, we included uniquely barcoded wild-type, sir2Δ, and SIR2-overexpressing (SIR2-OE) strains in the histone mutant pool. The sir2Δ and SIR2-OE strains are known to have shorter and longer life spans, respectively (Kaeberlein et al. 1999).
Figure 1.
A high-throughput screen for histone mutations altering life span. (A) Illustration of screening strategy. (B) Scatter plots showing changes in relative abundance of each histone mutant in the oldest population (four rounds of old cell sorting) compared with the young and intermediate old cell fractions. Values in parentheses represent mean bud scar counts. (C) Performances of internal reference strains in the screen. Six short-lived sir2Δ strains and four long-lived SIR2-OE strains were depleted and enriched, respectively, in the oldest population compared with six wild-type strains. All reference strains carry distinct barcodes. Error bars are standard error of the mean among strains bearing different barcodes. Values in parentheses are mean bud scar counts. (D) Sequence display of histones H3 and H4, with mutations showing differential enrichment in the oldest fraction highlighted in color as indicated. The α helices of the histone proteins are indicated above the sequence.
The histone mutant screen was performed in four biological replicates, and the biotin labeling, aging, and affinity purification of labeled mother cells (sorting) were performed for four consecutive rounds for progressively aged fractions. The young cell fraction from the first, second, third, and fourth round of sorting and old cell fractions from the second, third, and fourth rounds of sorting were saved for analysis. Genomic DNA was prepared from all samples, and the relative abundance of each barcoded strain was determined by yeast barcode microarray analysis.
The results showed general consistency among the four replicates, with 85% of mutants (337 out of 443) having good correlations (correlation coefficient >0.6) (Supplemental Fig. S1). Changes in relative enrichments for histone mutants were obvious between the oldest fraction and the young (Fig. 1B, left), and the changes were much smaller between the oldest and intermediate fractions (Fig. 1B, middle and right), indicating gradual and progressive changes in the mutant enrichment in the pool during aging. The built-in controls (uniquely barcoded wild type, sir2Δ, and SIR2-OE) showed the predicted enrichment changes; i.e., sir2Δ strains became depleted, and SIR2-OE strains became enriched in the oldest fraction (Fig. 1C; Supplemental Fig. S2). These data suggest that this screen approach is able to distinguish strains that have different replicative life spans.
Histone mutations that alter replicative life span
Differential enrichments for histone mutants were obvious when comparing between the young and old samples (Fig. 1B, left). Histone mutants depleted from the old cell fraction were predicted to have a shorter life span, and those enriched in the old cell fraction were predicted to have a longer life span. It should be noted that changes in cell doubling time may also affect relative abundance in old cell fractions, and differences in doubling times can be inferred by the relative abundance of each mutant in the young fractions. Therefore, we first identified mutants showing changes in doubling time >20% based on their changes in relative abundance in the young fractions, and then life span predictions were made for each mutant based on their growth rate-corrected relative enrichments in old cells compared with young cells. An overview of predicted life span changes for each mutant is shown in Figure 1D.
To confirm the screen prediction based on the barcode microarray analysis, we carried out another biological replicate of the screen with three rounds of old cell sorting and quantified the barcodes by next-generation sequencing. The results generally agreed, with a correlation coefficient of 0.78 between the two sets of experiments (Supplemental Fig. S3).
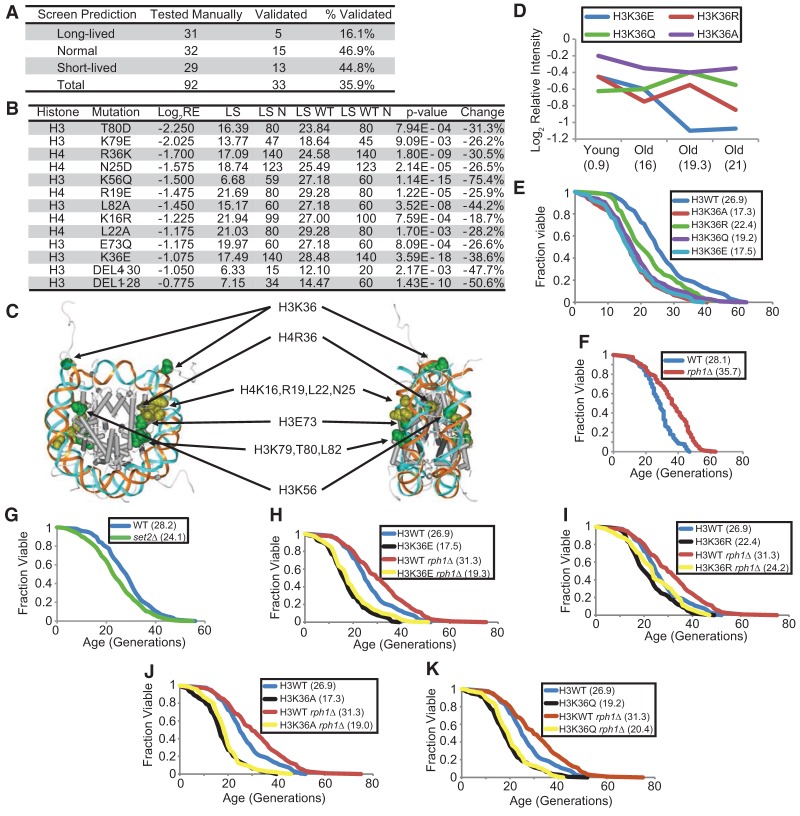
To verify the life span screen predictions, we selected mutants predicted to be long-lived, normal (like wild type), and short-lived to determine their replicative life span in the dissection-based yeast life span assay. Among all of the 92 mutants tested, 33 showed life spans matching the screen predictions (35.9% replication rate). Although seemingly lower than expected, this is a success rate comparable with a previously published yeast chronological life span screen using the same histone mutant library (Matecic et al. 2010). However, we do emphasize that the results from the screen need to be validated by independent life span assays before proceeding to mechanistic studies. Nevertheless, it has generated interesting hypotheses that need to be further tested. Mutants predicted to be short-lived and normal-lived have higher rates of validation by life span assays—44.8% and 46.9%, respectively, as shown in Figure 2A—whereas a large majority of long-lived predictions are false positives, as determined by standard dissection-based life span assays. One cause of false positives for long-lived predictions is that many of them showed a moderate to severe slow growth phenotype. Despite the relatively low prediction success rate, the screen results are valuable because they narrow the search for mutants with a truly longer or shorter life span. For example, among the 437 histone mutant strains, we called 38 short-lived, representing 8.7% of the mutants; hence, a 44.8% validation rate would represent a more than fivefold increase in short-lived mutant discovery probability.
Figure 2.
Histone mutations that alter replicative life span. (A) Table listing the number of histone mutants selected from the life span screen and the validation results showing the success rate of the screen. (B) Table listing all validated short-lived mutants, their locations (histone and mutation), relative enrichments in the screen, life span (LS), number of cells used for life span assay (LS N), experimentally matched wild-type life span (LS WT), number of wild-type cells used for life span assay (LS WT N), Wilcoxon rank sum P-value (P-value), and percent change in life span of the mutant (change). (C) Location of the life span-shortening histone residues on the nucleosome crystal structure. (D) Performance of the H3K36 mutants in the screen. The relative intensity of a particular mutant in the population was traced over the cell sorting assay. Values in parentheses are mean bud scar counts. (E) Replicative life span assays with wild type (blue) and H3K36 mutants. (F,G) Replicative life span assay with wild-type (blue), rph1Δ (red), and set2Δ (green). (H–K) Replicative life span assays with wild type (blue), rph1Δ (red), H3K36 mutants (black), and H3K36 mutant with rph1Δ (yellow). Mean life spans are indicated in parentheses. All life span P-values are listed in Supplemental Table S3.
All 13 validated short-lived mutants are listed and highlighted in the structure of the nucleosome core particle (Fig. 2B,C). Short-lived histone mutants tended to cluster at certain areas in the nucleosome core particle. One group of short-lived mutants is the LRS (loss of rDNA silencing) mutants, including H3E73, K79, T80, and L82 (Park et al. 2002; Fry et al. 2006). This is consistent with the previous findings that rDNA silencing and stability are critical for longevity and that accumulation of ERCs is a dominant factor for yeast aging (Sinclair and Guarente 1997; Stumpferl et al. 2012). Another cluster of short-lived mutants is located near the histone H4 tail, ranging from K16 to R36 (Fig. 2C). This region has been shown to mediate internucleosomal interactions initially observed in the first high-resolution crystal structure of the nucleosome core particle and later verified in the crystal structure of tetranucleosomes (Luger et al. 1997; Schalch et al. 2005). The short-lived mutants located in this patch very likely interfere with internucleosomal interactions and hence may prohibit the formation or maintenance of higher-order chromatin structure. This is consistent with the observations that open and dysregulated chromatin structures are associated with old cells (Feser and Tyler 2011). These results suggest that the high-throughput screen method can not only verify predictable effects of histone mutations on life span but also provide critical insights into how chromatin and epigenetic changes influence replicative aging.
Perturbing H3K36me3 levels alters life span
Several lines of evidence indicate an important role for H3K36—and particularly its trimethylation—in regulating yeast life span. First, results from the histone mutant screen revealed two H3K36 mutants (H3K36R and K3K36E) to be significantly depleted in the old cell fractions and predicted to have a life span among the shortest 16% of all screened mutants. Two other mutants, H3K36A and H3K36Q, were also depleted but to a lesser extent (Fig. 2D). Second, to further explore this pathway in aging, the K36 demethylase mutant rph1Δ was found to extend replicative life span by ∼30%, whereas the corresponding methyltransferase mutant, set2Δ, shortened life span by 15% (Fig. 2F,G). Rph1 removes methyl groups from dimethylated and trimethylated H3K36, whereas Jhd1 does so from dimethyl and monomethyl states. Deletion of only RPH1, and not JHD1 (Supplemental Fig. S4), extends life span, suggesting that trimethylated H3K36 specifically may regulate longevity. Supporting the high-throughput histone mutant screen data, all of the H3K36 substitution mutants showed significantly shorter life spans (Fig. 2E). Consistent with predictions that it is these methylation sites that are critical to life span extension by rph1Δ, these mutants were refractory to life span extension by deletion of RPH1 (Fig. 2H–K). Therefore, we ascertained that the life span extension by rph1Δ required its demethylase action on H3K36.
Life span extension in an rph1Δ strain is not due to suppression of ERC production or desilencing of subtelomeric loci
The rDNA locus in yeast is a highly repetitive region in chromosome XII made up of ∼150 copies of a 9.1-kb rDNA unit. The repetitive nature of this region makes it particularly susceptible to recombination events that increase in frequency as cells age. The accumulation of ERCs in old cells, formed by recombination events within the rDNA locus, has been directly implicated in limiting life span (Sinclair and Guarente 1997). To test whether the rph1Δ strain was long-lived due to suppression of ERC formation, we performed quantitative PCR (qPCR) at the three rDNA genes RDN18, RDN58, and NTS2. The qPCR validated the increased rDNA content in wild-type aged cells and showed similar levels in the rph1Δ aged cells in two independent biological replicates (Supplemental Fig. S5A). We also detected increased transcription from the NTS2 locus in old cells; however, deletion of RPH1 did not suppress formation of these transcripts (Supplemental Fig. S5B). Thus, the life span extension by rph1Δ does not seem to occur through suppression of ERC formation or transcription regulation at the rDNA locus.
The loss of Rph1 itself or the consequent increase in H3K36me3 in subtelomeric regions may deregulate the silencing of genes located at those sites. Misregulation of subtelomeric gene silencing is a hallmark of aging cells and is one of the consequences of the loss of Sir2 with age (Dang et al. 2009). We compared the ability of wild-type, set2Δ, and rph1Δ strains to silence a URA3 reporter inserted at various locations near TEL11L29 (Supplemental Fig. S6). URA3 insertions close to the telomere at or near the X core (XC) and X repeat (XR) elements (positions #1, #2, and #5) showed effective silencing in all three strains. This is in contrast to a control sir2Δ strain containing a URA3 insertion at the XC element (position #1) that is known to affect silencing (Supplemental Fig. S6, bottom right). Insertions further away from the telomere (positions #3 and #4) showed silencing defects as expected. Importantly, however, there was no difference in the ability of the wild-type, set2Δ, and rph1Δ strains to silence the URA3 reporter at positions #3 and #4. These results suggest that the loss of Set2 or Rph1 does not affect silencing of subtelomeric genes and thus is unlikely to affect life span through this mechanism.
Cryptic transcription increases in old cells
In yeast, the presence of H3K36me3 over gene bodies plays a vital role in closing chromatin structure after the passage of Pol II. H3K36me3 does so by preventing histone exchange and hyperacetylation, spacing nucleosomes over the gene body, and inhibiting spurious transcript initiation at intragenic cryptic promoters (Smolle and Workman 2013). Since deletion of the demethylase Rph1 extends life span, we hypothesized that, in wild-type yeast, the H3K36me3 levels are likely lowered as cells age and that rph1Δ mitigates this effect. However, we could not reproducibly detect this loss by Western blot in aging yeast cells before the general depletion of histones in older cells that we and others have previously observed (Dang et al. 2009; Feser et al. 2010; data not shown). It is therefore likely that there are local rather than global losses in this mark, as further investigated below.
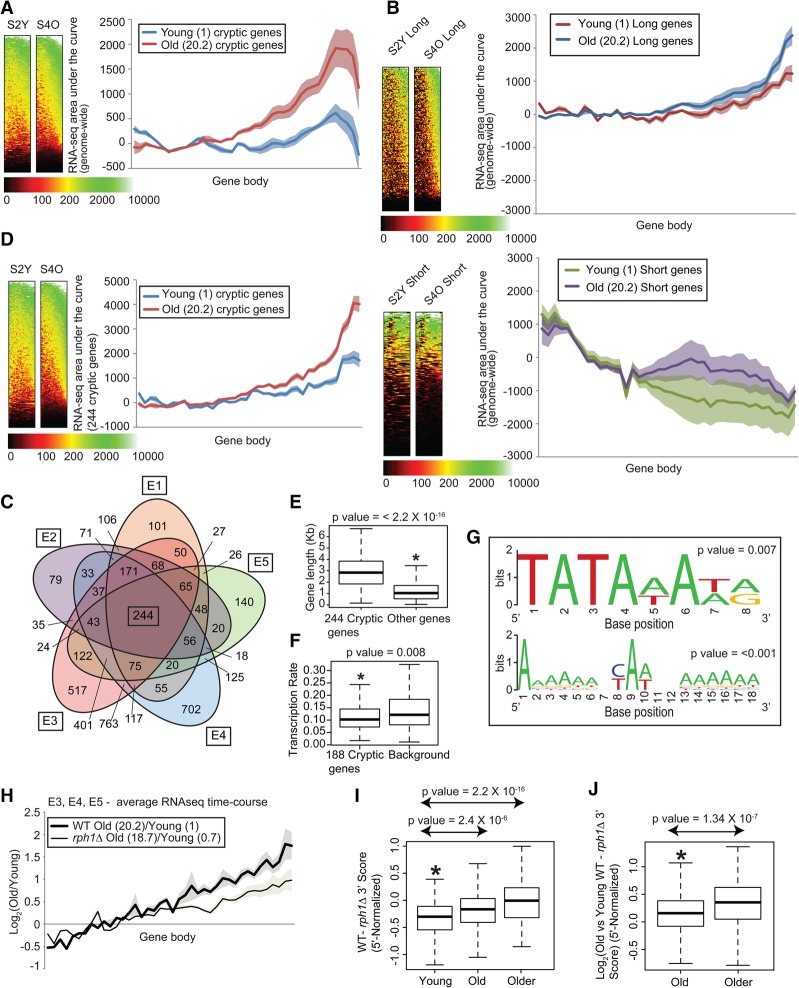
We noted that H3K36me3 levels have been anti-correlated with the production of cryptic transcripts (Smolle and Workman 2013). We sought to address whether cryptic transcription increased in old cells genome-wide by next-generation sequencing. We did not use Northern blot because: (1) the overall yield of total RNA and mRNA from old cells is very limited, (2) the isolation of old cells is not easily scaled up, (3) Northern blotting is low-throughput, and (4) the identities of potentially age-specific cryptic transcripts are unknown. We thus prepared strand-specific libraries from polyA-selected RNA from young and old cells. Since yeast genes are generally short, we modified the traditional RNA sequencing (RNA-seq) protocol by randomly fragmenting the RNA into 40- to 50-base-pair (bp) fragments rather than 200- to 300-bp fragments (Nagalakshmi et al. 2008). This provides higher-resolution analysis over gene bodies to enable detection and mapping of shorter transcripts. Indeed, short intragenic transcripts at the 3′ ends of genes were detected by specifically using this higher-resolution protocol (short fragment) (Supplemental Table S4, data sets E1–E5) but not the traditional protocol (long fragment) (Supplemental Table S5 [data sets E7–E9]; Supplemental Fig. S7). Metagene plots of RNA-seq reads over gene bodies (pooled from five independent RNA-seq data sets) (Supplemental Tables S4 [E1–E5], S5 [alignment and reproducibility parameters for each data set]) indicated high levels of transcripts from 3′ ends of genes in old cells after correcting for gene length (Fig. 3A). In addition, the effect was more pronounced for genes ranking in the top 10% in length than for those in the bottom 10% (Fig. 3B).
Figure 3.
Cryptic transcription increases in old cells, and suppression of cryptic transcripts extends life span. (A,B) Metagene plots and heat maps of normalized tag counts over all genes (A) and the top 10% longest and shortest genes (672 genes each) (B) in young and old wild-type cells; values in parentheses are mean bud scar counts. Shaded error bands represent standard error of the mean. (C) Five-way Venn diagram of genes with age-related cryptic transcription in five RNA-seq data sets (Supplemental Table S4, E1–E5). (D) Metagene plots and heat maps of normalized tag counts over gene bodies of 244 cryptic genes. (E,F) Box plots showing that the gene length distribution (E) and transcription frequency (F) of the 244 cryptic genes are higher and lower, respectively, than those of a background set of noncryptic genes. (G) WebLogo outputs of regular expression pattern searches for TATA (top) and Initiator (bottom) sequences in the cryptic genes. P-values for the number of motifs are indicated. (H) Metagene plots of normalized tag counts over cryptic gene bodies plotted as a log ratio of old versus young. (I,J) Box plot showing progressive cryptic transcription suppression measured as the difference (I) or log2 (old/young) difference (J) between wild-type and rph1Δ tag counts at the 3′ end of the cryptic genes after normalizing to the 5′ half. P-values are indicated at the top.
Genes were identified as having age-related cryptic transcription if (1) the tag counts at the 3′ ends of genes were at least twofold higher than those at the 5′ ends, (2) the 3′/5′ ratio of tag counts in old cells was at least 1.5-fold higher than in young cells, and (3) the area under the curve of the gene (total transcripts from the gene) in old cells exceeded five percentile of all expressed genes (to exclude lowly expressed genes that may be affected by background noise). Using these parameters, we identified 1300 genes in E1, 1118 genes in E2, 2759 genes in E3, 2931 genes in E4, and 1454 genes in E5. Genes identified as cryptic in any one experiment often showed age-related increases in cryptic transcript levels in the other experiments as well, even if they failed to meet our strict definition of cryptic transcription (Supplemental Fig. S8). We were able to consistently identify 244 genes with age-related intragenic transcript up-regulation from the five independent RNA-seq data sets (Fig. 3C,D; Supplemental Fig. S9, see the Supplemental Material for the gene list). Although cryptic transcript up-regulation with age is likely a genome-wide phenomenon, at least for the 244 genes, we noted that they tend to be longer and infrequently transcribed compared with a randomly selected set of genes (Fig. 3E,F; the transcription rates were acquired from Pelechano et al. 2010). Note that, due to lack of transcription rate information on 56 of the 244 cryptic genes, only 188 genes were included in the analysis. It is likely that we detected these correlations because cryptic transcripts are more easily detected in long and infrequently transcribed genes (Li et al. 2007b,c).
A confounding question is whether the cryptic transcripts in old cells accumulate due to increased internal RNA initiation or decreased degradation. We took two approaches to answer this question. First, we performed Northern blotting for a known cryptic gene, STE11, in cells harboring a deletion in the major subunits of the nuclear exosome complex or cytoplasmic mRNA degradation machineries. None of these strains generated a shorter stable cryptic transcript, and this was in stark contrast to the deletion of SET2, EAF3, RCO1, and other genes encoding elongation complexes like SPT6 and SPT21 published by others (Supplemental Fig. S10; Li et al. 2007b; Cheung et al. 2008). Thus, generation of cryptic transcripts does not involve the nuclear exosome RNA degradation complex. Second, we found strong evidence of promoter-like sequences such as TATA and Initiator in the bodies of these age-associated cryptic genes (Fig. 3G). Scanning from the start to the stop codon, ∼140 TATA-like motifs [TATA(A/T)A(A/T)(A/G)] were found within the ORFs of 107 of the 244 cryptic genes, and 410 Initiator-like sequences [A-(A-rich)5NYA(A/T)NN(A-rich)6] were found in 171 of the 244 cryptic genes. Both the number of cryptic genes with internal TATA motifs and the total number of motifs in cryptic genes were significant when compared with randomly sampled noncryptic genes (P-values were based on a permutation test running 1000 iterations) (Supplemental Table S6; see the Supplemental Material). Collectively, these lines of evidence suggest that the internal transcripts are primarily regulated at the level of increased initiation rather than degradation.
Suppression of cryptic transcripts is associated with longer life span
Since there is an increase of intragenic spurious transcripts in aged wild-type yeast, we investigated whether the generation of these transcripts was partially suppressed in the old rph1Δ mutants, given the reciprocal association between H3K36me3 levels and cryptic transcripts. Genome-wide suppression of cryptic transcription in old cells harboring a deletion of RPH1 was not obvious (data not shown). However, the transcription profiles in the core group of 244 cryptic genes showed significant suppression of cryptic transcription at the 3′ ends of gene bodies in the old rph1Δ cells (Fig. 3H–J). This implies that the deletion of RPH1 maintains transcriptional fidelity by suppressing intragenic cryptic transcripts, potentially extending life span.
Genes showing cryptic transcript up-regulation with age lose H3K36me3 over gene bodies
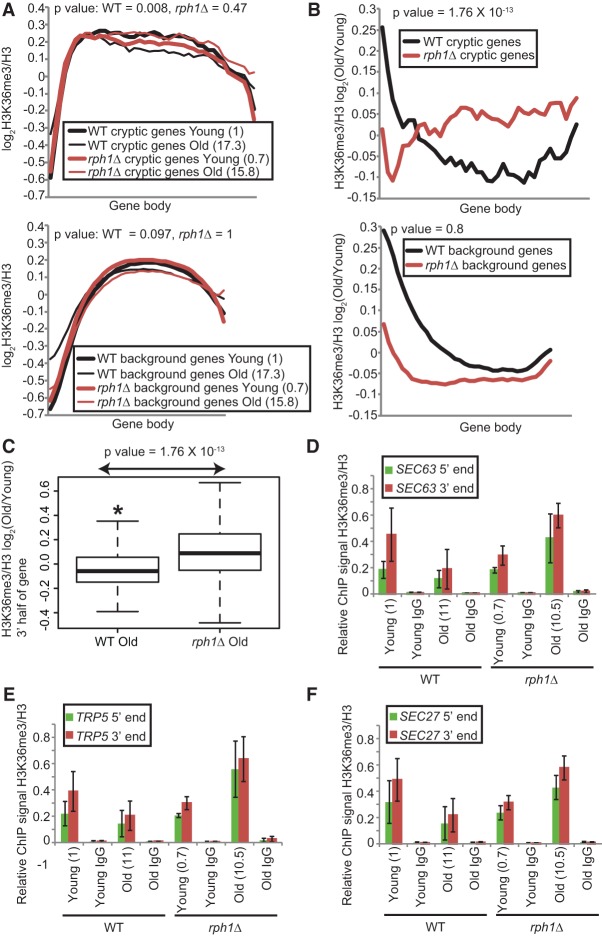
We then returned to the question of local changes in H3K36me3. To determine whether cryptic genes identified in old cells show appreciable changes in H3K36me3 status in their retained histones over gene bodies, we performed chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) of H3K36me3 and H3 in sorted wild-type and mutant cells. Two independent biological replicates were processed and sequenced (Supplemental Tables S4 [F1 and F2], S5 [alignment parameters for each data set]). The reads were normalized to H3, averaged across the two samples, and plotted across all protein-coding genes and over the 244 cryptic gene list. We noted that, genome-wide, the H3K36me3 levels were stably maintained across all ages after normalizing to H3 levels (Fig. 4A, bottom; data not shown). However, the 244 cryptic genes lost the modification with age in wild type, most obviously in the gene bodies (Fig. 4A, top). Strikingly, they retained H3K36me3 in rph1Δ old cells (Fig. 4A, top). This retention was apparent by directly examining the ChIP-seq signal across gene bodies between young and old in rph1Δ compared with wild-type old cells (Fig. 4A, top) or in the H3K36me3 signal ratio of old to young in rph1Δ compared with wild type (Fig. 4B, top). No such H3K36me3 retention was observed for randomly sampled background genes (Fig. 4A,B, bottom). A simpler and more quantitative view is to compare the H3K36me3 signal summed across the 3′ half of old cell-associated cryptic genes in the rph1Δ with wild-type strains, which similarly shows increased H3K36me3 old/young in rph1Δ compared with wild-type (Fig. 4C). We then validated the ChIP-seq observations by H3K36me3 ChIP-qPCR on two independent sorted populations of old cells. We observed that H3K36me3 was reduced in wild-type old cells at three cryptic genes (SEC63, TRP5, and SEC27) but was retained or was even higher in old cells harboring rph1Δ (Fig. 4D–F). Taken together, there is a clear increase in H3K36me3 at the 3′ end of cryptic genes in the rph1Δ strain in old cells.
Figure 4.
Genes showing up-regulation of cryptic transcription with age lose H3K36me3 over gene bodies. (A) Metagene plots of H3K36me3 tag counts normalized to H3 over the gene bodies of 244 cryptic genes (top) and all noncryptic genes designated as background (bottom). (B) Same as in A except that the tag counts are expressed as a ratio of old to young. P-values were calculated from the 3′ half of gene bodies. (C) Box plots depicting the significant reduction of H3K36me3 signal over the 3′ half of the gene. Tag counts are expressed as a ratio of old to young. (D–F) qPCR of H3K36me3 ChIP plotted relative to H3 at the 5′ and 3′ ends of three cryptic genes: SEC63, TRP5, and SEC27. Error bars are standard error of the mean of two independent biological repeats of cell sorting. Values in parenthesis are mean bud scar counts.
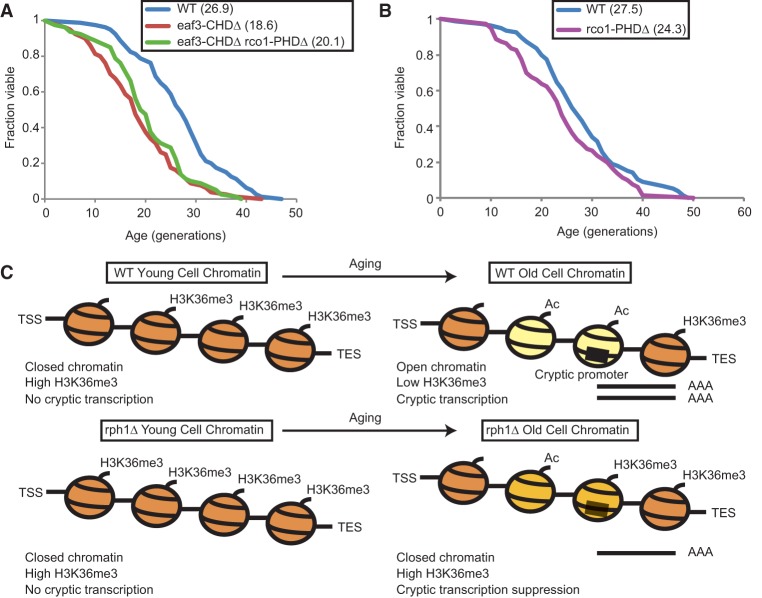
H3K36me3 recruits the deacetylase complex Rpd3S as well as the chromatin remodeling enzymes ISW1b and Chd1 (Smolle et al. 2012). We reasoned that if the short life span effect of H3K36me3 loss is mediated through one of these pathways, disruption of the recruited enzyme should phenocopy the aging phenotype of the set2Δ and H3K36 mutants. Since deletion of ISW1 or CHD1 does not shorten life span (Dang et al. 2014), we focused on the Rpd3S complex. The loss of H3K36me3 over the gene bodies of the cryptic genes likely causes inefficient recruitment of the Rpd3S complex and hyperacetylation, leading to spurious initiation of internal transcripts (Carrozza et al. 2005). Eaf3 and Rco1 are subunits specific to the Rpd3S complex that is recruited to gene bodies to remove acetyl groups laid down by histone acetyltransferases, and specific recruitment domains in the Rpd3S complex have been shown to bind directly to H3K36me3 (Li et al. 2007b). Hence, to evaluate the role of this pathway in regulating life span, we tested three strains for life span defects: one containing a deletion in the chromodomain of the Eaf3 subunit (eaf3-CHDΔ), another containing a deletion of the plant homeodomain (PHD) finger in the the Rco1 subunit (rco1-PHDΔ), and a third strain harboring deletions in both domains (eaf3-CHDΔ rco1-PHDΔ). Loss of the CHD and/or PHD shortened life span, suggesting that the association between the Rpd3S complex and H3K36me3 is important for proper longevity (Fig. 5A,B; Li et al. 2007b). Interestingly, the eaf3-CHDΔ had a stronger effect on life span compared with the rco1-PHDΔ, mirroring the relative levels of cryptic transcription previously observed in these mutants (Li et al. 2007b).
Figure 5.
Overall model of regulation of life span by the Set2/Rpd3S pathway. (A) Replicative life span assay with wild type (blue), a yeast strain lacking the chromodomain of Eaf3 (eaf3-CHDΔ; red), or both the chromodomain of Eaf3 and the PHD finger of Rco1 (eaf3-CHDΔ rco1-PHDΔ; green). (B) Replicative life span assay with wild type (blue) and a strain lacking the PHD finger of Rco1 (rco1-PHDΔ; purple). Mean life spans are indicated in parentheses. (C) Model showing the aging in wild-type and rph1Δ cells. Wild-type cells lose H3K36me3 from gene bodies with age, thus resulting in hyperacetylation, increased chromatin accessibility (marked by yellow nucleosomes), and high levels of cryptic transcription that is detrimental to life span. In contrast, rph1Δ cells retain moderate levels of H3K36me3, promoting deacetylation and closing down chromatin over intragenic cryptic promoters (marked by light-orange nucleosomes), resulting in suppression of cryptic transcription and life span extension.
Overall, we propose that, during aging, the loss of H3K36me3 leads to “open” chromatin formation at intragenic regions, exposing cryptic promoters and hence generating cryptic transcripts. Deletion of RPH1 retains H3K36me3 over the same set of genes in old cells, thus restoring “closed” chromatin, suppressing spurious initiation within gene bodies, and enhancing longevity (Fig. 5C).
Cryptic transcript up-regulation in old age is conserved in higher eukaryotes
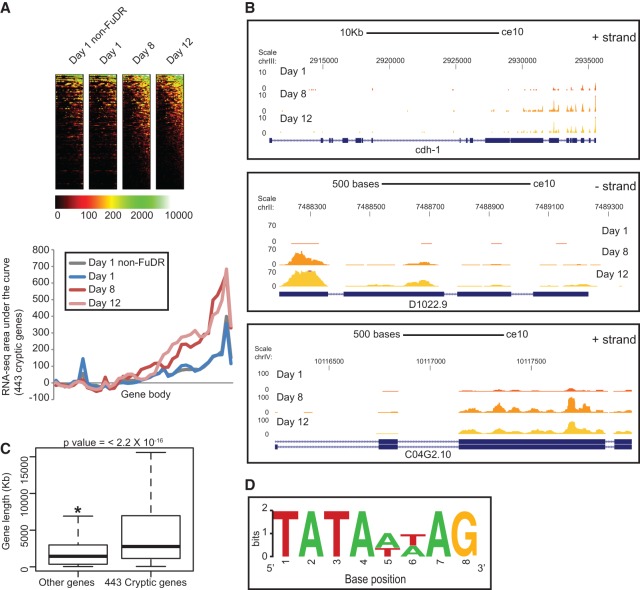
In order to investigate whether transcription fidelity is lost with age in higher eukaryotes as well, we monitored age-related cryptic transcript up-regulation in the soil nematode C. elegans (Supplemental Tables S4 [E6], S5 [alignment parameters]). Much like yeast, C. elegans serves as a suitable model for whole-organism aging in addition to having a rapid life span (mean life span of 14 d), small genome size, powerful genetics, conserved epigenetic pathways, and known single mutations that alter life span. Therefore, we performed RNA-seq in young (day 1), middle aged (day 8), and old (day 12) worms following the same short fragment protocol as used in yeast (see Supplemental Tables S4, S5). To simplify the collection of worms of different ages, they were grown in the presence of 5-fluoro-2′-deoxyuridine (FUdR), which suppresses the development of eggs and hence the birth of progeny. In contrast to our observations in yeast (Fig. 3A), metagene plots of normalized tag counts over all gene bodies did not show evident cryptic transcription genome-wide (data not shown). However, using the same parameters to define cryptic transcripts in yeast (see above), we discovered that 443 worm genes had higher accumulation of cryptic transcripts at the 3′ end of genes in middle-aged and old worms (Fig. 6A,B). The use of FUdR did not seem to have any obvious effect on the RNA-seq profiles. Similar to yeast (Fig. 3E), these genes also tended to be longer than an equal-numbered, randomly selected background set of genes from the worm genome (Fig. 6C). In keeping with our hypothesis that age-specific intragenic transcripts accumulate due to increased production and not degradation, we noted that there are 145 TATA-like sequences in 110 of the 414 cryptic genes with proper annotation (Fig. 6D; Supplemental Table S6), and this intragenic TATA-like motif is very similar to the canonical C. elegans TATA motif [(GC)TATA(AT)(AT)AG], missing only the leading (G/C) (Grishkevich et al. 2011). Both the number of cryptic genes with internal TATA-like motifs and the total number of motifs in cryptic genes were significant (Supplemental Table S6) when compared with randomly sampled noncryptic genes. These observations suggested that, like old yeast, old worms also suffer a loss of proper regulation of transcription in intragenic regions. Interestingly, loss of the Set2 homolog met-1 shortens life span, while the knockdown of the Rph1 homolog jmjd-2, which is also the only known H3K36me3 demethylase in worms, extends worm life span in a germline-dependent manner (Ni et al. 2012). Hence, it is likely that a similar aging regulation mediated through cryptic transcription and possibly involving H3K36me3 may be conserved in worms.
Figure 6.
Cryptic transcript up-regulation in old age is conserved in higher eukaryotes. (A) Metagene plots and heat maps of normalized tag counts over 443 genes in worm that show up-regulation of cryptic transcripts at day 8 and day 12. (B) Browser track views of cdh-1, D1022.9, and C04G2.10 showing the increase in 3′ end tag counts at day 8 and day 12. (C) Box plot showing that the gene length distribution of the cryptic genes is significantly higher than that of a background set of noncryptic genes. (D) WebLogo output of regular expression pattern searches for a modified TATA sequence in the worm cryptic genes.
Discussion
The study of epigenetic regulation of longevity has led to a general model of aging in which old cells show increased levels of active histone modifications and global loss of histones leading to chromatin relaxation and misregulation of gene expression (Feser et al. 2010; Das and Tyler 2012; Hu et al. 2014). These findings suggest that manipulation of the enzymes regulating these modifications could potentially stabilize chromatin structure by impacting the retained histones and extending life span (Dang et al. 2009; Greer et al. 2010, 2011; Han and Brunet 2012).
In this study, we performed a large-scale population-based screen of histone H3 and H4 substitution mutants that affect yeast replicative life span. We identified two classes of histone mutants that shorten life span: those disrupting chromatin compaction and those compromising rDNA silencing. This is consistent with recent observations that a pervasive open and active chromatin conformation results in a shorter life span (Das and Tyler 2012). Moreover, our screen not only detects histone residues known to affect life span but also identifies novel residues that were previously not known to have effects on longevity. This high-throughput approach, when combined with various yeast genetic libraries, can expedite mechanistic studies of aging, provided that life span assays can be validated by other established approaches.
We found that mutations to Lys36 on H3 resulted in shorter life span. On the nucleosome structure, H3K36 is at the base of the long H3 N-terminal tail at the site where it protrudes from the core. This site is modified by the methyltransferase Set2 that can add up to three methyl groups cotranscriptionally. Interestingly, only one known methyltransferase is known to modify this site across all eukaryotes, whereas methylation at other sites is often carried out by more than one enzyme. Consistent with enzyme nonredundancy and in support of the histone substitution screen results, deletion of Set2 in yeast resulted in a shorter life span (Fig. 2G). In contrast, we found that deletion of the only H3K36me3 demethylase, Rph1, extended life span (Fig. 2F), suggesting that this modification has longevity benefits. Our conclusions are based on the fact that deletion of Jhd1 in the matα background did not have an effect on life span. However, pending life span data in the mata background, we cannot completely rule out the impact of H3K36 monomethylation and dimethylation on life span. H3K36 methylation is also influenced by Set2 recruitment regulatory factors like Ctk1 and Bur1 kinases that phosphorylate Ser2 of the Rpb1 C-terminal domain and the PAF complex (Bartkowiak et al. 2011). However, these factors have no reported role in aging. H3K36me3 marks active genes, and the modification is primarily spread over the gene body, peaking at 3′ ends (Butler and Dent 2012; Smolle and Workman 2013). The modification provides a docking platform for specific transcription cofactor complexes and blocks access to others; the combined action of these factors restores “closed” chromatin structure over gene bodies to prevent cryptic transcription (Li et al. 2007a; Butler and Dent 2012).
Thus, in young cells, this general mechanism to preserve chromatin structure over gene bodies after Pol II passage is likely to be crucial to maintain transcriptional fidelity and precision. The gross change in chromatin structure evident in old cells is caused by loss of histones (Feser et al. 2010), local reductions in modifications such as H3K36me3 that we observed here, and local increases in activating modifications such as H4K16ac (Dang et al. 2009) and has a marked impact on transcription. Hu et al. (2014) recently described global transcriptional amplification in old yeast cells as a consequence of global nucleosome loss. Our high-resolution RNA-seq method has for the first time enabled us to see short cryptic intragenic transcripts that are normally undetectable by traditional RNA-seq methods. We showed the up-regulation of these transcripts with age in two model organisms: yeast and worms (Figs. 3D, 6A). Given these observations, it is possible that similar up-regulation of intragenic short transcripts may also occur during mammalian aging and cellular senescence. Single gene deletions in yeast (such as set2Δ, eaf3-CHDΔ, and rco1-PHDΔ) or point mutations in H3K36 that lead to accumulation of these transcripts result in shorter life spans (Fig. 5A,B). Interestingly, there is a significant overlap of genes that show age-dependent cryptic transcript up-regulation and transcription elongation mutants (set2Δ, spt6, and spt16 mutants) that show cryptic transcript up-regulation (Supplemental Fig. S11; Cheung et al. 2008; Lickwar et al. 2009).
Very recently, Pu et al. (2015) reported that H3K36me3 patterns are correlated (1) directly with 3′ untranslated region (UTR) lengths, (2) inversely with age-dependent variation in gene expression, and (3) indirectly with longevity in worms. Although we did not focus on gene expression variation, there were few similarities in our observation. First, similar to Pu et al. (2015), we did not find an appreciable change in H3K36me3 with age in yeast. Second, the variation in gene expression with age may be due to the spurious initiation of cryptic transcription, as some genes lose H3K36me3 with age. Third, loss of somatic methyltransferase met-1 resulted in a shorter life span, much like a set2Δ strain in yeast (Pu et al. 2015).
The cause of accumulation of these transcripts with age is due to either increased production, impaired degradation, or both. Nascent RNA-seq techniques like NET-seq and GRO-seq are challenging in old cells given (1) limited quantities of RNA isolated from old cells and (2) the disproportionate transcription from coding and rDNA templates. Without this information, however, it is not possible to completely eliminate the possibility of RNA degradation. However, the degradation machinery for these intragenic transcripts has not been unambiguously defined, and mutants of RNA degradation machineries like the exosome complex do not lead to accumulation of this transcript class (Supplemental Fig. S10; Smolle and Workman 2013). Additionally, our discovery of promoter-like motifs within the age-specific cryptic genes in yeast and worms (Figs. 3G, 6D) further argues for an initiation mechanism rather than degradation.
It is important to note that we did not analyze nonpolyadenylated sense or antisense transcripts in our data sets. It is known that H3K36me3 represses a number of antisense transcripts regulated by Spt6 and Set2 (DeGennaro et al. 2013). We focused on the analysis of sense polyadenylated cryptic transcripts for the following two reasons. (1) We believe that polyadenylated stable transcripts could have long term effects on aging, and (2) the level of antisense transcription is much lower than sense transcription. However, we do not rule out the role of these transcripts in life span regulation.
Taken together, the current evidence supports an epigenetic aging model encompassing the breakdown of chromatin structure via the accumulation of activating modifications, loss of repressing modifications, and reduction of nucleosome numbers to initiate a cascade of spurious transcriptional events (including gene-internal cryptic transcription) that promote the aging process. Given that cryptic transcription increases with age and many mutants in the Set2/Rpd3S pathway have short life spans, as shown here, it is important to consider the impact of cryptic transcription in limiting life span. Cryptic transcripts may (1) increase the amount of “junk” transcripts in the cell, thus engaging the translation machinery in fruitless activities; (2) be translated or mistranslated into proteins that might be toxic, including misfolded proteins that would elicit an unfolded protein response; and/or (3) titrate important transcription factors, including Pol II and general transcription factors, away from more appropriate genomic locations. In support of the first model, overloading the cell with templates for translation entails production and consumption of overly large amounts of ATP, increasing the generation of reactive oxygen species that can be detrimental to life span (Balaban et al. 2005). In general, reduced translation has proven beneficial for longevity in multiple model systems (Johnson et al. 2013).
Finally, we found that cells lacking Rph1 maintain higher H3K36me3 levels and have reduced levels of spurious cryptic transcripts with age, commensurate with a 30% extension of mean replicative life span. Similar effects on longevity have also been reported in worms following knockdown of Rph1 homolog jmjd-2 (Ni et al. 2012). Although, in higher eukaryotes, the Rph1 homolog targets both H3K9 and H3K36, it is exciting to envision the design of specific small molecule or other inhibitors targeting H3K36 demethylation activity to ameliorate age-associated human disease.
Materials and methods
Yeast strains
The yeast strains used in this study are listed in Supplemental Table S1. Standard YPD medium was used in all experiments.
Purification of old yeast mother cells
Up to 4 × 108 replicatively aged yeast mother cells (20–25 generations old on average) were purified as described (Dang et al. 2014) for the life span screen, RNA-seq, or ChIP-seq.
Yeast replicative life span assay
Life span assays were done by micromanipulation under a dissection scope as described (Kaeberlein et al. 1999). Statistical significance of life span differences were calculated by Wilcoxon rank sum test.
Total RNA extraction from yeast, real-time qPCR, and Northern blotting
Total RNA was extracted from frozen cell pellets using the Qiagen RNeasy kit following the instructions, except that after resuspending in QIAzol lysis reagent, cells were subjected to bead beating. Bead beating was performed with 1.0-mm-diameter zirconia/silica beads (BioSpec) four times, 1 min each time, with 2-min pauses on ice. On-column DNase I digestion was performed to remove residual genomic DNA. RNA purity and concentration were estimated by Nanodrop. For qPCR, 1 µg of total RNA was converted to cDNA using the high-capacity RNA to cDNA kit (Applied Biosystems). A 1:100 dilution of cDNA was then subjected to real-time PCR with genomic DNA standards using Fast SYBR Green master mix (Applied Biosystems).
To estimate rDNA copy number, a 1:100 dilution of ChIP input DNA was used against genomic DNA standards to do qPCR. All qPCR quantitation was done using the absolute (standard curve) method.
Northern blotting was performed using a NorthernMax kit (Life Technologies). Briefly, 10 µg of total RNA was subjected to polyA selection using the Dynabeads mRNA Direct kit (Ambion) and loaded on a 1% formaldehyde agarose gel, blotted on a positively charged nylon membrane (Hybond N+, GE Healthcare), UV cross-linked, and then probed with radiolabeled PCR products designed against the 5′ end of STE11 and the 3′ end of STE11 and ACT1.
Worm strains, media, and aging
N2 (C. elegans variant Bristol) obtained from the Caenorhabditis Genetics Center was used in the aging experiment. Worms were staged by bleaching gravid adults and synchronized to L1 in the M9 buffer. L1 worms were grown on NGM plates seeded with OP50 Escherichia coli until L4, which corresponds to day 0, when they were transferred to NGM plates supplemented with 100 µM FUdR and seeded with OP50. At days 1, 8, and 12 of adulthood, 500–1000 worms and 10,000 worms were washed in the M9 buffer and collected for RNA-seq and ChIP experiments, respectively.
Total RNA extraction from worms
Worms were lysed in QIAzol (Qiagen) by bead beating with 1.0-mm-diameter zirconia/silica beads (BioSpec) four times, 1 min each time, with 2-min pauses on ice. Total RNA was then extracted and purified with miRNeasy minikit (Qiagen).
High-throughput life span screen with the histone H3/H4 mutant library
All strains from the Boeke histone H3/H4 mutant library were pooled together with additional uniquely barcoded wild-type, sir2Δ, and SIR2-OE strains. The pool of mutants was cultured and subjected to four successive rounds of old mother sorting to save ∼108 old cells after sorts 2, 3, and 4 (S2O, S3O, and S4O). Progressively aged cells were indicated by the increasing number of bud scar counts. Equivalent amounts of young cells after each round of sorting (S1Y, S2Y, S3Y, and S4Y) as well as an aliquot prior to aging and sorting (USC) were also saved. Genomic DNA was extracted from young and old fractions using standard methods (Current Protocols in Molecular Biology), and the barcodes UPTAG and DNTAG were quantified by either yeast barcode microarray (Agilent) as described (Matecic et al. 2010) or next-generation sequencing. To prepare next-generation sequencing libraries, primers U1h-F1 (CCCTACACGACGCTCTTCCGATCTATGTCCACGAGGTCTCT) and U2h-R1 (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCCTCGACCTGCAGCGTA) were used to amplify UPTAG, and primers D1h-F1 (CCCTACACGACGCTCTTCCGATCTCCCAGCTCGAATTCATC) and D2h-R1 (GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCGGTGTCGGTCTCGTAG) were used for DNTAG. All next-generation sequencing libraries were indexed and sequenced in 1/10 of a lane in HiSeq 2000 (Illumina). All data were converted to fold change compared with USC and designated as relative enrichment. All mutant strains were ranged by S4O relative enrichment. Both doubling time and replicative life span may affect S4O relative enrichment. However, relative enrichment changes of young fractions compared with the fraction prior to the overnight culturing were affected only by doubling time. Therefore, S1Y/USC, S3Y/S2O, S4Y/S3O values were used to estimate relative enrichment effects caused by doubling time difference. This growth effect on S4O relative enrichment was considered only for mutants with an estimated doubling time 20% different from that of the wild type. S2O and S3O relative enrichment data were used to filter out mutants that showed significant variations. Longevity calls were based on S4O relative enrichment after correcting for difference in doubling time and filtered for variation outliers, with a cutoff at a standard deviation from the mean of the log (S4O relative enrichment) values.
RNA-seq library preparation for quantifying cryptic transcripts
To generate libraries for RNA-seq, mRNA was purified from 5 µg of total RNA using the Dynabeads mRNA Direct kit (Ambion). Since yeast genes are relatively short (average gene size ∼1.3 kb), we chose to fragment the mRNA to ∼40–50 bp rather than the conventional 200- to 300-bp size using RNA fragmentation reagents from Ambion. We believe that this gives better resolution over gene bodies to quantify 5′ and 3′ reads for estimation of cryptic transcripts. To do this, the fragmented RNA was phosphorylated by T4 PNK (New England Biolabs) and then size-selected on a 15% polyacrylamide gel-containing urea. Following size selection, the entire RNA was used as input in the TruSeq small RNA sample preparation protocol (Illumina). Final size selection of the amplified library was done using 6% polyacrylamide gels. For the sake of consistency, RNA-seq libraries from worms were prepared using the same protocol. The quantity and quality of the libraries were assessed by BioAnalyzer (Agilent) and qPCR (Kapa Biosystems). Eight multiplexed libraries were pooled and sequenced on a single lane on the Illumina HiSeq or NextSeq 500 platform.
ChIP and preparation of ChIP-seq libraries
ChIP was performed as described previously from 4 × 108 cells (Dang et al. 2009). Briefly, cross-linked cells were lysed by bead beating in FA lysis buffer (50 mM HEPES-KOH at pH 7.5, 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100), and the chromatin was sheared using a Diagenode Bioruptor or Covaris Ultrasonicator to achieve an average size of <500 bp. Equal aliquots of sonicated chromatin were used per immunoprecipitation reaction (H3K4me3 [Abcam, ab8580], H3K36me3 [Active Motif, 61101], and H3 [Abcam, ab1791]), and 10% of the amount was saved as input. Immunoprecipitation was performed using protein A Dynabeads (Life Technologies). ChIP-qPCR was performed against standards made from a dilution series of an input DNA mix. Enrichment was ascertained by calculating signal over IgG. To ensure that we were getting H3K4me3 and H3K36me3 enrichment at 5′ ends and 3′ ends, respectively, ChIP-qPCR validation was performed with 5′ and 3′ end primer sets over long genes before library preparation for ChIP-seq. Primer sequences are available on request.
For ChIP-seq, 1 ng of DNA from immunoprecipitation and input was used to prepare libraries using the Microplex library preparation kit (Diagenode). Twelve multiplexed libraries were sequenced in a single lane on the Illumina HiSeq or NextSeq 500 platform.
List of RNA-seq and ChIP-seq data sets and computational methods
For a list of RNA-seq and ChIP-seq data sets and computational methods, see the Supplemental Material. The raw data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=yhylgoigxrwtfuh&acc=GSE65767.
Supplementary Material
Acknowledgments
We thank Jerry Workman for the strains YBL619, YBL634, and YBL694; the University of Pennsylvania Next-Generation Sequencing Core (NGSC) for sequencing our ChIP-seq and RNA-seq libraries; and members of the Berger laboratory for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH)/National Institute on Aging (NIA) grant P01AG031862 to S.L.B., American Heart Association grant 15POST21230000 to P.S., NIH/NIA grant K99/R00AG037646 and Cancer Prevention Research Institute of Texas scholar award R1306 to W.D., NIH Training grant T32ES007032 to B.M.W., NIH grant R01AG039390 and the University of Washington Nathan Shock Center of Excellence in the Basic Biology of Aging (NIH P30AG013280) to M.K., NIH/NIA grant R01AG043080 to B.K.K., and NIH Common Fund grant U54GM103250 to J.D.B.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.263707.115.
References
- Adams PD, Ivanov A, Pawlikowski J, Manoharan I, Van Tuyn J, Nelson DM, Singh Rai T, Shah PP, Hewitt G, Korolchuk VI, et al. 2013. Lysosome-mediated processing of chromatin in senescence. J Cell Biol 202: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. 2005. Mitochondria, oxidants, and aging. Cell 120: 483–495. [DOI] [PubMed] [Google Scholar]
- Bartkowiak B, Mackellar AL, Greenleaf AL. 2011. Updating the CTD story: from tail to epic. Genet Res Int 2011: 623718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander G, Guarente L, Words K. 2004. The Sir2 family of protein deacetylases. Annu Rev Biochem 73: 417–435. [DOI] [PubMed] [Google Scholar]
- Butler JS, Dent SYR. 2012. Chromatin ‘resetting’ during transcription elongation: a central role for methylated H3K36. Nat Struct Mol Biol 19: 863–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia W-J, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. [DOI] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD, Jef D. 2008. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell 134: 1066–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Steffen KK, Perry R, Dorsey JA, Johnson FB, Shilatifard A, Kaeberlein M, Kennedy BK, Berger SL. 2009. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459: 802–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Sutphin GL, Dorsey JA, Otte GL, Cao K, Perry RM, Wanat JJ, Saviolaki D, Murakami CJ, Tsuchiyama S, et al. 2014. Inactivation of yeast Isw2 chromatin remodeling enzyme mimics longevity effect of calorie restriction via induction of genotoxic stress response. Cell Metab 19: 952–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Tyler JK. 2012. Histone exchange and histone modifications during transcription and aging. Biochim Biophys Acta 1819: 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro CM, Alver BH, Marguerat S, Stepanova E, Davis CP, Bahler J, Park PJ, Winston F. 2013. Spt6 regulates intragenic and antisense transcription, nucleosome positioning, and histone modifications genome-wide in fission yeast. Mol Cell Biol 33: 4779–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Tyler J. 2011. Chromatin structure as a mediator of aging. FEBS Lett 585: 2041–2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feser J, Truong D, Das C, Carson JJ, Kieft J, Harkness T, Tyler JK. 2010. Elevated histone expression promotes life span extension. Mol Cell 39: 724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry CJ, Norris A, Cosgrove M, Boeke JD, Peterson CL. 2006. The LRS and SIN domains: two structurally equivalent but functionally distinct nucleosomal surfaces required for transcriptional silencing. Mol Cell Biol 26: 9045–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giblin W, Skinner ME, Lombard DB. 2014. Sirtuins: guardians of mammalian healthspan. Trends Genet 30: 271–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, Han S, Banko MR, Gozani O, Brunet A, et al. 2010. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 125: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishkevich V, Hashimshony T, Yanai I. 2011. Core promoter T-blocks correlate with gene expression levels in C. elegans. Genome Res 21: 707–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Brunet A. 2012. Histone methylation makes its mark on longevity. Trends Cell Biol 22: 42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Chen K, Xia Z, Chavez M, Pal S, Seol J-H, Chen C-C, Li W, Tyler JK. 2014. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 28: 396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Rabinovitch PS, Kaeberlein M. 2013. mTOR is a key modulator of ageing and age-related disease. Nature 493: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR Jr, Zhang J, Guarente L. 1995. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80: 485–496. [DOI] [PubMed] [Google Scholar]
- Kim T, Buratowski S. 2007. Two Saccharomyces cerevisiae JmjC domain proteins demethylate histone H3 Lys36 in transcribed regions to promote elongation. J Biol Chem 282: 20827–20835. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Gardner KE, Liang G, Erdjument-Bromage H, Tempst P, Zhang Y. 2007. Demethylation of histone H3K36 and H3K9 by Rph1: a vestige of an H3K9 methylation system in Saccharomyces cerevisiae? Mol Cell Biol 27: 3951–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DW, Ahn SH. 2011. Role of yeast JmjC-domain containing histone demethylases in actively transcribed regions. Biochem Biophys Res Commun 410: 614–619. [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. 2007a. The role of chromatin during transcription. Cell 128: 707–719. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. 2007b. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316: 1050–1054. [DOI] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. 2007c. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. 2009. The set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PLoS One 4: e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, Smith JS. 2010. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet 6: e1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Sanderson BW, Delventhal KM, Bradford WD, Staehlinghampton K, Shilatifard A. 2008. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat Struct Mol Biol 15: 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Ebata A, Alipanahiramandi E, Lee SS. 2012. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 11: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. 2010. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 17: 1218–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-HH, Cosgrove MS, Youngman E, Wolberger C, Boeke JD. 2002. A core nucleosome surface crucial for transcriptional silencing. Nat Genet 32: 273–279. [DOI] [PubMed] [Google Scholar]
- Pelechano V, Chávez S, Pérez-Ortín JE. 2010. A complete set of nascent transcription rates for yeast genes. PLoS One 5: e15442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu M, Ni Z, Wang M, Wang X, Wood JG, Helfand SL, Yu H, Lee SS. 2015. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev 29: 718–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Dowen RH, Lourenco GF, Kirienko NV, Heimbucher T, West JA, Bowman SK, Kingston RE, Dillin A, Asara JM, et al. 2013. DAF-16 employs the chromatin remodeller SWI/SNF to promote stress resistance and longevity. Nat Cell Biol 15: 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Duda S, Sargent DF, Richmond TJ. 2005. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436: 138–141. [DOI] [PubMed] [Google Scholar]
- Shah PP, Donahue G, Otte GL, Capell BC, Nelson DM, Cao K, Aggarwala V, Cruickshanks HA, Rai TS, McBryan T, et al. 2013. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev 27: 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. 1997. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell 91: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sinclair D, Mills K, Guarente L. 1998. Aging in Saccharomyces cerevisiae. Annu Rev Microbiol 52: 533–560. [DOI] [PubMed] [Google Scholar]
- Smeal T, Claus J, Kennedy B, Cole F, Guarente L. 1996. Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84: 633–642. [DOI] [PubMed] [Google Scholar]
- Smolle M, Workman JL. 2013. Transcription-associated histone modifications and cryptic transcription. Biochim Biophys Acta 1829: 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 19: 884–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Grant PA, Briggs SD, Sun Z-W, Bone JR, Caldwell JA, Mollah S, Cook RG, Shabanowitz J, Hunt DF, et al. 2002. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpferl SW, Brand SE, Jiang JC, Korona B, Tiwari A, Dai J, Seo J, Jazwinski SM. 2012. Natural genetic variation in yeast longevity. Genome Res 22: 1963–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. 2012. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489: 452–455. [DOI] [PubMed] [Google Scholar]
- Wasko BM, Kaeberlein M. 2013. Yeast replicative aging: a paradigm for defining conserved longevity interventions. FEMS Yeast Res 14: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Helfand SL. 2013. Chromatin structure and transposable elements in organismal aging. Front Genet 4: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.