Burman et al. find enrichment of several histone modifications over clinically relevant translocation-prone genome regions. Experimental modulation of histone marks sensitized genome regions to breakage by endonuclease challenge or irradiation and promoted formation of chromosome translocations.
Keywords: chromosome translocations, histone modifications, chromatin structure
Abstract
Chromosome translocations are well-established hallmarks of cancer cells and often occur at nonrandom sites in the genome. The molecular features that define recurrent chromosome breakpoints are largely unknown. Using a combination of bioinformatics, biochemical analysis, and cell-based assays, we identify here specific histone modifications as facilitators of chromosome breakage and translocations. We show enrichment of several histone modifications over clinically relevant translocation-prone genome regions. Experimental modulation of histone marks sensitizes genome regions to breakage by endonuclease challenge or irradiation and promotes formation of chromosome translocations of endogenous gene loci. Our results demonstrate that histone modifications predispose genome regions to chromosome breakage and translocations.
Chromosome translocations are among the most common genetic aberrations found in human cancers (Mitelman et al. 2007). The initiating molecular event in the formation of any chromosome translocation is the occurrence of persistent DNA double-strand breaks (DSBs) (Roukos and Misteli 2014), frequently induced by cellular and genotoxic stress (Mani and Chinnaiyan 2010; Lin et al. 2012). Translocations recur at nonrandom genomic sites, and breakpoints tend to cluster in the translocating genes, often within introns (Zhang and Rowley 2006). The molecular features that determine which regions of the genome are particularly susceptible to breakage and translocation are largely unknown (Roukos et al. 2013a). While DNA sequence features such as CpG content, repetitive elements, and secondary non-B DNA structures have been implicated in promoting breakage of some genome regions (Nambiar and Raghavan 2011), large-scale sequencing of translocation junctions indicates considerable sequence diversity over translocation-prone genome regions (Talkowski et al. 2011). Given the integral role of chromatin in DNA accessibility and repair (Price and D'Andrea 2013) and the fact that DSBs occur in the context of chromatin, we sought to probe the role of histone modifications in the formation of chromosome translocations using a combined computational, biochemical, and cell-based approach.
Results
Computational analysis of chromatin features at recurrent breakpoints
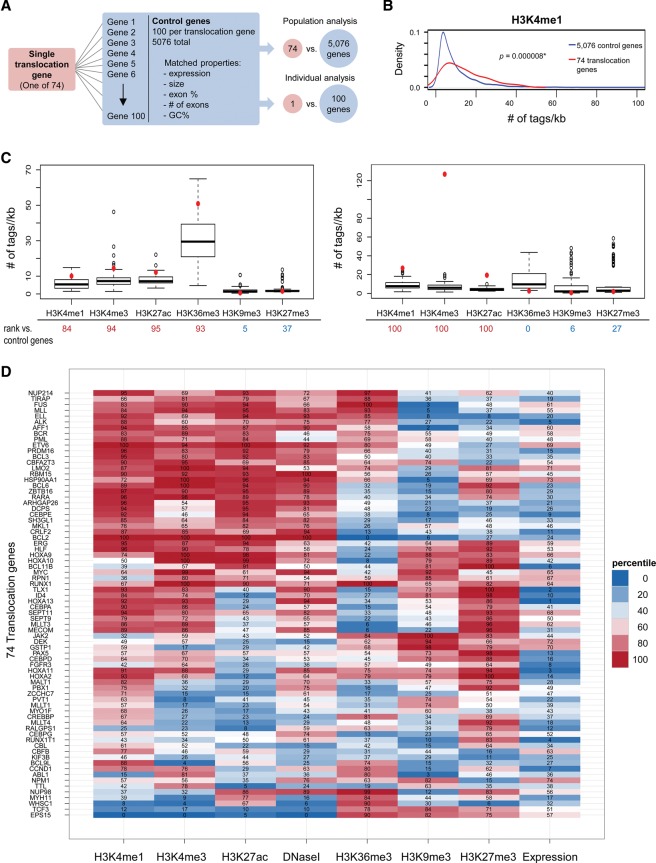
To identify candidate chromatin features that may mark human translocation breakpoints, we computationally analyzed histone modifications and chromatin structure in a large set of frequent translocation genes in primary CD34+ hematopoietic cells using genome-wide chromatin immunoprecipitation (ChIP) sequencing (ChIP-seq) and DNase I sequencing (DNase I-seq) data available through the National Institutes of Health Roadmap Epigenomics Project. CD34+ cells are ideally suited for this analysis, since many translocation events in leukemias and lymphomas are thought to occur within the hematopoietic stem cell population or in early progenitors derived from this population (Bonnet and Dick 1997; Bernt and Armstrong 2009). A set of 74 translocation genes, defined as translocated in at least 10 clinical cases of hematologic malignancies, was selected using the Mitelman Database of Chromosome Aberrations (translocation genes) (Supplemental Table S1). Each translocation gene was paired with a set of 100 control RefSeq genes not implicated in any translocations (control genes) (Fig. 1A; Supplemental Table S1). Each set of control genes shared closely matching properties with its translocation gene, including quantitative expression status based on mRNA-seq data, gene length, exon percentage, number of exons, and GC content (Fig. 1A; see the Materials and Methods). The histone modification ChIP-seq tag density in the gene body ±2 kb of the pooled set of 74 translocation genes was first compared with the aggregate population of 5076 control genes (<7400 due to overlapping control genes). While most histone modifications were similar among the compared gene sets, H3K4me1 was significantly enriched in the translocation genes (P < 0.01) (Fig. 1B; Supplemental Fig. S1A–F). To achieve higher sensitivity and eliminate the possibility of not detecting histone modifications enriched or depleted in subpopulations of genes, each translocation gene was individually compared with its set of 100 matched control genes and ranked relative to them (Fig. 1C,D; Supplemental Fig. S1G). This analysis confirmed that H3K4me1 was strongly enriched in a majority of translocation genes (41 of 74 translocation genes, >75th percentile). By hierarchical clustering, most H3K4me1-enriched translocation genes showed mutual enrichment of H3K4me3, H3K27ac, and DNase I hypersensitivity (Fig. 1D). While several of these marks are commonly associated with transcriptionally active regions, the enrichments in translocation-prone genes were not due to their elevated transcriptional activity, since the control genes were selected based on closely matching expression levels (Supplemental Fig. S2). A distinct set of translocation genes was characterized by depletion of the H3K9me3-repressive mark compared with control genes (14 of the 37 bottom genes according to gene expression) despite their transcriptional silencing based on mRNA-seq data (Supplemental Fig. S3). Fifteen translocation genes, including RUNX1, CEBPA, SEPT11, and four out of five Hox genes, featured bivalent enrichment of H3K4me3 and H3K27me3, which is commonly seen in genes poised for transcription in stem cells (Bernstein et al. 2006). Taken together, these data suggest the presence of specific histone modification patterns at translocation-prone genes in hematologic malignancies.
Figure 1.
Computational analysis of chromatin features at recurrent translocation genes. (A) Strategy for analysis of chromatin features using a population or individual gene approach. Seventy-four frequent translocation genes were identified, and, for each translocation gene, 100 control genes were selected with closely matched properties: expression, size, exon percentage, number of exons, and GC percentage. (B) For population analysis, a kernel density plot comparing H3K4me1 signal density distribution in 74 translocation genes versus 5076 control genes is shown. Signal density was measured over each annotated gene body ±2 kb, and the average read count (number of tags per kilobase per 1 million reads) in each gene is represented on the X-axis. The distribution of H3K4me1 in translocation genes is significantly enriched compared with control genes. (*) P < 0.01, Wilcoxon test. (C) For individual gene analysis, the average read count (number of tags per kilobase per 1 million reads) for each histone modification of each translocation gene (red dot) was compared with its 100 control genes (box plot). The percentage of control genes whose tag densities were lower than that of the translocation gene was calculated (rank listed below the X-axis). (D) Heat map of individual analyses of histone modifications and DNase I hypersensitivity for 74 translocation genes. The percentiles of each translocation gene ranked amongst its control genes are indicated. The rank of expression for translocation genes from 1 to 74 is shown at the right. Translocation genes were hierarchically clustered using the overall ranking matrix.
Altered levels of histone modification at breakpoints in anaplastic large cell lymphoma
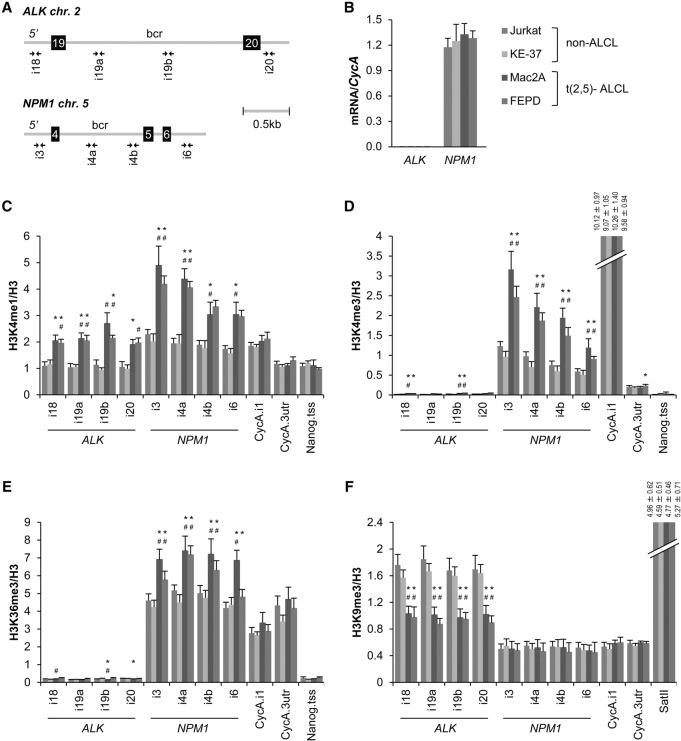
To specifically test the association between histone modifications and translocations in a well-defined clinically relevant translocation system, we used anaplastic large cell lymphoma (ALCL). ALCL is characterized by recurrent translocations between nucleophosmin 1 (NPM1) on chromosome 5 and anaplastic lymphoma kinase (ALK) on chromosome 2, leading to constitutive activation of the ALK tyrosine kinase (Tabbo et al. 2013). However, a significant fraction of symptomatically indistinguishable patients lacks the t(2;5) translocation (Tabbo et al. 2013). t(2;5)-negative ALCL cell lines derived from these patients are poised to undergo t(2,5) translocations, since NPM1 and ALK form translocations at high frequency upon irradiation (Mathas et al. 2009). To identify chromatin features that may contribute to predisposing ALK and NPM1 toward breakage and translocation, we probed the histone modification landscape over the breakpoint regions in t(2;5)-negative cells. Histone modifications were mapped using specific primer sets around the most frequent breakpoints located in the 910-base-pair (bp) intron 4 of NPM1 and the 1923-bp intron 19 of ALK (Fig. 2A), and histone modification levels were normalized to unmodified H3. As previously reported, the NPM1 gene was expressed in all four cell lines, whereas the ALK gene transcript was undetectable in all lines (Fig. 2B; Mathas et al. 2009). Comparative mapping by quantitative ChIP (qChIP) of a set of histone modifications across the breakpoint regions in two t(2,5)-negative ALCL cell lines, FEPD and Mac2A, showed twofold enrichment of H3K4me1, H3K4me3, and H3K36me3 at the NPM1 locus compared with control Jurkat and KE37 cells that do not form NPM1–ALK translocations upon irradiation (P < 0.05) (Fig. 2C–E; Mathas et al. 2009). As observed in the global computational analysis, histone modification enrichments were not related to expression level, as NPM1 was similarly expressed across all four lines (Fig. 2B). Similarly, the ALK locus was marked by a 1.5-fold enrichment of H3K4me1 and a 1.5-fold reduction in H3K9me3 across the breakpoint region in t(2,5)-negative ALCL lines compared with control cell lines (P < 0.05) (Fig. 2C,F), again uncorrelated with transcription status (Fig. 2B). These alterations did not represent a global alteration of H3 methylation marks in ALCL cells, since the levels of these modifications were unchanged at the constitutively active cyclophilin A (CycA) gene, the constitutively inactive Nanog locus, and pericentric-heterochromatin-specific SatII regions in all cell lines (Fig. 2C–F). Similarly, lower levels of nucleosome density across NPM1 and ALK regions were detected in t(2;5)-negative ALCL cells, as indicated by the reduced ratio of immunoprecipitated H3 DNA to total DNA input (Supplemental Fig. S4A). Several chromatin features, including active (H3K9ac, H3K27ac, H3K79me2, and H4K16ac) and inactive (H3K9me1 and H3K27me3) histone marks as well as those found to mark both active and inactive genes (H3K56ac and H4K20me1), did not differ between cell lines (Supplemental Fig. S4B–I).
Figure 2.
Altered levels of histone modifications at translocation breakpoints in t(2,5)− ALCL. (A) Schematic representation of the human ALK and NPM1 breakpoint cluster regions (bcr). (Squares) Exons; (gray lines) introns; (paired arrows) primer pairs used in ChIP-qPCR (quantitative PCR) experiments. (B) mRNA levels of ALK and NPM1 relative to CycA in Jurkat (dark blue), KE37 (light blue), Mac2A (dark red), and FEPD (light red) cell lines determined by qPCR. (C–F) Mapping of H3K4me1 (C), H3K4me3 (D), H3K36me3 (E), and H3K9me3 (F) at breakpoint and control regions in cell lines indicated in B by ChIP-qPCR. The percentage of input was normalized to unmodified H3. (B–F) values represent means ± SEM from three to four independent experiments. (*) P < 0.05 to Jurkat cells; (#) P < 0.05 to KE37 cells, Student's t-test.
H3K4 methylation facilitates DSB formation by endonucleases
We next sought to directly test the role of specific histone modifications in chromosome breakage and translocation formation. As a first approach, we tested DSB formation by endonucleases, since several translocation events in hematopoietic and solid tumors result from off-target and sequence-independent DSBs induced by endogenous endonucleases (Lin et al. 2012). Using a previously characterized Lac repressor/operator protein–DNA-tethering system containing an integrated I-SceI restriction site (Soutoglou and Misteli 2008; Burgess et al. 2014), we created chromatin domains enriched in specific histone modifications by tethering fusion proteins between the LacR and histone-modifying enzymes responsible for candidate translocation-relevant histone modifications (H3K4 methyltransferases ASH2L and SET7/9, H3K36 methyltransferase SET2, and H3/4 lysine acetyltransferase TIP60) to the LacO array in U2OS cells (Fig. 3A). DSB formation by I-SceI in the chromatin domain was assessed after expression for 12 h of the glucocorticoid receptor I-SceI (GR-I-SceI), which localizes to the cytoplasm with trace amounts in the nucleus due to the leakiness of cytoplasmic retention (Fig. 3A; Soutoglou et al. 2007). The catalytic activity and deposition of the correct histone modifications on the LacO array was confirmed by immunofluorescence (IF) and ChIP (Fig. 3B; Supplemental Fig. S5A–E). Tethering of ASH2L or SET7/9 resulted in visible expansion of the array and reduced recovery of immunoprecipitated H3 at the array, suggesting local chromatin decondensation (P < 0.05) (Fig. 3B,C; Supplemental Fig. S5F). When probed by ligation-mediated PCR (LM-PCR), I-SceI-induced DSB formation was elevated ∼20-fold after tethering the positive control remodeler BRG1 compared with LacR alone and five- and fourfold in H3K4 methylated domains created by ASH2L and SET7/9, respectively (P < 0.05) (Fig. 3D). Similarly, hyperacetylated domains created by TIP60 and H3K36 methylated domains created by SET2 were ∼2.5-fold more sensitive to DSBs by I-SceI than LacR alone (P < 0.05) (Fig. 3D). In contrast, SUV3-9H1-mediated H3K9 methylation, which was found depleted over translocation-prone genome regions, allowed twofold fewer DSBs than LacR alone, indicating that H3K9 methylation impairs I-SceI-induced DSB formation (P < 0.05) (Fig. 3D). We conclude that H3K4 methylation and, to a lesser extent, H3K36 methylation and histone acetylation decondense chromatin and facilitate DSB formation when challenged by an endonuclease.
Figure 3.
H3K4 methylation induces chromatin decondensation and facilitates DSB formation by endonucleases. (A) Schematic representation of chromatin-protein-tethering system. Lac repressor (LacR) fusions to either mCherry alone or to histone-modifying enzymes are tethered to the LacO-I-SceI-TetO array in U2OS cells after transient expression. DSBs are formed by transient expression of GR-I-SceI, which localizes to the cytoplasm with trace amounts in the nucleus due to leakiness of cytoplasmic retention. (B, left panel) Visualization of mCherry-LacR-ASH2L (ASH2L) or mCherry-LacR (Lac) 20 h after expression. Values in the left panel show median array size ± median absolute deviation from >50 nuclei. Enrichment of H3K4me3 at ASH2L- or Lac-tethered arrays (arrows) when cells were fixed and immunostained for H3K4me3 is shown in the middle and right panels (merge). Values in the right panel show the percentage of positive colocalizations between tethered array and histone modification foci from >100 nuclei. Images are the maximum intensity projections of representative cells. Bar, 5 µm. (C) H3 density calculated from ChIP-qPCR of unmodified H3 relative to total DNA input at primers for the array in cells expressing mCherry-LacR fusions to the indicated histone-modifying enzymes or mCherry-LacR alone (Lac). (D) LM-qPCR detecting the quantity of DSBs in cells expressing mCherry-LacR fusions to the indicated histone-modifying enzymes or mCherry-LacR alone (Lac) and GR-I-SceI. (C,D) Values represent means ± SD from three independent experiments. (*) P < 0.05, Student's t-test.
H3K4 methylation facilitates breakage of endogenous loci
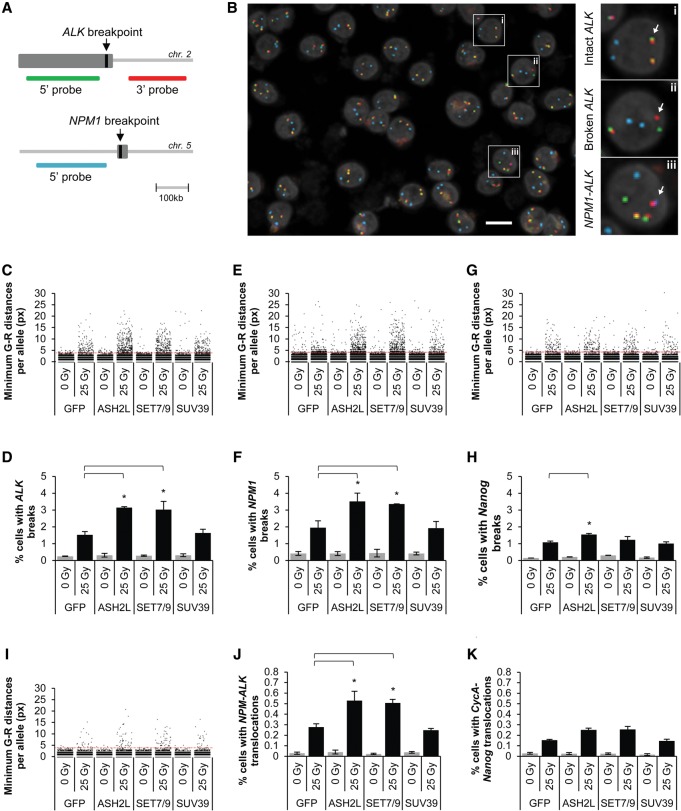
To address whether translocation-associated histone modifications affect DNA breakage at endogenous loci in cells, we generated t(2,5)-negative ALCL FEPD and non-ALCL control Jurkat cell lines stably overexpressing GFP-ASH2L or GFP-SET7/9. Elevated H3K4 methylation at ALK and NPM1 breakpoints was confirmed by ChIP (Supplemental Fig. S6). Breakage frequency was measured 24 h after exposure to 25 Gy of ionizing radiation (IR) by high-throughput fluorescence in situ hybridization (hiFISH) of 8000–10,000 cells per experimental condition using break-apart probe sets for the ALK and NPM1 loci (Fig. 4A). These probe sets report on chromosome breakage by measuring separation (more than four pixels; equivalent to a distance of 1.28 μm) of two probes targeted to either side of the ALK and NPM1 breakpoint regions (Fig. 4A,B, panels i,ii; Supplemental Fig. S7A; see the Materials and Methods). As a positive technical control, at least one ALK and one NPM1 breakage event were detected in 95.4% (95% CI: 94.5–96.1, 2695 cells) and 96.1% (95% CI: 95.2%–96.8%, 2448 cells) of cells, respectively, in the t(2;5)-positive ALCL cell line Karpas299 (Fischer et al. 1988; data not shown). Cells expressing either H3K4 methyltransferase exhibited twofold higher breakage frequency (ALK: ASH2L 3.15% ± 0.05% [SD], SET7/9 3.02% ± 0.49%, P < 0.05; NPM1: ASH2L 3.52% ± 0.49%, SET7/9 3.36% ± 0.01%) than GFP-expressing control cells (ALK: 1.53% ± 0.19%; NPM1: 1.95% ± 0.41%) after irradiation (Fig. 4C–F). As a control for a histone-modifying enzyme not observed to increase DSB formation by endonucleases (Fig. 3D), cells expressing SUV3-9H1 caused no significant increase in breakage frequency (ALK: 1.64% ± 0.22%; NPM1: 1.93% ± 0.40%; Fig. 4C–F). Similar, albeit weaker, effects were observed at an ALCL-unrelated locus (Nanog) that, like ALK, is not expressed in t(2,5)-negative FEPD cells (Fig. 4G,H). In all cases, breakage frequencies in irradiated cells were severalfold higher than the background breakage frequency in nonirradiated cells (0.31% ± 0.10% across all conditions) (Fig. 4C–H). The extent of breakage correlated with H3K4 methylation levels, which were considerably higher at ALK and NPM1 than at Nanog (Supplemental Fig. S6A,B), suggesting a locus-independent effect of H3K4 methylation on breakage susceptibility. Along the same lines, expression of either H3K4 methyltransferase caused an increase in DNA breaks at ALK in Jurkat cell lines but to a lesser degree than in FEPD cells (Supplemental Fig. S7B,C), in line with the lower levels of H3K4 methylation at breakpoints in Jurkat cells (Fig. 2C,D). These observations suggest that increased H3K4 methylation promotes breakage susceptibility to irradiation.
Figure 4.
H3K4 methylation increases breakage and translocation formation by irradiation. (A) Schematic representation of the ALK probe set. (Green probe) 5′ ALK region; (red probe) 3′ ALK region; (far red [represented as cyan] probe) 5′ NPM1 region. (B) Merged maximum intensity projection image (seven Z-planes, four channels per plane) of irradiated FEPD cells stained with the ALK break-apart probe set using hiFISH. (Green) 5′ ALK; (red) 3′ ALK; (cyan) 5′ NPM1; (gray) DAPI set to 30% transparency. The three dashed boxes indicate representative cells (magnified in the right panels) with an intact ALK allele (panel i), a broken ALK allele (panel ii), or an NPM1–ALK translocation (panel iii), respectively (arrows). Bar, 10 µm. (C,E,G) Jitter plots of minimum hiFISH green–red distances in FEPD-derived cell lines stably expressing the indicated GFP fusion proteins. An ALK (C), NPM1 (E), or Nanog (G) breakage event was defined as a minimum green–red distance of more than four pixels (red dashed line). For each experimental condition, at least 20,000 minimum distances are represented. (D,F,H) Percentages of cells treated as in C, E, and G, respectively, with at least one ALK (D), NPM1 (F), or Nanog (H) breakage event per cell. (I,J,K) Determination of translocation frequencies. (I) Jitter plot of minimum hiFISH green–red (5′ALK–3′ALK) distances in the subset of cells from C that also contain proximal events between 5′NPM1 and 3′ALK. An NPM1–ALK translocation event was defined on a per-red-allele basis as a minimum red–far red distance of four pixels or less and a minimum green–red distance of more than four pixels (dashed red line). For each experimental condition, at least 3000 minimum distances are represented. (J,K) Percentages of cells treated as in J with at least one NPM1–ALK translocation event (J) or CycA–Nanog translocation event (K) per cell. (D,F,H,J,K) Values represent means ± SD from three independent experiments. (*) P < 0.05, one-way ANOVA followed by Tukey's post-test for multiple comparisons between all irradiated samples.
H3K4 methylation facilitates translocation of endogenous loci
To finally determine whether H3K4 methylation increased the frequency of ALCL translocations, we detected NPM1–ALK translocation events in intact FEPD cells by hiFISH as the colocalization of a separated 3′ALK break-apart probe (more than four pixels from the nearest 5′ ALK probe) with a 5′NPM1 probe (four pixels or less) (Fig. 4A,B, panel iii; Supplemental Fig. S7A; see the Materials and Methods). The frequency of NPM1–ALK translocations increased approximately twofold in cells expressing either ASH2L (0.53% ± 0.09%, P < 0.05) or SET7/9 (0.51% ± 0.03%, P < 0.05) when compared with cells expressing GFP alone (0.28% ± 0.03%) or SUV3-9H1 (0.25% ± 0.02%) 24 h after challenge by irradiation (Fig. 4I,J). The increased translocation frequency in ASH2L- and SET7/9-expressing cells is consistent with the increased number of DNA breaks seen in these cells (Fig. 4C–F). As a control and as expected due to the lower incidence of breakage at the Nanog locus, the level of translocations between the normally nontranslocating 3′ Nanog and 5′ CycA was significantly lower than NPM1–ALK translocations in FEPD cells expressing ASH2L (0.25% ± 0.02%, P < 0.05) or SET7/9 (0.26% ± 0.03%, P < 0.05) (Fig. 4K). Translocation frequencies in irradiated cells were severalfold higher than the background frequency of translocations in nonirradiated cells (0.03% ± 0.008% across all conditions) (Fig. 4J,K). We conclude that H3K4 methylation enhances translocation frequency in intact cells.
Discussion
We provide here evidence that the levels of histone modifications in translocation-prone genome regions influence chromosome breakage and translocation frequency. Using several independent approaches, we identified H3K4 monomethylation and trimethylation as enriched histone modifications in translocation-prone genome regions. We found elevated H3K4 methylation levels in a computational analysis of 74 translocation-prone genome regions and by directed biochemical analysis of the ALK and NPM1 translocation breakpoints in ALCL. While these modifications are generally associated with transcriptionally active, open genome regions (Bannister and Kouzarides 2011), we show that their enrichment is not due to elevated transcriptional activity of translocation-prone regions. Our findings at naturally occurring translocation sites extend recent correlative observations from genome-wide studies that mapped I-SceI-induced translocation junctions in B cells to within or near transcriptionally active regions of the genome (Chiarle et al. 2011; Klein et al. 2011) and in translocation-negative prostate cancer cells, where liganded androgen receptor (AR) binds near the TMPRSS2 translocation gene and increases its transcriptional activity, leading to elevated H3K4me3 levels across the breakpoint region (Lin et al. 2009; Yu et al. 2010). Furthermore, H3K4me3 has been implicated in DSBs generated by endonucleases during class switch and V(D)J recombination processes in lymphocytes (Daniel and Nussenzweig 2012), and both RAG2 and AID target extrachromosomal sites that are highly enriched in H3K4me3 in a sequence-independent manner (Shimazaki et al. 2009; Ji et al. 2010; Stanlie et al. 2010).
The increased formation of DSBs by endonucleases in regions of high H3K4 methylation is likely due to chromatin decondensation, resulting in higher chromatin accessibility (Mostoslavsky et al. 2003). In line with this interpretation, tethering of ASH2L or SET7/9 to chromatin leads to its visible expansion, similar to the effects of tethering chromatin remodelers such as BRG1 or VP16 (Tumbar et al. 1999), as well as increased I-SceI-induced breaks. Decondensation by H3K4 methylation may also account for the observed increase in DSB formation upon irradiation, as suggested by the finding of increased levels of DSBs after globally decondensing chromatin by inhibition of histone deacetylases (Camphausen and Tofilon 2007) or by placing nuclei in hypotonic conditions and subjecting them to irradiation (Warters and Lyons 1992; Takata et al. 2013). In addition, increased γ-H2AX foci formation occurs at a defined region of transcribed genes relative to a gene-poor region after irradiation (Falk et al. 2008). Changes in histone modifications may also affect the progression of the DNA damage response (DDR) by interfering with the restoration of baseline chromatin structure after repair (Ayrapetov et al. 2014; Burgess et al. 2014; Khurana et al. 2014), since, in ASH2L-expressing cells, recondensation at the site of DNA damage as well as upstream DDR signaling is dampened (Burgess et al. 2014), and γ-H2AX foci formation after irradiation is associated with loss of H3K4me3 (Seiler et al. 2011; Lafon-Hughes et al. 2013; Maroschik et al. 2014). These observations suggest that H3K4 hypermethylation may impair DDR progression by counteracting condensation and signaling required for efficient DDR, thus resulting in persistent DSBs and enhancing the likelihood of formation of translocations. Taken together, our findings point to an important role of the local chromatin environment, specifically histone modifications, in determining the susceptibility of genome regions to breakage and translocation formation.
Materials and methods
Chromatin tethering
Transient transfections of LacR constructs were carried out by electroporation of 2 µg of construct per 1 million LacO–I-SceI–TetO U2OS cells using Amaxa Nucleofector kit V (Lonza) according to the manufacturer's protocol. For IF and ChIP experiments, cells were fixed 20 h after transfection. For I-SceI experiments, 2 million cells were transfected first with 4 µg of a chromatin-tethering factor and, 12 h later, trypsinized, counted, and transfected with 5 µg of CFP–I-SceI–GR per 1 million cells using the same protocol. After 12 h (24 h total), cells were harvested for LM-PCR. For these experiments, cells were maintained in DMEM with 10% charcoal-dextran-treated serum (Atlanta Biologicals) starting 48 h prior the first transfection.
FISH
The following BAC clones were used for generation of break-apart probes: NPM-ALK probes used were ALK 5′ break apart (labeled in 488; RP11-119L19), ALK 3′ break apart (labeled in 568 or Cy5; RP11-100C1), NPM1 5′ (labeled in Cy5 or 568; RP11-1072I20), and NPM1 3′ (labeled in 488; CTD-2336JI). CycA-Nanog probes used were Nanog 5′ break apart (labeled in 488; RP11-298G5), Nanog 3′ break apart (labeled in 568; RP11-141A17), and CycA 5′ (labeled with Cy5; RP11-829C16).
The ALK break-apart probes were located 32 kb upstream of and 65 kb downstream from the ALK breakpoint in intron 19 (Fig. 4A). The NPM1 break-apart probes mapped 55 kb upstream of and 39 kb downstream from the NPM1 breakpoint in intron 4. The control Nanog break-apart probes mapped 55 kb upstream of and 40 kb downstream from the center of the Nanog gene and were chosen with a similar distance in between them to resemble the ALK and NPM1 break-apart probes. The 5′ CycA probe mapped 45 kb upstream of the CycA gene. The specificity of all probes was verified by PCR.
High-throughput imaging
Mounted coverslips were imaged using a slide holder adaptor on an Opera QEHS high-throughput confocal microscope (PerkinElmer) running Opera 2.0.1 software. All image acquisition was performed using a planar apochromatic 40×, NA 0.9 water immersion lens (Olympus) using 1.3-Mp CCD cameras with pixel binning of 2. The pixel size in this imaging configuration was 320 nm. Four channels (DAPI, Alexa488, Alexa568, and Cy5) were acquired in three separate exposures. For each coverslip, multichannel images were acquired in seven Z-planes (1.5 µm apart) per field over a total of 150 fields per coverslip. More than 8000 cells were imaged per experimental condition.
Automated image analysis
Image analysis was performed using Acapella 2.6 (PerkinElmer) using a modified version of a previously described custom Acapella script (Roukos et al. 2013b). Briefly, images from different Z-planes were maximally projected. For each field, the image in the maximally projected DAPI channel was used for nuclear segmentation. Nuclear area and roundness were then calculated. These two nuclear attributes were used to filter out small or irregular nuclear objects, likely representing nuclear debris and/or nuclear segmentation errors. The filtered population of nucleus ROIs was then used to sequentially detect FISH signals in different channels using a previously described spot detection algorithm (Roukos et al. 2013b). Euclidean two-dimensional distances in pixel units between the spot ROI centers of all of the possible combinations of FISH signals of different colors (Alexa488/Alexa568, green/red; Alexa568/Cy5, red/far red) in the same nucleus were then calculated. All of the single-cell-level and single-spot-distance-level data were individually indexed and exported as independent text files.
Data analysis
hiFISH data analysis was performed using the statistical analysis software R (version 3.1.2; http://www.R-project.org). Single-cell-level and single-distance-level data were read from the text files and concatenated, and experimental annotations (cell line, IR treatment, transfected construct, repeat, and FISH probe mix) were extracted from the files names. Single-cell-level information, such as the number of detected FISH signals per cell in each color, was added to the single-spot-distance-level data by an inner join operation based on common indexes in the two data sets. In order to exclude possible FISH and spot detection artifacts from the subsequent analysis, the single-spot-distance-level data set was filtered to retain only spot distances relative to cells that conformed to both of the following two criteria: (1) at least two green, two red, and two far red FISH spots, and (2) the same number of green and red FISH spots. Minimum per-red-allele spot distances were calculated for the green/red and red/far red data sets. The minimum red/far red distances were then added to the corresponding minimum green/red distances by an inner join using matching Red Spot indexes. A spot center-to-center distance proximity threshold of four pixels (1.28 mm) was empirically determined based on negative control Jurkat cells (Supplemental Fig. S6B). A breakage event on a per-red-spot basis was defined as a minimum red/green distance of more than four pixels. A translocation event on a per-red-spot basis was defined as a minimum red/green distance of more than four pixels and a minimum red/far red distance of four pixels or less. Cells containing at least one breakage or translocation event were classified as positive for the respective event class. The original single-cell data sets, single-spot-distance data sets, and R analysis script are available on request.
Genome-wide data analysis
For each translocation gene and each control gene, the number of mapped sequencing tags in the gene body (defined as transcribed region ±2 kb) from each ChIP-seq and DNase-seq data set was calculated and normalized by the length of the region and the number of all mapped tags in each data set. This normalized number of mapped sequencing tags was referred to as tag density per kilobase and was used to represent the level of histone modification or chromatin accessibility.
Translocation genes and control genes were compared as both population (74 translocation genes vs. 5076 control genes) and individual (one translocation gene vs. 100 control genes) analyses (Fig. 1A). For the population analysis, the distribution of tag densities for each histone modification and DNase I hypersensitivity was compared between gene bodies (defined above) of 74 translocation genes and 5076 control genes. Density plots were generated using function density available in R Project. For the individual analysis, the levels of histone modifications and chromatin accessibility in the gene body of each control gene were compared with those of its selected control genes. For each histone modification or DNase I, the tag density of the translocation gene was compared with the tag densities of its control genes in box plots using function boxplot in R. The percentage of control genes whose tag densities were lower than that of the translocation gene was calculated, and the translocation gene was given a rank (0–100) equal to this percentage. The 74 translocation genes were clustered by their ranks for each histone modification and DNaseI hypersensitivity using function hclust available in R, and heat maps were generated using R package ggplot2 version 0.8.9.
Statistical analysis
ChIP and RT–PCR results are presented as means ± SEM from a minimum of three independent experiments. LM-PCR and hiFISH results are presented as means ± SD from three independent experiments. Student's t-test was used for comparisons between two experimental groups, and one-way ANOVA followed by Tukey's post-test was used for comparisons of more than two groups. P < 0.05 was considered statistically significant for these analyses. The Wilcoxon test was used for comparison between two pooled gene populations in the computational analysis, since they did not follow normal distributions. P < 0.01 was considered statistically significant. Statistical tests were performed using Microsoft Excel or the GraphPad Prism software package (version 6.0). Additional details are described in the figure legends.
Supplementary Material
Acknowledgments
We thank Katherine McKinnon (National Cancer Institute) for help with FACS experiments, Tatiana Karpova (National Cancer Insitute Fluorescence Imaging Microscopy Facility) for help with microscopy, and Stephan Mathas (Charite-Berlin) for technical assistance. We also thank Grace Gill (Tufts University), Terry Furey (University of North Carolina), and members of the Misteli laboratory for helpful feedback. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and a Howard Hughes Medical Institute-National Institutes of Health fellowship to B.B. B.B. and T.M. designed the study. B.B. performed the majority of experiments and data analysis, Z.Z.Z. and J.D.L. performed bioinformatics analysis, and B.B. and G.P. performed the hiFISH data analysis. B.B. and T.M. wrote the manuscript, the other authors read and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.262170.115.
References
- Ayrapetov MK, Gursoy-Yuzugullu O, Xu C, Xu Y, Price BD. 2014. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc Natl Acad Sci 111: 9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21: 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326. [DOI] [PubMed] [Google Scholar]
- Bernt KM, Armstrong SA. 2009. Leukemia stem cells and human acute lymphoblastic leukemia. Semin Hematol 46: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. 1997. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med 3: 730–737. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Burman B, Kruhlak MJ, Misteli T. 2014. Activation of DNA damage response signaling by condensed chromatin. Cell Rep 9: 1703–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camphausen K, Tofilon PJ. 2007. Inhibition of histone deacetylation: a strategy for tumor radiosensitization. J Clin Oncol 25: 4051–4056. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, et al. 2011. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell 147: 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Nussenzweig A. 2012. Roles for histone H3K4 methyltransferase activities during immunoglobulin class-switch recombination. Biochim Biophys Acta 1819: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk M, Lukasova E, Kozubek S. 2008. Chromatin structure influences the sensitivity of DNA to γ-radiation. Biochim Biophys Acta 1783: 2398–2414. [DOI] [PubMed] [Google Scholar]
- Fischer P, Nacheva E, Mason DY, Sherrington PD, Hoyle C, Hayhoe FG, Karpas A. 1988. A Ki-1 (CD30)-positive human cell line (Karpas 299) established from a high-grade non-Hodgkin's lymphoma, showing a 2;5 translocation and rearrangement of the T-cell receptor β-chain gene. Blood 72: 234–240. [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S, Kruhlak MJ, Kim J, Tran AD, Liu J, Nyswaner K, Shi L, Jailwala P, Sung MH, Hakim O, et al. 2014. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep 8: 1049–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein IA, Resch W, Jankovic M, Oliveira T, Yamane A, Nakahashi H, Di Virgilio M, Bothmer A, Nussenzweig A, Robbiani DF, et al. 2011. Translocation-capture sequencing reveals the extent and nature of chromosomal rearrangements in B lymphocytes. Cell 147: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon-Hughes L, Di Tomaso MV, Liddle P, Toledo A, Reyes-Abalos AL, Folle GA. 2013. Preferential localization of γH2AX foci in euchromatin of retina rod cells after DNA damage induction. Chromosome Res 21: 789–803. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Rosenfeld MG. 2012. Molecular logic underlying chromosomal translocations, random or non-random? Adv Cancer Res 113: 241–279. [DOI] [PubMed] [Google Scholar]
- Mani RS, Chinnaiyan AM. 2010. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet 11: 819–829. [DOI] [PubMed] [Google Scholar]
- Maroschik B, Gurtler A, Kramer A, Rossler U, Gomolka M, Hornhardt S, Mortl S, Friedl AA. 2014. Radiation-induced alterations of histone post-translational modification levels in lymphoblastoid cell lines. Radiat Oncol 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathas S, Kreher S, Meaburn KJ, Johrens K, Lamprecht B, Assaf C, Sterry W, Kadin ME, Daibata M, Joos S, et al. 2009. Gene deregulation and spatial genome reorganization near breakpoints prior to formation of translocations in anaplastic large cell lymphoma. Proc Natl Acad Sci 106: 5831–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Alt FW, Bassing CH. 2003. Chromatin dynamics and locus accessibility in the immune system. Nat Immunol 4: 603–606. [DOI] [PubMed] [Google Scholar]
- Nambiar M, Raghavan SC. 2011. How does DNA break during chromosomal translocations? Nucleic Acids Res 39: 5813–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D'Andrea AD. 2013. Chromatin remodeling at DNA double-strand breaks. Cell 152: 1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Misteli T. 2014. The biogenesis of chromosome translocations. Nat Cell Biol 16: 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Burman B, Misteli T. 2013a. The cellular etiology of chromosome translocations. Curr Opin Cell Biol 25: 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. 2013b. Spatial dynamics of chromosome translocations in living cells. Science 341: 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler DM, Rouquette J, Schmid VJ, Strickfaden H, Ottmann C, Drexler GA, Mazurek B, Greubel C, Hable V, Dollinger G, et al. 2011. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res 19: 883–899. [DOI] [PubMed] [Google Scholar]
- Shimazaki N, Tsai AG, Lieber MR. 2009. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell 34: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Misteli T. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science 320: 1507–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. 2007. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. 2010. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proc Natl Acad Sci 107: 22190–22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabbo F, Ponzoni M, Rabadan R, Bertoni F, Inghirami G. 2013. Beyond NPM–anaplastic lymphoma kinase driven lymphomagenesis: alternative drivers in anaplastic large cell lymphoma. Curr Opin Hematol 20: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H, Hanafusa T, Mori T, Shimura M, Iida Y, Ishikawa K, Yoshikawa K, Yoshikawa Y, Maeshima K. 2013. Chromatin compaction protects genomic DNA from radiation damage. PLoS One 8: e75622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Ernst C, Heilbut A, Chiang C, Hanscom C, Lindgren A, Kirby A, Liu S, Muddukrishna B, Ohsumi TK, et al. 2011. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am J Hum Genet 88: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J Cell Biol 145: 1341–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warters RL, Lyons BW. 1992. Variation in radiation-induced formation of DNA double-strand breaks as a function of chromatin structure. Radiat Res 130: 309–318. [PubMed] [Google Scholar]
- Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, et al. 2010. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 17: 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rowley JD. 2006. Chromatin structural elements and chromosomal translocations in leukemia. DNA Repair (Amst) 5: 1282–1297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.