Here, Ni et al. determined the crystal structures of a key Hippo pathway regulator, Mob1, bound to either Mst2 (human Hippo) or Lats1, two core pathway kinases. Further mechanistic and functional studies provide novel insight into the conserved mechanism by which the Hippo kinase activates Lats through stepwise phosphorylation.
Keywords: phosphorylation, autoinhibition, Mst2, Lats1, Mob1, X-ray crystallography, NMR
Abstract
The Mst–Lats kinase cascade is central to the Hippo tumor-suppressive pathway that controls organ size and tissue homeostasis. The adaptor protein Mob1 promotes Lats activation by Mst, but the mechanism remains unknown. Here, we show that human Mob1 binds to autophosphorylated docking motifs in active Mst2. This binding enables Mob1 phosphorylation by Mst2. Phosphorylated Mob1 undergoes conformational activation and binds to Lats1. We determine the crystal structures of phospho-Mst2–Mob1 and phospho-Mob1–Lats1 complexes, revealing the structural basis of both phosphorylation-dependent binding events. Further biochemical and functional analyses demonstrate that Mob1 mediates Lats1 activation through dynamic scaffolding and allosteric mechanisms. Thus, Mob1 acts as a phosphorylation-regulated coupler of kinase activation by virtue of its ability to engage multiple ligands. We propose that stepwise, phosphorylation-triggered docking interactions of nonkinase elements enhance the specificity and robustness of kinase signaling cascades.
The Hippo tumor suppressor pathway controls cell numbers and organ size in multicellular organisms by coordinately regulating cell proliferation and apoptosis. The Hippo pathway was originally discovered in Drosophila by genetic screens designed to identify overgrowth mutants and was later shown to be conserved in mammals and also essential for animal development (Edgar 2006; Harvey and Tapon 2007; Badouel et al. 2009; Pan 2010; Zhao et al. 2010a; Halder and Johnson 2011; Staley and Irvine 2012; Barry and Camargo 2013). In mammals, the core components of the Hippo pathway include the Ste-20 family kinases Mst1/2, the nuclear Dbf2-related (NDR) family kinases Lats1/2, and the adaptor proteins Mob1 and Salvador (Sav). All four proteins are potential tumor suppressors, and dysfunction of Hippo signaling has been linked to human cancers (Zhao et al. 2008a; Harvey et al. 2013; Johnson and Halder 2014).
In the canonical mammalian Hippo pathway, activated Mst1/2 in conjunction with Sav phosphorylate and activate the Lats1/2–Mob1 complex, which in turn phosphorylates and inactivates the transcriptional coactivator YAP (Yes-associated protein) (Huang et al. 2005; Dong et al. 2007; Zhao et al. 2007; Hao et al. 2008; Hong and Guan 2012). YAP is a major downstream effector of the Hippo pathway. It associates with the TEAD family of transcription factors to stimulate the transcription of Hippo-responsive genes that promote cell proliferation and inhibit apoptosis (Zhao et al. 2008b; Luo 2010; Sudol et al. 2012). Lats1/2-mediated phosphorylation inhibits YAP by promoting its cytoplasmic sequestration (Dong et al. 2007; Zhao et al. 2007; Hao et al. 2008) and ubiquitination-dependent degradation (Zhao et al. 2010b), thereby inhibiting the expression of Hippo-responsive genes, halting proliferation, and promoting apoptosis. Although recent genetic screens and biochemical studies have identified many upstream regulators of the Hippo pathway (Boggiano and Fehon 2012; Enderle and McNeill 2013), the activation mechanisms of the core Mst–Lats kinase cascade are still poorly understood.
Mst1/2 can be activated through autophosphorylation at their activation loop, and this autophosphorylation requires Mst1/2 homodimerization through their C-terminal SARAH (Sav/Rassf/Hpo) domain (Avruch et al. 2012; Jin et al. 2012; Ni et al. 2013). Activated Mst1/2 then phosphorylate and activate Lats1/2 (Chan et al. 2005). Similar to other NDR family kinases, Lats1/2 activation requires sequential phosphorylation of two conserved key regulatory sites (Bichsel et al. 2004; Chan et al. 2005; Hergovich et al. 2006; Hergovich and Hemmings 2009). Mst1/2 first phosphorylate Lats1 at T1079, located in the hydrophobic motif (HM) C-terminal to the kinase domain. This phosphorylation strongly stimulates Lats1 autophosphorylation at S909 in the activation loop (T loop), leading to its activation. Phosphorylation of both residues is essential for Lats1 activation (Chan et al. 2005; Hergovich et al. 2006; Hergovich 2013).
The highly conserved Mob proteins are important kinase regulators in organisms from yeast to humans. Among the four human Mob proteins (Mob1–4), only Mob1 functions as an activator of Lats1/2 (Hergovich 2011). Mst1/2 can phosphorylate Mob1 at its N-terminal residues, T12 and T35. These phosphorylation events are thought to be critical for the assembly of the Lats1–Mob1 complex, as the T12A/T35A (2TA) mutant of Mob1 no longer binds to Lats1 in vitro (Praskova et al. 2008). Phosphorylated Mob1 binds to the conserved Mob1-binding domain (MBD) of Lats1 that lies just N-terminal to its kinase domain and regulates Lats1 activation (Bothos et al. 2005; Hergovich et al. 2006). How Mst1/2-dependent Mob1 phosphorylation promotes Mob1 binding to Lats1 is not understood. In addition, the role of Mob1 in Mst1/2-dependent activation of Lats1/2 is unclear.
In this study, using in vitro reconstitution, X-ray crystallography, and functional cellular assays, we defined the mechanisms by which Mob1 mediates Mst2-dependent Lats1 activation, a central event during Hippo signaling. We show that Lats1 activation requires sequential, multistep Mst2-dependent phosphorylation of all partners in this process, including Mst2 itself, Mob1, and Lats1. First, active Mst2 autophosphorylates multiple residues in the linker between its kinase and SARAH domains, creating phospho-peptide-docking motifs for Mob1. Second, phospho-Mst2 (pMst2) binding converts Mob1 from an autoinhibited, closed conformation to an active, open conformation capable of Lats1 binding. Formation of the Mst2–Mob1–Lats1 ternary complex promotes both Mst2-dependent Lats1 HM phosphorylation and Mob1 phosphorylation. Third, the phosphorylated N-terminal tail of Mob1 competes with the pMst2 linker for the same phospho-peptide-binding site on Mob1, releasing phospho-Mob1 (pMob1)–Lats1 from Mst2. Fourth, pMob1 retains the active conformation and acts in concert with the Lats1 phospho-HM to allosterically promote Lats1 autophosphorylation in its T loop, leading to Lats1 kinase activation. We propose that the highly regulated, dynamic scaffolding by Mob1 effectively couples the activation of two core kinases (Mst2 and Lats1) in the Hippo pathway. This type of interdependent, multistep phosphorylation by an upstream kinase to transduce signals downstream ensures signaling specificity.
Results
Mst2 autophosphorylates its linker to create phospho-docking motifs for Mob1
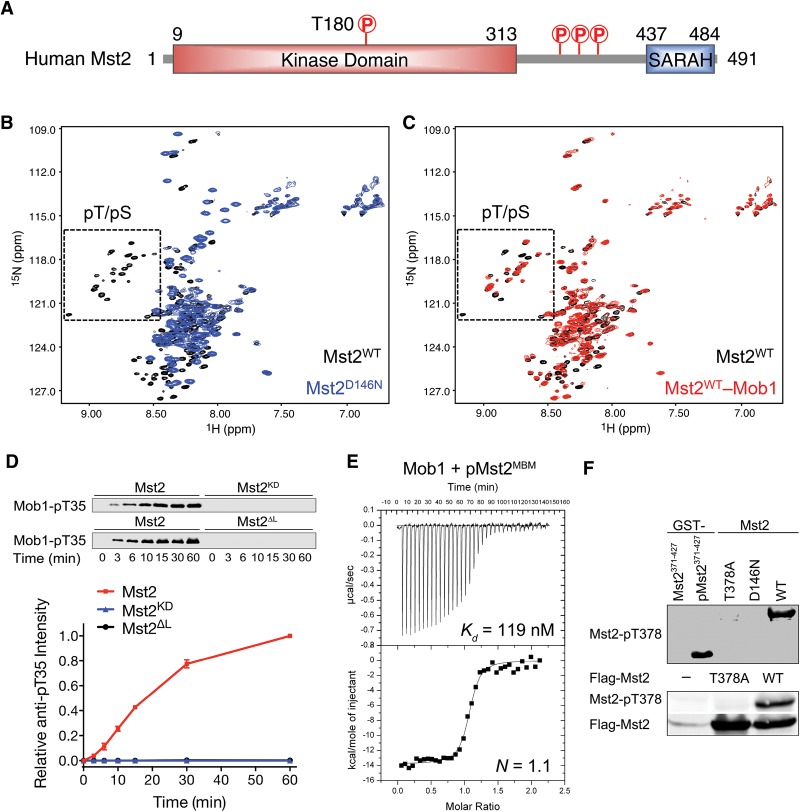
The full-length human Mst2 kinase contains an N-terminal kinase domain, a long flexible linker, and a C-terminal SARAH dimerization domain (Fig. 1A). To characterize potential structural differences between active and inactive Mst2, we analyzed 15N-labeled human wild-type Mst2 and kinase-inactive Mst2D146N proteins with nuclear magnetic resonance (NMR) spectroscopy. We showed previously that the linker region of human Mst2 is flexible (Ni et al. 2013). Strikingly, we observed a new cluster of peaks a ∼8.5–9.0 ppm in the spectrum of Mst2WT but not Mst2D146N (Fig. 1B). Their unique chemical shifts suggested that this cluster of peaks belonged to phosphoserine/threonine (pS/pT) residues. Mass spectrometry analysis then confirmed that recombinant Mst2WT indeed underwent autophosphorylation at multiple S/T residues in the linker (Supplemental Fig. S1A). More importantly, addition of recombinant purified human Mob1 to Mst2WT caused the disappearance of several pS/pT peaks (Fig. 1C), indicating that Mob1 bound to a subset of pS/pT residues in Mst2. Consistent with the NMR data, recombinant purified Mob1 cofractionated with Mst2WT, but not Mst2D146N or the Mst2 mutant with the linker-deleted (Mst2ΔL), during gel filtration chromatography (Supplemental Fig. S1B). Therefore, Mob1 binds to the autophosphorylated linker of Mst2.
Figure 1.
Human Mob1 binds to the autophosphorylated Mst2 linker. (A) Domain organization of human Mst2. (B) Overlay of 1H/15N TROSY-HSQC spectra of 15N-labeled Mst2WT (black) and kinase-inactive Mst2D146N (blue). Peaks belonging to pT/pS residues are boxed. (C) Overlay of 1H/15N TROSY-HSQC spectra of 15N-labeled Mst2WT (black) and 15N-labeled Mst2WT bound to unlabeled Mob1 (red). Peaks belonging to pT/pS residues are boxed. (D) Quantitative immunoblotting with antibody against Mob1-pT35 of kinase reactions containing human Mob1 and the indicated Mst2 proteins at the indicated times. (KD) Kinase domain; (ΔL) linker deletion. The relative Mob1-pT35 signal intensities, normalized to those of wild-type Mst2 reactions at 60 min (100%), are plotted against time. Means with range for two independent experiments are plotted. (E) Isothermal titration calorimetry (ITC) curves of the binding between Mob1 and the pMst2MBM peptide, with Kd and binding stoichiometry (N) indicated. (F) Mst2 undergoes autophosphorylation at T378. Anti-Mst2-pT378 immunoblots of the indicated purified recombinant Mst2 proteins (top panel) and lysates of HEK293 cells transfected with the indicated plasmids (bottom panel).
Because Mob1 T12 and T35 are two functionally important Mst1/2 phosphorylation sites in human cells, we tested whether the Mob1–pMst2 interaction regulated Mob1 phosphorylation by Mst2. As revealed by quantitative immunoblotting with antibodies to pMob1, full-length Mst2 phosphorylated Mob1 efficiently at both T12 and T35 (Fig. 1D; Supplemental Fig. S1C). In contrast, Mst2ΔL or the Mst2 kinase domain (Mst2KD) alone was inactive in T12/T35 phosphorylation. Therefore, the linker of Mst2 is required for Mob1 phosphorylation, possibly through recruiting Mob1. Because phosphorylation of Mob1 at T12 and T35 displayed similar kinetics, we only monitored Mob1 phosphorylation at T35 in all subsequent experiments.
To define the Mob1-binding motif (MBM) in the Mst2 linker, we made glutathione-S-transferase (GST)-Mst2 fusion proteins that contained various linker fragments. We phosphorylated these GST-Mst2 linker proteins with Mst2KD and repurified them with glutathione beads to remove Mst2KD. The Mst2 linker fragment containing residues 376–400 was the smallest fragment that retained phosphorylation-dependent Mob1 binding (Supplemental Fig. S2A), indicating that this region constituted a functional MBM. Sequence alignment revealed that among the four S/T residues in this MBM, only T378 was conserved among Mst and Hippo proteins (Supplemental Fig. S2B). By individually mutating each S/T residue, we found that only the T378A mutation abolished the gel mobility shift of phosphorylated GST-Mst2371–427 and its phosphorylation-dependent binding to Mob1 (Supplemental Fig. S2C), demonstrating the importance of T378 phosphorylation in Mob1 binding. Furthermore, a synthetic phospho-T378 Mst2 MBM peptide (pMst2MBM; residues 371–401) bound to Mob1 with a dissociation constant (Kd) of 119 nM, as measured by isothermal titration calorimetry (ITC) (Fig. 1E). Finally, addition of pMst2MBM caused chemical shift changes of >60 peaks in the 15N/1H HSQC spectrum of 15N-labeled full-length Mob1 (Supplemental Fig. S2D), indicative of an extensive binding interface between the two.
To confirm that T378 was phosphorylated in full-length Mst2, we raised a phospho-specific antibody against phospho-T378 Mst2. This antibody specifically recognized phosphorylated GST-Mst2371–427 (Fig. 1F). This antibody also recognized the full-length Mst2 wild type, but not T378A or the kinase-inactive D146N, purified from bacteria or expressed in human cells. These results indicate that full-length Mst2 undergoes autophosphorylation at T378 in vitro and in human cells. Taken together, Mst2 autophosphorylates T378 in its linker to create a functional phospho-docking motif for Mob1.
Structure of Mob1 bound to a pMst2 peptide
To define the minimal Mob1 domain capable of binding to pMst2MBM, we constructed two N-terminal truncation mutants of Mob1 (ΔN32 and ΔN50) and tested their binding to pMst2. Both mutants retained pMst2 binding, albeit with reduced affinities (Supplemental Fig. S3A). We obtained crystals of human Mob1ΔN50 bound to pMst2MBM that diffracted to 2.65 Å resolution and solved the structure of this Mob1–pMst2 complex using molecular replacement (Supplemental Table S1).
In the Mob1–pMst2 structure, the Mob1 core (residues 52–211) adopts a conformation virtually identical to that of free human Mob1 (Fig. 2A), indicating that binding of pMst2 does not appreciably alter the conformation of the Mob1 core. In Mob1, helices H2, H4, H5, and H7 form a four-helix bundle, with one end capped by helices H3 and H6 and a Zn2+ ion coordinated by C79, C84, H161, and H166 (Fig. 2A). The bound Zn2+ ion is also present in the free Mob1 structure and is required for Mob1 folding. On one face of the helix bundle, helices H8 and H9 cover the central regions of H5 and H7, respectively. The long loop connecting H3 and H4, including the short S1/S2 β hairpin, packs against helices H4 and H5.
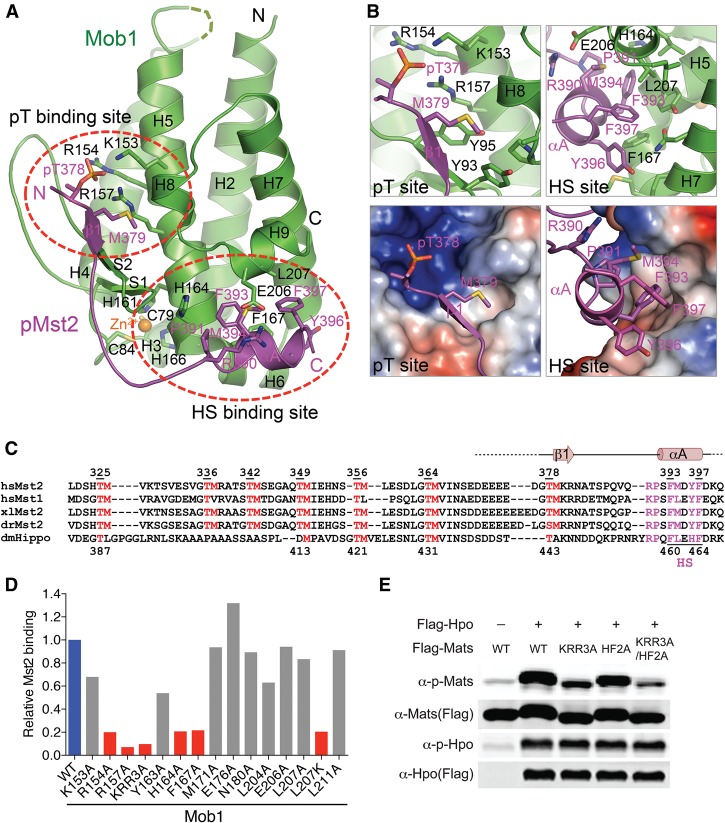
Figure 2.
Crystal structure and binding interface of human Mob1–pMst2. (A) Cartoon drawing of the crystal structure of human Mob1ΔN50 in complex with the pMst2MBM peptide. The pT-binding site and hydrophobic sequence (HS)-binding site are circled. Mob1 is colored green. pMst2 is colored magenta. Mob1 and pMst2 residues are shown as green and magenta sticks, respectively, and pMst2 residues are labeled in magenta. All structural figures were generated with PyMol (https://www.pymol.org). (B) Zoomed-in views of pMst2 and Mob1 interactions at the pT- and HS-binding sites, respectively. (C) Sequence alignment of the Mst2 linker (residues 321–400) from humans to flies. Conserved TM dipeptide motifs are colored red. The HS motif is underlined, and conserved residues in direct contact with Mob1 at the HS region are colored magenta. The secondary structure elements of pMst2MBM are above the sequences and colored salmon. (Hs) Homo sapiens; (Xl) Xenopus laevis; (Dr) Danio rerio; (Dm) Drosophila melanogaster. Human Mst2 and Drosophila Hippo residue numbers are indicated. (D) Relative Mst2 binding of the indicated Mob1 mutant normalized against Mob1 wild type (100%). Mob1 mutants that lost or retained phosphorylation-dependent Mst2 binding are colored red and gray, respectively. (E) Immunoblots of lysates of Drosophila S2R+ cells transfected with the indicated plasmids.
The Mob1–pMst2 interface consists of two spatially separated contacting sites (Fig. 2A,B). At the first site, residues 378–384 of pMst2 bind at a surface formed by the S1/S2 β hairpin and helices H5 and H8 of Mob1. Residues 379–381 of pMst2 fold into strand β1 that augments the S1/S2 β hairpin in Mob1 to form a small anti-parallel β sheet. pT378 binds at a highly positively charged pocket formed by three basic residues on H5; namely, K153, R154, and R157. M379 occupies the +1 position relative to pT378 and inserts into an adjacent hydrophobic pocket on Mob1. The side chains of the +2/+3 residues K380 and R381 do not make strong contact with Mob1. We refer to this site of Mob1 as the pT-binding site. The major interactions at this site involve the pTM dipeptide motif of pMst2 and are highly similar to those observed in the crystal structure of Mob1 bound to an unnatural phosphopeptide ligand (Rock et al. 2013).
At the second site, the C-terminal part of pMst2 (residues 390–398) binds to a shallow groove between helices H6 and H9 (Fig. 2A,B). R390 of pMst2 forms favorable electrostatic interactions with E206 of Mob1. The FMDYF hydrophobic sequence (HS) of pMst2 folds into a short helix (αA) and contacts Mob1 through extensive hydrophobic interactions. The HS motif is highly conserved among the Mst and Hippo proteins (Fig. 2C). This second pMst2 contact site of Mob1 is thus termed the HS-binding site.
The loop between the pTM and HS motifs is less conserved and makes minimal contacts with Mob1 (Fig. 2A). In fact, only one of the six molecules of Mob1–pMst2 in the asymmetric unit has clear electron density for this loop (Supplemental Fig. S3B), indicating that this region of pMst2 is not well ordered. Thus, pMst2 engages Mob1 at two distinct sites through the bipartite pTM and HS motifs, which are separated by a flexible loop.
The HS motif is not present in the unnatural phosphopeptide ligand of Mob1 reported earlier (Rock et al. 2013). The HS–Mob1 interactions revealed in our structure are thus missing in the structure of Mob1 bound to that pS peptide. However, Mob1 bound to that shorter pS peptide with a Kd of 174 nM, which is comparable with that of Mob1–pMst2 (119 nM). Thus, the bulk of the binding energy between Mob1 and phosphopeptides is contributed by the pS/T motif. The HS motif has an auxiliary role and may augment the binding affinities between Mob1 and suboptimal pS/T motifs.
We created 15 Mob1 mutants targeting conserved residues in direct contact with pMst2 at the pT- or HS-binding site and tested their binding to both unphosphorylated and phosphorylated GST-Mst2371–427 (Fig. 2D; Supplemental Fig. S3C,D). For the pT-binding site, Mob1 R154A, R157A, and KRR3A (K153A/R154A/R157A) lost binding to pMst2. For the HS-binding site, Mob1 H164A, F167A, and L207K had weakened pMst2 binding. These results validate the importance of both the pT- and HS-binding sites of Mob1 in pMst2 binding.
Because many pMst2-binding residues of Mob1 are conserved in Mats (Mob1 in Drosophila), we created corresponding mutants in Mats—the pT-binding site mutant MatsKRR3A and the HS-binding site mutant MatsHF2A (H164A/F167A)—and tested their phosphorylation by Hippo (Hpo; Mst1/2 in Drosophila). When Mats and Hpo were coexpressed in Drosophila S2R+ cells, the wild-type Mats was efficiently phosphorylated by Hpo (Fig. 2E). In contrast, MatsKRR3A and MatsHF2A were less efficiently phosphorylated. The MatsKRR3A/HF2A mutant with both sites mutated had even further reduced phosphorylation. These results indicate that the Mats–Hpo interaction is required for Mats phosphorylation by Hpo in Drosophila cells.
L307 in the budding yeast Mob1 (corresponding to L207 in human Mob1) has been reported to be critical for yeast viability (Mrkobrada et al. 2006), indicating that the HS-binding site is critical for Mob1 function in that organism. The functional importance of the pT-binding site in yeast Mob1 has also been demonstrated (Rock et al. 2013). Taken together, these results validate the functional relevance of the conserved pT- and HS-binding sites of Mob1 proteins in diverse organisms.
Multiple phospho-TM motifs in the Mst2 linker redundantly recruit Mob1
To determine whether the MBM (residues 371–400) of Mst2 was necessary and sufficient for Mob1 binding, we created two Mst2 linker mutants in the context of the full-length Mst2 protein: (1) ΔL/MBM (Mst2ΔL/MBM, which contains only the MBM but lacks all other linker residues) and (2) T378A (Mst2T378A, with T378 mutated to alanine). Mst2ΔL/MBM bound to Mob1 with a Kd of 230 nM, as measured by ITC (Supplemental Fig. S4A), and efficiently phosphorylated Mob1 (Fig. 3A), indicating that the MBM motif alone is sufficient for the recruitment and phosphorylation of Mob1 by Mst2. Surprisingly, Mst2T378A retained Mob1 binding, as they cofractionated on gel filtration columns (Supplemental Fig. S4B). It also efficiently phosphorylated Mob1 (Fig. 3A). Therefore, there are multiple functional Mob1-binding elements in the Mst2 linker. The MBM containing pT378M is not the only Mob1-binding element in this region.
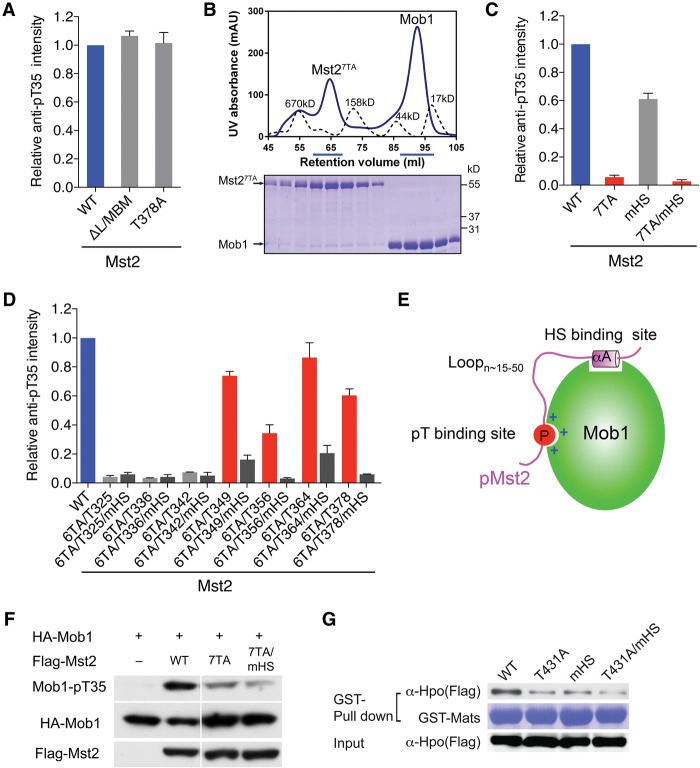
Figure 3.
The Mst2 linker contains multiple pTM motifs for Mob1 binding. (A) Quantification of the relative Mob1-pT35 signals in kinase reactions containing the indicated Mst2 proteins. Means with range for two independent experiments are plotted. (B) UV traces of molecular weight standards (dashed line) and Mst27TA and Mob1 mixed at a 1:2 molar ratio (solid line) fractionated on a Superdex 200 gel filtration column. The underlined fractions were separated on SDS-PAGE and stained with Coomassie. (C) Quantification of the relative Mob1-pT35 signals in kinase reactions containing the indicated Mst2 proteins. Means with range for two independent experiments are plotted. (D) Quantification of the relative Mob1-pT35 signals in kinase reactions containing the indicated Mst2 proteins. Means with range for two independent experiments are plotted. (E) Schematic drawing of the bipartite Mob1–pMst2 interaction, which involves the binding of pTM and HS motifs connected by a flexible loop with variable length. (F) Immunoblots of lysates of HEK293 cells transfected with the indicated plasmids. (G). Immunoblots (top and bottom panels) of lysates of Drosophila S2R+ cells transfected with the indicated Flag-Hpo plasmids (Input) and proteins bound to GST-Mats beads. The GST-Mats protein bound to beads was stained with Coomassie (middle panel).
Our Mob1–pMst2 structure highlighted the importance of the phospho-T378M motif in Mob1 binding and also demonstrated that only one phospho-TM motif can bind to Mob1 at any given time. Intriguingly, there are six additional TM motifs in the Mst2 linker (Fig. 2C). Mass spectrometry revealed that all seven TM motifs in the Mst2 linker were indeed autophosphorylated in recombinant purified full-length Mst2 (Supplemental Fig. S1A). We hypothesized that other phospho-TM motifs could also independently mediate Mob1 binding. To test this hypothesis, we mutated all seven threonine residues in the TM motifs to alanines in full-length Mst2. The resulting Mst27TA mutant was defective in Mob1 binding and phosphorylation (Fig. 3B,C). Although Mst27TA still has an intact HS motif, it no longer binds to Mob1, indicating that the HS motif alone is not sufficient for Mst2 binding to Mob1. Mutating the Mst2 HS motif from FMDYF to AADAA (mHS) reduced Mob1 phosphorylation by ∼40%, however (Fig. 3C). Taken together, these results indicate that phosphorylation of at least one of the seven TM motifs is required for Mst2 binding to Mob1. The HS motif contributes to Mob1 binding and phosphorylation by Mst2.
To determine which TM motifs are functional in Mob1 binding, we made seven Mst2 6TA mutants by restoring a single intact TM motif in the 7TA mutant and compared their relative activities in phosphorylating Mob1. Restoring T349, T356, T364, and T378 individually rescued Mob1 phosphorylation to varying degrees, whereas restoring T325, T336, and T342 individually had little effect (Fig. 3D). Thus, only T349, T356, T364, and T378 are functional in Mob1 binding, with the most conserved T364M motif being the most effective. Mutating the HS motif decreased Mob1 phosphorylation by all functional, single-TM-containing 6TA mutants, indicating that the HS motif facilitates Mob1 binding by each of the functional phospho-TM motifs.
Our structure of Mob1–pMst2 has established that Mob1 binds to Mst2 through a bipartite interaction, with an N-terminal phospho-TM moiety of the Mst2 ligand occupying the pT-binding site and the C-terminal HS moiety engaging the HS-binding site. Our mutagenesis results suggest that four pTM motifs in the Mst2 linker can act redundantly to bind the pT-binding site of Mob1. The sole HS motif can pair with any given one of the pTM motifs to form a functional bipartite ligand for Mob1. We note that the four functional Mst2 TM motifs are located more closely (in sequence) to the HS motif than the three nonfunctional ones. A simple explanation is that the loop connecting the TM and HS motifs in a functional Mob1-binding ligand cannot exceed certain lengths (Fig. 3E).
We next tested whether the Mst2–Mob1 interaction was functionally important in living human cells. Flag-Mst2 wild type greatly stimulated HA-Mob1 phosphorylation in HEK293 cells (Fig. 3F). Mst27TA and Mst27TA/mHS (7TA and mHS double mutant) phosphorylated Mob1 much less efficiently, indicating that the Mob1–pMst2 interaction is critical for Mob1 phosphorylation in human cells. Furthermore, the phosphorylation-dependent Mst2–Mob1 interaction appears to be conserved between Drosophila Hpo and Mats, as the Hpo mutant HpoT431A (equivalent to Mst2T364A), HpomHS (F460A/L461A/F464A), or HpoT431A/mHS (Fig. 3G) bound less efficiently to GST-Mats as compared with Hpo wild type. These results suggest that, similar to human Mst2, the pT431M and HS motifs in Hpo constitute a functional bipartite Mats-binding ligand. Taken together, our results thus far establish an evolutionarily conserved, phospho-dependent interaction between Mst and Mob proteins.
Structure of the pMob1–Lats1 complex
When Hippo signaling is on, Lats1 activation occurs through a two-step process (Chan et al. 2005; Hergovich 2013). First, Mst1/2 phosphorylate Lats1 at T1079 in the HM. Second, Lats1 undergoes autophosphorylation at S909 in its T loop, leading to its activation (Fig. 4A). Mst1/2-dependent T1079 phosphorylation is required for Lats1 autophosphorylation at its T loop. Mob1 is required for both activating phosphorylation events of Lats1 (Praskova et al. 2008).
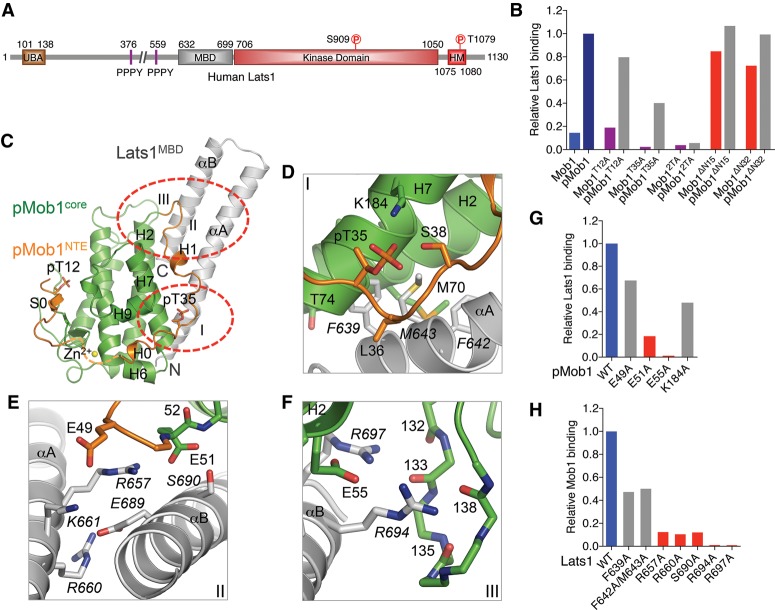
Figure 4.
Crystal structure and binding interface of human pMob1–Lats1. (A) Domains and motifs of human Lats1 with their boundaries indicated. (B) Quantification of Lats1 binding of the indicated Mob1 and pMob1 proteins. The relative Lats1 intensities were normalized to that of wild-type GST-pMob1 (100%). (C) Overall structure of pMob1–Lats1. Three pMob1–Lats1 interfaces (I–III) are circled. The Mob1 N-terminal extension (NTE) is colored orange. The Mob1 core is colored green. Lats1 is colored gray. pT12 and pT35 are shown as sticks. (D–F) Zoomed-in views of pMob1–Lats1 interactions at interfaces I–III. Residues from Lats1 are labeled in italic. Residues from the Mob1 core (green), Mob1 NTE (orange), and Lats1 (gray) are shown as sticks. (G) Quantification of Lats1 binding of the indicated pMob1 proteins. The relative Lats1-binding intensities were normalized to that of wild-type GST-pMob1 (100%). (H) Quantification of GST-pMob1 binding of the indicated Lats1 proteins. The relative amounts of Lats1 bound to GST-pMob1 were normalized to that of wild-type Lats1 (100%).
To understand how Mob1 promotes Lats1 activation, we first characterized the Lats1–Mob1 interaction in vitro (Fig. 4B; Supplemental Fig. S5A). GST-Mob1 bound weakly to 35S-labeled full-length Lats1. GST-Mob1 phosphorylated by Mst2 (pMob1) bound to Lats1 much more efficiently. Mutation of both T12 and T35, the two Mst2 phosphorylation sites in Mob1, abolished phosphorylation-dependent binding between Mob1 and Lats1. Mutation of either site weakened the pMob1–Lats1 interaction, suggesting that both phosphorylation events contributed to this interaction. Surprisingly, unphosphorylated Mob1ΔN15 (deletion of residues 1–15) and Mob1ΔN32 (deletion of residues 1–32) bound more tightly to Lats1 than the unphosphorylated full-length Mob1 did (Fig. 4B). Phosphorylation by Mst2 only slightly enhanced the binding of these truncation mutants. Thus, these results suggest that the N-terminal region of Mob1, especially the first 15 residues, interferes with Lats1 binding in an autoinhibited conformation. Phosphorylation of Mob1 might stimulate Lats1 binding in part through relieving this autoinhibition.
To investigate how pMob1 interacted with Lats1, we mapped the minimal MBD of Lats1 to residues 632–699 (Supplemental Fig. S5B), purified the complex of full-length pMob1 bound to Lats1602–704 (encompassing the MBD) (Supplemental Fig. S5C), and determined its crystal structure at 2.3 Å resolution with the single anomalous dispersion (SAD) method (Fig. 4C; Supplemental Table S2). The Mob1 core (residues 52–212) in the pMob1–Lats1 complex adopts a conformation nearly identical to that of free Mob1. The N-terminal extension (NTE; residues 10–51) forms three secondary structural elements: β strand S0, 310 helix H0, and α helix H1. All three elements associate intramolecularly with the Mob1 core. Phospho-T12 (pT12) in the NTE binds to the pT-binding site of Mob1. Helix H0 of the NTE binds to the HS-binding site of Mob1. Helix H1 associates with helix H7 through both hydrophobic and charged interactions.
The Lats1 MBD folds into two anti-parallel helices and binds at a flat surface of Mob1 formed by H2, H7, and residues 35–51 of the NTE. The Lats1-binding and pMst2-binding surfaces of Mob1 do not overlap, suggesting that Lats1 and Mst2 can bind to Mob1 simultaneously. There are three major interfaces between pMob1 and Lats1 (Fig. 4D–F). Interface I involves the N-terminal part of αA in Lats1 and the H0–H1 loop in Mob1. Interfaces II and III are close to each other and mostly involve electrostatic interactions between the middle part of αA and the C-terminal half of αB from Lats1 and helices H2 and H7 and the H4–H5 loop from Mob1.
To validate the pMob1–Lats1-binding interface observed in our structure, we made GST-Mob1 and 35S-labeled Lats1 mutants targeting residues at or near this interface. All four GST-Mob1 mutants exhibited weaker binding to Lats1, with E55A at interface III being most defective (Fig. 4G; Supplemental Fig. S6A). Interestingly, the corresponding yeast Mob1 mutant E151K is deficient in binding to the kinase Dbf2, indicating that the Mob1–Lats1 interaction is conserved (Komarnitsky et al. 1998). All Lats1 mutants also exhibited weaker binding to pMob1 (Fig. 4H; Supplemental Fig. S6B). In particular, two mutants targeting residues at interface III (R694A and R697A) completely lost pMob1 binding. Similar Lats1 mutations at interfaces II and III have previously been shown to affect the Lats1–Mob1 interaction and the Lats1 kinase activity in human cells (Hergovich et al. 2006). Interestingly, although R660 is not in direct contact with Mob1, it forms an intramolecular charged interaction with E689 (Fig. 4E). Therefore, the weakened binding between Lats1 R660A and Mob1 is likely due to the disruption of Lats1 structure by this mutation. Collectively, these results validate the importance of the pMob1–Lats1 interface. Many critical interface residues in the Lats1 MBD are conserved in human NDR1 (Supplemental Fig. S6C), and mutations in this region of NDR1 have been shown to disrupt its binding to Mob1 (Bichsel et al. 2004). Therefore, Lats1 and NDR likely share a similar Mob1-binding mechanism.
Phosphorylation-dependent conformational activation of Mob1
Phosphorylation of Mob1 at T12 and T35 promotes Lats1 binding. However, neither pT12 nor pT35 makes direct contact with Lats1 in the pMob1–Lats1 structure. Moreover, as shown in Figure 4B and Supplemental Figure S5A, deletion of the N-terminal 15 residues (including T12) of Mob1 permits Lats1 binding. These results suggest that the first 15 residues of Mob1 blocks Lats1 binding in an autoinhibited conformation and that phosphorylation of T12 relieves this autoinhibition. The previously reported crystal structure of free human Mob1ΔN32 lacked the first 15 residues. We could not crystallize the unphosphorylated full-length human Mob1 despite repeated attempts. Fortunately, residues 3–13 of metazoan Mob1 proteins contained a conserved FxFx5KxF motif, which was also present in yeast Mob1 at residues 102–112 (FxYx5KxF; note that yeast Mob1 has a much longer NTE) (Fig. 5A). In the crystal structure of yeast Mob1, this motif binds intramolecularly at the conserved Lats1-binding surface, with Y104 and F112 making extensive hydrophobic contacts with the Mob1 core (Fig. 5B). In particular, F112 binds to a deep conserved hydrophobic pocket corresponding to the pocket in human Mob1 that binds to Lats1 R697 (Fig. 5C), providing a possible explanation for why the N-terminal FXFx5KxF motif can inhibit Lats1 binding. We note that human Mob1 E55 forms a favorable charged interaction with Lats1 R697. The corresponding E155 in yeast Mob1 points to an opposite direction and is not in close contact with F112. Instead, F112 packs against other hydrophobic residues at this site in yeast Mob1. Thus, unphosphorylated human Mob1 likely also adopts an autoinhibited, closed conformation, with its N-terminal FXFx5KxF motif occupying a part of the Lats1-binding surface.
Figure 5.
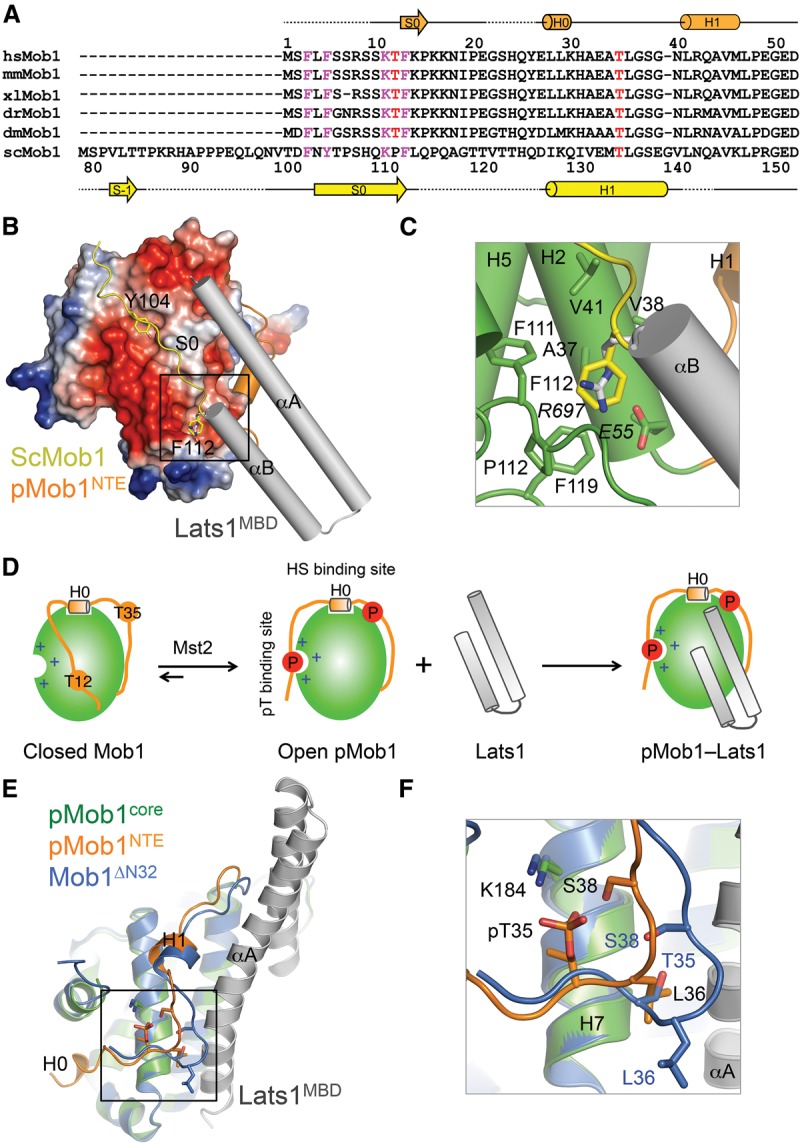
Phosphorylation-dependent conformational activation of Mob1. (A) Sequence alignment of the N-terminal region of Mob1. Secondary structural elements of pMob1 are drawn above the sequences. Secondary structural elements of S. cerevisiae (Sc) Mob1 are drawn below the sequences. The conserved FxFx5KxF motif is colored magenta. T12 and T35 are colored red. (B) Superposition of ScMob1 (Protein Data Bank [PDB] code 2HJN) and pMob1–Lats1 structures (this study). Only the FxYx5KxF motif-containing fragment from ScMob1 is shown. The Mob1-binding site of ScMob1 F112 is boxed. (C) Zoomed-in view of the boxed region in B to show that ScMob1 F112 (yellow sticks) and Lats1 R697 (gray sticks) occupy the same Mob1-binding site. (D) Model of phosphorylation-triggered conformational activation and Lats1 binding of Mob1. (E) Superposition of unphosphorylated Mob1ΔN32 (PDB code 1PI1) and pMob1–Lats1 structures (this study). Mob1ΔN32 is colored blue. The pMob1 core is colored green, and pMob1 NTE is colored orange. Lats1 is colored gray. The H0–H1 loop around pT35 is boxed. (F) Zoomed-in view of the boxed region in E to show the reorganization of the H0–H1 loop in pMob1 by T35 phosphorylation. Residues of Mob1ΔN32 and pMob1 are shown as blue and orange sticks, respectively.
The Lats1-binding surface on human Mob1 is negatively charged (Fig. 5B). The N-terminal 20 residues of Mob1 contain several positively charged residues. Phosphorylation of T12 within the FXFx5KxF motif by Mst2 is expected to introduce electrostatic repulsion between the N-terminal region and the negatively charged surface of Mob1. More importantly, phosphorylation of T12 creates a pT12F motif (similar to a pTM motif) that is capable of binding to the pT-binding site located at the opposite face of Mob1. Therefore, the combination of these destabilizing and stabilizing interactions involving pT12 is capable of reorganizing the conformation of the N-terminal region of Mob1, peeling it away from the Lats1-binding surface and anchoring it at the pT- and HS-binding sites (Fig. 5D). This conformational transition model of Mob1 from the closed conformer to the open conformer largely accounts for phosphorylation-dependent Lats1 binding. Because residues in the H0 helix or in the HS-binding site of Mob1 are not modified, the H0 helix likely remains bound at the HS-binding site in both closed and open Mob1 conformers.
Phosphorylation of Mob1ΔN15 and Mob1ΔN32 still enhanced their binding to Lats1 (Fig. 4B; Supplemental Fig. S5A), indicating that pT35 has an additional role in Lats1 binding aside from relieving autoinhibition. Although pT35 does not directly contact Lats1, comparison between the unphosphorylated Mob1ΔN32 structure and our pMob1–Lats1 structure reveals that phosphorylation of T35 reorganizes the conformation of neighboring residues from A34 to G39 (Fig. 5E,F). pT35 develops a favorable electrostatic interaction with K184 on Mob1 H7 and forms hydrogen bonds with S38. As a result, the H0–H1 loop is more tightly anchored to H7, positioning L36 for hydrophobic interactions with residues from Lats1 αA. Therefore, phosphorylation of Mob1 by Mst2 enhances Lats1 binding through two mechanisms: releasing autoinhibition by the N-terminal FXFx5KxF motif and reorganizing the H0–H1 loop.
Since unphosphorylated Mob1 had detectable Lats1 binding (Fig. 4B; Supplemental Fig. S5A), the autoinhibition by the N-terminal FXFx5KxF motif is incomplete. In the absence of phosphorylation, Mob1 might undergo the closed–open conformational transition to some degree. Phosphorylation or ligand binding simply shifts this equilibrium toward the open state (Fig. 5D).
Mst2 binding to Mob1 suffices to activate Mob1 for Lats1 binding
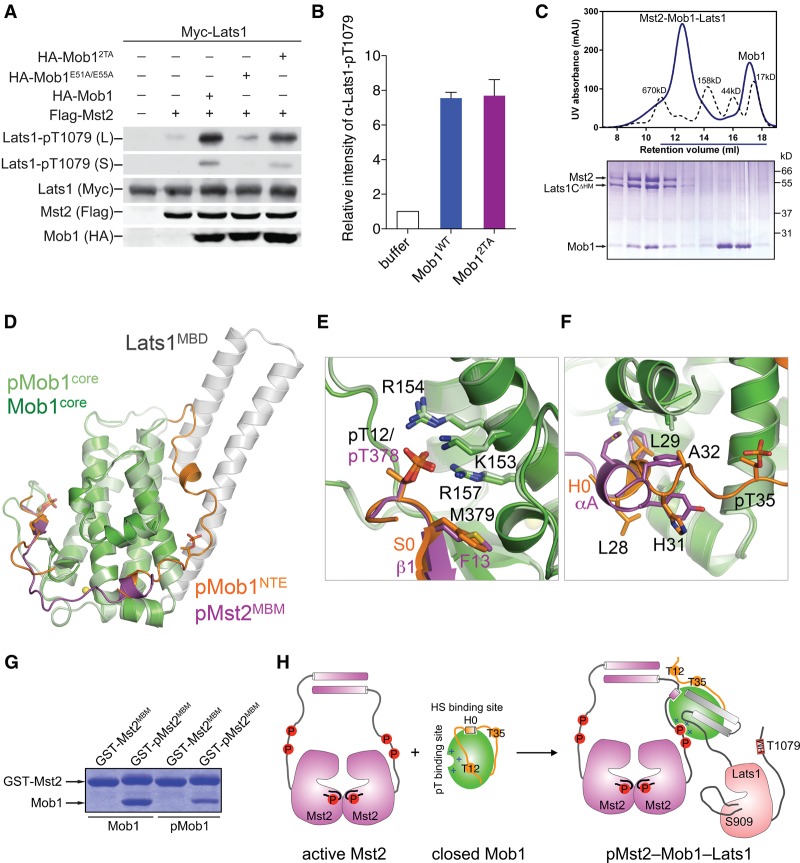
We next investigated the functions of the Mob1–Lats1 interaction. When coexpressed in HEK293 cells, Flag-Mst2 weakly phosphorylated Myc-Lats1 at T1079 in its HM (Fig. 6A). Coexpression of HA-Mob1 with Flag-Mst2 and Myc-Lats1 greatly enhanced Lats1 T1079 phosphorylation. The simplest explanation for this Mob1-dependent stimulation of Lats1 T1079 phosphorylation by Mst2 is that Mob1 simultaneously binds to Mst2 and Lats1, forming a ternary Mst2–Mob1–Lats1 complex in which Mst2 phosphorylates Lats1 more efficiently. Consistent with this hypothesis, the Mob1E51A/E55A mutant deficient for Lats1 binding failed to stimulate Lats1 T1079 phosphorylation by Mst2 (Fig. 6A). Surprisingly, another Lats1-binding-deficient Mob1 mutant, Mob12TA, still stimulated Mst2-mediated T1079 phosphorylation. Furthermore, Mob1WT or Mob12TA greatly stimulated Lats1 T1079 phosphorylation by Mst2 in an in vitro kinase assay with purified recombinant proteins (Fig. 6B). These results suggest that Mob1 phosphorylation per se might be dispensable for the formation of the Mst2–Mob1–Lats1 ternary complex.
Figure 6.
Mst2 binding to Mob1 is sufficient to activate Mob1 for Lats1 binding. (A) Immunoblots of lysates of HEK293 cells transfected with the indicated plasmids. Both long (L) and short (S) exposures of the Lats1-pT1079 blots are shown. (B) Quantification of Lats1C (residues 602–1130) T1079 phosphorylation by Mst2 in the presence of Mob1 wild type or 2TA. Means with range for two independent experiments are plotted. (C) UV traces of molecular weight standards (dashed line) and the Mst2–Mob1–Lats1CΔHM complex (solid line) fractionated on a Superose 6 gel filtration column. The underlined fractions were separated on SDS-PAGE and stained with Coomassie. (D) Superposition of pMst2–Mob1 and pMob1–Lats1 structures. (E,F) Zoomed-in view of the pT- and HS-binding sites of the superimposed structures in D. Residues of the Mob1 core (green), pMob1 (pale green), the Mob1 NTE (orange), and pMst2 (magenta) are shown as sticks. S0 and H0 of pMob1 NTE are colored orange. αA and β1 of pMst2 are colored magenta. (G) Coomassie-stained gel of Mob1 or pMob1 bound to GST-Mst2MBM (residues 371–401) or GST-pMst2MBM beads. (H) Model for pMst2-dependent activation of Mob1 for the formation of the pMst2–Mob1–Lats1 ternary complex.
Mob1 phosphorylation promotes Lats1 binding mainly through relieving autoinhibition rather than through creating phospho-recognition motifs. We tested whether Mst2 binding to unphosphorylated Mob1 was sufficient to activate it for Lats1 binding. Indeed, addition of active full-length Mst2 (pMst2) to unphosphorylated Mob1 greatly stimulated its ability to bind Lats1, as evidenced by the formation of the stoichiometric pMst2–Mob1–Lats1 complex during gel filtration chromatography (Fig. 6C). This result indicates that binding of pMst2 to Mob1 alone (without Mob1 phosphorylation) suffices to activate Mob1 for Lats1 binding, promoting the formation of the pMst2–Mob1–Lats1 ternary complex.
This surprising result nicely explains a major conundrum, as revealed by the comparison of our Mob1–pMst2 and pMob1–Lats1 structures (Fig. 6D). The phosphorylated NTE of pMob1 (pMob1NTE; residues 10–51) and the phosphorylated pT378-Mst2MBM peptide (pMst2MBM; residues 371–401) bind to the same sites on Mob1. For example, pT12 in the Mob1 NTE and pT378 in the Mst2 MBM bind to the same pT-binding site on Mob1 (Fig. 6E). The H0 helix of Mob1 and the Mst2 HS motif (αA) bind to the same HS-binding site on Mob1 (Fig. 6F). Thus, the intramolecular association of the phosphorylated NTE with the Mob1 core is expected to weaken pMst2 binding and dissociate the pMst2–Mob1 complex. Indeed, pMob1 bound to pMst2 more weakly than unphosphorylated Mob1 did (Fig. 6G). On the one hand, this phenomenon is consistent with the enzyme–substrate relationship of Mst2 and Mob1 and can explain the release of the product (pMob1) from the enzyme (Mst2). On the other hand, Lats1 prefers to bind to phosphorylated Mob1, which can no longer associate with Mst2. The competition between phospho-NTE of Mob1 and pMst2 thus hinders the formation of the pMst2–Mob1–Lats1 complex.
The fact that pMst2 binding to Mob1 (without Mob1 phosphorylation) stimulates Mob1 binding to Lats1 resolves this conundrum. Binding of the HS motif in pMst2 to Mob1 is expected to displace the H0 helix of the unphosphorylated Mob1 NTE from the HS-binding site (Fig. 6H). The dissociation of the H0 helix shifts the conformational equilibrium of Mob1 toward the open state, allowing Lats1 binding and the formation of the pMst2–Mob1–Lats1 ternary complex. In this complex, Mst2 can phosphorylate both Mob1 at T12 and T35 and Lats1 at T1079. Mob1 T12/T35 phosphorylation then triggers the release of the pMob1–Lats1 complex already phosphorylated at T1079. Thus, Mob1 acts as a dynamic scaffold module to transiently bring Mst2 and Lats1 into close proximity, enhancing Lats1 T1079 phosphorylation. We stress that we did not definitively show the existence of a transient complex among unphosphorylated Mob1, Lats1, and Mst2 in vivo.
Mob1 allosterically promotes Lats1 autophosphorylation at the activation loop
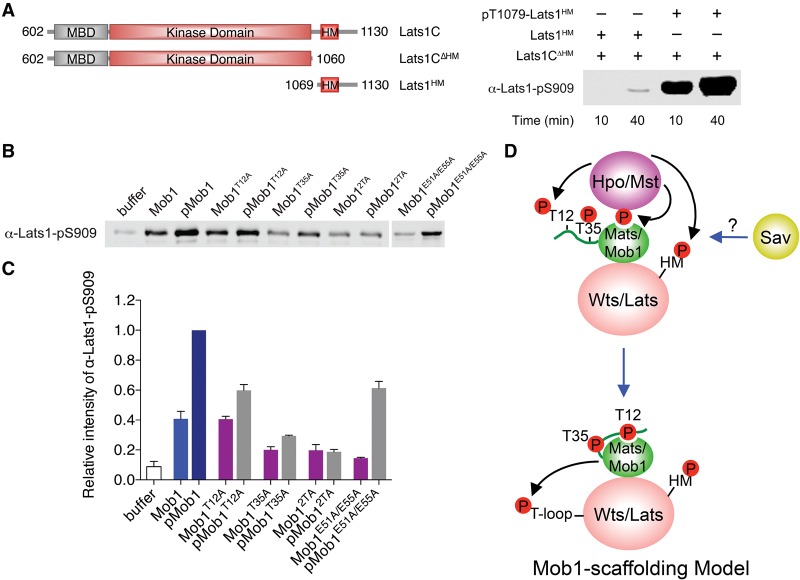
Mst2-mediated Lats1 T1079 phosphorylation is a prerequisite for Lats1 autophosphorylation at S909 in its activation loop (T loop), which ultimately activates Lats1. We tested whether Mob1 in the context of the binary pMob1–Lats1 complex stimulated Lats1 autophosphorylation. Addition of pT1079-containing Lats1HM (Lats1 residues 1069–1130) pretreated with recombinant Mst2 greatly stimulated S909 autophosphorylation of Lats1CΔHM (a Lats1 kinase fragment that lacked the HM motif) (Fig. 7A). As a control, the unphosphorylated Lats1HM had little effect in this assay. Thus, addition of pT1079-Lats1HM alone can bypass the need for Mst2 and activate Lats1 S909 autophosphorylation in trans. How pT1079-Lats1HM activates Lats1 is not understood and will need to be addressed by additional structural studies. Regardless, the fact that the phosphorylated HM fragment could activate Lats1 autophosphorylation at S909 in trans enabled us to separate the effects of Mob1 in stimulating Mst2-dependent Lats1 T1079 phosphorylation from any potential effects in stimulating Lats1 S909 autophosphorylation.
Figure 7.
Mob1 allosterically promotes Lats1 autoactivation. (A) Lats1-pS909 blot (shown in the right panel) of kinase reactions containing the indicated Lats1 proteins depicted in the left panel. (B) Lats1-pS909 blot of kinase reactions containing 2 µM Lats1CΔHM, 4 µM pT1079-Lats1HM, and the indicated Mob1 proteins (2 µM). (C) Quantification of Lats1-pS909 levels in B. The relative α-Lats1-pS909 intensities were normalized to wild-type pMob1 (100%). Means with range for two independent experiments are plotted. (D) Mob1 scaffolding model for Mst-dependent Lats activation during Hippo signaling.
Strikingly, addition of Mob1 further stimulated S909 autophosphorylation of Lats1CΔHM in the presence of pT1079-Lats1HM (Fig. 7B,C). Mob1 pretreated with Mst2 (pMob1) was even more active in this assay. As expected, the phosphorylation-deficient Mob1T12A, Mob1T35A, and Mob12TA mutant blunted this phosphorylation-dependent effect. The Mob1T35A and Mob12TA mutants were less active than the unphosphorylated Mob1, possibly due to a structural role of T35 (in the unphosphorylated form) in Lats1 binding. The Lats1-binding-deficient Mob1E51A/E55A mutant was also deficient in stimulating Lats1 S909 phosphorylation (Fig. 7B,C), indicating that Mob1 binding to the Lats1 MBD was required for this stimulation.
Discussion
Despite the tremendous progress in the discovery of upstream regulators of the Hippo pathway, the activating mechanism of the core Hippo–Lats kinase cascade is still not understood. Our present study addresses this pressing issue and provides the following model for Mob1-dependent Lats1 activation by Mst2 (Fig. 7D). In this model, Mst2 autophosphorylates multiple TM sites in its linker, creating phospho-docking motifs to recruit Mob1. Mst2 binding triggers the conformational activation of Mob1 from the closed state to the open state. Mst2-bound, activated Mob1 further recruits Lats1, forming a pMst2–Mob1–Lats1 ternary complex. In this complex, active Mst2 phosphorylates Mob1 at T12 and T35 in the NTE and Lats1 at T1079 in the HM. The phosphorylated Mob1 NTE folds back to bind its core at the pMst2-binding sites, releasing the primed pMob1–Lats1 complex. In the absence of Mst2, phosphorylation of Mob1 at T12 and T35 maintains the open state of Mob1 and preserves the pMob1–Lats1 complex. Phosphorylated HM and pMob1 then collaborate to stimulate Lats1 autophosphorylation at the activation loop, leading to its activation.
In the case of human Mst2, multiple pTM motifs act redundantly to engage the pT-binding site in Mob1. The physiological meaning of this redundancy is unknown at present. One possibility is that this multisite autophosphorylation by Mst2 can more effectively buffer against the actions of phosphatases in vivo and produce more robust Mob1 activation in response to certain upstream signals.
The mechanism by which Mob1 binding to Lats1 MBD allosterically activates Lats1 autophosphorylation at its T loop is not understood. All NDR kinases known to interact with Mob proteins, including Lats1, have an insertion of 30–60 residues in their T loop. Although this insertion is not well conserved, it is rich in basic residues. This basic insertion has been shown to inhibit the autophosphorylation of human NDR in its T loop (Bichsel et al. 2004). Binding of Mob1 to the conserved MBD of NDR has been proposed to release the inhibitory effect of this basic insertion. We speculate that Mob1 stimulates Lats1 autophosphorylation through a similar allosteric mechanism.
Another core component of the Hippo pathway, Sav, has been proposed to promote Lats1 activation through acting as a scaffold to recruit Lats/Wts to Mst/Hpo (Harvey et al. 2003). Besides contributing to Mst/Hpo activation by binding to Mst/Hpo through its SARAH domain (Hwang et al. 2007), Sav was reported to bind the PPXY motifs in Lats/Wts through its WW domains, therefore promoting Lats/Wts activation (Tapon et al. 2002). However, direct evidence for the formation of an Mst–Sav1–Lats ternary complex and Sav-dependent activation of Lats in vitro is lacking (Chan et al. 2005). We found that human Sav WW domains bound to a PPXY-containing peptide of Lats1 (AGTVNRQPPPYPNGQS) with a Kd of 156 µM, which is much weaker as compared with the pMob1–Lats1 binding. On the other hand, because we failed to obtain full-length recombinant Lats1 protein that contains the N-terminal PPXY motifs, we were unable to directly test the role of Sav in mediating Mst/Mob1-dependent activation of Lats1.
Our study thus establishes a major framework of core Mst–Lats kinase activation in the Hippo pathway, in which Mob1 acts as the key scaffold. Mob1 alone is sufficient to promote Lats1 activation in the absence of Sav in vitro. Furthermore, activated Hpo can functionally bypass Sav under several circumstances in flies (Pantalacci et al. 2003; Ribeiro et al. 2010). In contrast, Mats/Mob1 is indispensable for Wts/Lats activation in all cases (Wei et al. 2007; Nishio et al. 2012). Our new model with Mob1 as the central scaffolding protein is thus more consistent with the available genetic evidence, which supports the essential role of Mob1 in Mst–Lats kinase activation. Our model is also consistent with recent phylogenetic analysis showing that Sav appeared later during evolution than Hpo, Mats, Sd, and Yki (Sebe-Pedros et al. 2012). For example, the single-cell organism Capsaspora does not encode a Sav homolog but contains an Mst/Hpo homolog with conserved pTM and HS motifs capable of activating Wts when overexpressed in Drosophila (Sebe-Pedros et al. 2012). Nevertheless, our model does not exclude the possibility that other regulators, such as Sav, can further enhance Mob1-dependent activation of Wts/Lats by Hpo/Mst in the context of the Mst–Mob1–Lats complex (Yin et al. 2013). Future experiments are needed to address this issue.
The Hippo pathway controls tissue homeostasis by regulating the balances between cell proliferation and quiescence and between cell death and survival. Dysregulation of the Hippo pathway has been linked to human cancers. The Mst–Lats kinase cascade forms the core of this pathway. Our study provides key structural and mechanistic insights into the Mob1-dependent activation of Lats1 by Mst2. Through dynamic scaffolding and allosteric mechanisms, Mob1 promotes a multistep, sequential process of Lats1 activation and effectively couples the activation of two kinases in the same cascade. The requirement for orderly phosphorylation of multiple targets by an upstream kinase to activate a downstream one ensures specificity in signal transduction.
Materials and methods
Plasmids and antibodies
The coding regions of human Mob1 and human Mst2 linker fragments were cloned into a modified pGEX-6P vector (GE Healthcare) that included a tobacco etch virus (TEV) cleavage site at the N terminus. The coding region of either human Lats1CΔHM (residue 602–1060) or Lats1C (residue 602–1130) was cloned together with wild-type Mob1 (residue 1–216) into a modified pRSF-Duet vector (EMD Millipore). The cDNAs of Lats1MBD (residue 602–704) and Lats1HM (residue 1069–1130) were cloned into a modified pET29 vector (EMD Millipore) that included an N-terminal His6 tag. The coding regions of human Lats1 fragments were cloned into a modified pCS2-Myc vector. HA-Mob1, Flag-Mst2, and Myc-Lats1 expression constructs have been described previously (Chan et al. 2005; Zhao et al. 2007). Flag-Hpo was generated from Myc-Hpo described previously (Wu et al. 2003). Flag-Mats expression plasmid was generated from a cDNA clone (LD47553) obtained from the Drosophila Genomics Resource Center. The ORF of mats was amplified by PCR and inserted into pGEX-6P-1 vector to generate the GST-Mats construct. All point mutations were generated using QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). All constructs were verified by DNA sequencing.
Primary antibodies used in this study include Flag and HA (Sigma-Aldrich), Myc (Calbiochem), Wts-pT1077 (Yu et al. 2010), Mob1-pT12 (Praskova et al. 2008), phospho-T12 or phospho-T35 of Mob1 (Cell Signaling), and phopho-T1079 or phopho-S909 of Lats1 (Cell Signaling).
Protein expression and purification
The plasmids were transformed into the bacterial strain BL21(DE3)-T1R cells (Sigma) for protein expression. Untagged Mob1 and Mob1 mutants were purified with glutathione-Sepharose beads (GE Healthcare) and cleaved with TEV overnight at 4°C to remove the GST moiety. GST-Mob1 and GST-Mst2 linker proteins were purified with the glutathione-Sepharose beads. Mob1, GST-Mob1, and GST-Mst2 linker proteins were further purified by size exclusion chromatography with a Superdex 75 column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. Lats1CΔHM and Lats1C were purified with Ni2+-NTA resin (Qiagen) and cleaved with TEV overnight at 4°C to remove the N-terminal His6 tag in a buffer containing 20 mM HEPES (pH 7.0), 100 mM NaCl, 5% glycerol, 2 mM MgCl2, and 1 mM DTT. Because the expression level of Mob1 was very low from pRSF-Duet, Lats1CΔHM and Lats1C were the major products eluted from Ni2+-NTA resin. Lats1CΔHM and Lats1C were further purified by a Mono S cation exchange column followed by a Superdex 200 gel filtration column (GE Healthcare). The expression and purification of Mst2WT, Mst2D146N, Mst2ΔL, and Mst2KD were described previously (Ni et al. 2013). The pMst2–Mob1 complex was obtained by mixing 30 mg/mL Mob1ΔN50 with the pMst2MBM peptide (DEEEEDGpTMKRNATSPQVQRPSFMDYFDKQ) at 1:2 molar ratio.
To obtain GST-pMob1, purified GST-Mob1 was dialyzed in the kinase reaction buffer (50 mM Tris at pH 7.5, 150 mM NaCl, 10 mM MgCl2). GST-Mob1 was then treated with purified Mst2KD for 1 h at room temperature in the presence of 2 mM ATP. GST-pMob1 was reloaded onto glutathione-Sepharose beads to remove Mst2KD. GST-pMob1 was either eluted with glutathione from glutathione-Sepharose beads or cleaved with TEV overnight at 4°C to remove GST moiety. GST-pMob1 and pMob1 were further purified by a Superdex 75 column equilibrated with a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl.
Lats1MBD was purified by the denaturation method using 8 M urea (Qiagen) according to the manufacturer's instructions. Lats1MBD was refolded overnight at 4°C in a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. The pMob1–Lats1MBD complex was purified by a resource-Q anion exchange column (GE Healthcare) after mixing pMob1 and Lats1MBD at a 1:4 molar ratio. The pMob1–Lats1MBD complex was further purified by a Superdex 200 column equilibrated with a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl. Purified pMob1–Lats1MBD was concentrated to 10 mg/mL for crystallization.
To obtain pT1079-Lats1HM, Lats1HM was purified by the denaturation method as described for Lats1MBD. After refolding in the kinase reaction buffer, Lats1HM was treated with Mst2KD for 1 h at room temperature in the presence of 2 mM ATP. It was reloaded onto Ni2+-NTA resin to remove Mst2KD and then eluted from Ni2+-NTA resin with imidazole. pT1079-Lats1HM was further purified by a Superdex 75 column equilibrated with a buffer containing 20 mM Tris (pH 8.0) and 100 mM NaCl.
The selenomethionine-labeled Mob1 (SeMet-Mob1) was produced using the methionine biosynthesis inhibition method (Van Duyne et al. 1993).
Crystallization, data collection, and structure determination
The pMst2–Mob1 complex was crystallized at 20°C using the hanging drop vapor diffusion method with a reservoir solution containing 2.4 M sodium malonate (pH 7.0). The crystals were cryoprotected with the reservoir solution supplemented with 20% (v/v) ethylene glycol and then flash-cooled in liquid nitrogen. Crystals diffracted to a resolution of 2.65 Å and contained six pMst2–Mob1 molecules per asymmetric unit.
The phosphorylated SeMet–Mob1 and Lats1MBD complex (pMob1–Lats1) was crystallized at 20°C using the hanging drop vapor diffusion method with a reservoir solution containing 0.2 M diammonium citrate (pH 5.0) and 20% (w/v) PEG 3350. The crystals were cryoprotected with the reservoir solution supplemented with 24% (w/v) sucrose and then flash-cooled in liquid nitrogen. Crystals diffracted to a resolution of 2.29 Å and contained one pMob1–Lats1 molecule per asymmetric unit.
All X-ray diffraction data were collected at 100 K at the Structural Biology Center (Beamline 19-ID) at Argonne National Laboratory and processed with HKL3000 (Otwinowski and Minor 1997). Initial phases for pMst2–Mob1 were obtained by molecular replacement with Phaser using the crystal structure of human Mob1 (PDB code 1PI1) as a searching model (McCoy et al. 2007). Iterative model building and refinement were performed with Coot and Phenix, respectively (Adams et al. 2010; Emsley et al. 2010). Data collection and refinement statistics are summarized in Supplemental Table S1.
Initial phases for the SeMet-labeled pMob1–Lats1 were obtained by SAD with Phenix AutoSol. Automated model building was done with Phenix AutoBuild. Subsequent model building was performed manually in Coot. The structure was refined with Phenix. Data collection and refinement statistics are summarized in Supplemental Table S2.
The atomic coordinates and structure factors for human pMst2–Mob1 and pMob1–Lats1 have been deposited in the PDB with the accession codes 5BRM and 5BRK, respectively.
NMR spectroscopy
All NMR spectra were acquired at 30°C on a Varian INOVA 600-MHz four-channel spectrometer equipped with pulsed-field gradient triple-resonance probe using H2O/D2O 95:5 (v/v) as the solvent. Samples typically contained 0.1 mM 15N-labeled protein in the NMR buffer containing 20 mM sodium phosphate (pH 6.8), 100 mM KCl, 2 mM MgCl2, and 1 mM DTT. The data were processed and analyzed with NMRPipe and NMRView (Johnson and Blevins 1994; Delaglio et al. 1995).
ITC
ITC was performed with a VP-ITC titration calorimeter (MicroCal, Inc.) at 20°C. For Mob1 and pMst2MBM, calorimetric measurements were performed with 20 μM purified Mob1 in the cell and 200 μM pMst2MBM peptide in the syringe. For Mst2ΔL/MBM and Mob1, calorimetric measurements were carried out with 20 μM purified Mst2ΔL/MBM in the cell and 200 μM purified Mob1 in the syringe. All samples were in the same buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, and 2 mM MgCl2. Mob1 or pMst2MBM was injected with 35 portions of 8 μL. Binding parameters were evaluated using the Origin package provided with the instrument.
In vitro kinase assays
For Mst2 kinase assays, 0.5 nM Mst2 or Mst2 mutants was incubated with 0.4 μM Mob1 or Mob1 mutants in the kinase reaction buffer containing 50 mM Tris (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM ATP, and 1 mM DTT at room temperature. At each time point, an aliquot of the reaction mixture was taken and quenched by the addition of the same volume of 2× SDS sample buffer. The phosphorylated Mob1 NTE was analyzed by quantitative Western blotting using specific antibodies against either pT12 or pT35 of Mob1 (Cell Signaling). The signals were quantified with the Odyssey LI-COR imaging system.
For Lats1 kinase assays, 2 μM Lats1C (residues 602–1130) was incubated with 2 μM either Mob1 or pMob1 or their corresponding mutants together with 20 nM Mst2WT (residue 9–491) in the kinase reaction buffer at room temperature. At each time point, an aliquot of the reaction mixture was taken and quenched by the addition of the same volume of 2× SDS sample buffer. The phosphorylated Lats1 HM was analyzed by quantitative Western blotting using a specific antibody against pT1079 of Lats1 (Cell Signaling).
To assay Lats1 autophosphorylation, 2 μM Lats1CΔHM was incubated with 2 μM either Mob1 or pMob1 or their corresponding mutants together with 4 μM phosphorylated Lats1HM in the kinase reaction buffer at room temperature. The reaction was terminated at the indicated time point. The phosphorylated Lats1 T loop was analyzed by quantitative Western blot using a specific antibody against pS909 of Lats1 (Cell Signaling).
In vitro binding assays
GST-Mst2 fusion proteins were treated by the Mst2KD in the kinase reaction buffer either with or without 2 mM ATP for 30 min. GST-Mst2 fusion proteins were bound to glutathione-Sepharose beads to remove Mst2KD and then incubated with Mob1 or Mob1 mutants. The beads were washed three times with TBS (50 mM Tris at pH 7.5, 150 mM NaCl). The proteins retained on the beads were analyzed by SDS-PAGE.
To assay the binding between human Lats1 and Mob1 proteins, Myc-Lats1 was in vitro translated in reticulocyte lysate in the presence of 35S-methionine according to the manufacturer's protocol (Pierce). Purified GST-Mob1, GST-pMob1, or their corresponding mutants were bound to glutathione-Sepharose beads and incubated with 35S-labeled Lats1 or Lats1 mutants. The beads were washed three times with TBS containing 0.05% Tween. The proteins retained on the beads were analyzed by SDS-PAGE followed by autoradiography.
GST-Mats protein was expressed in BL21-CodonPlus (DE3)-RIPL cells (Agilent Technologies) and purified using glutathione-Sepharose beads. GST-Mats-bound beads were then mixed with cell lysate made from S2R+ cells expressing wild-type or mutant Flag-Hpo proteins for 2 h at 4°C. The beads were washed three times in PBS and then boiled in 2× SDS buffer for 5 min.
Drosophila and mammalian cell culture
Drosophila S2R+ cells were cultured at 25°C in Schneider's medium supplemented with 10% FBS and antibiotics (Invitrogen). 293T cells were maintained at 37°C in DMEM medium supplemented with 10% FBS and antibiotics (Invitrogen). Transfection and Western blotting were carried out as described previously (Yu et al. 2010).
Supplementary Material
Acknowledgments
We thank Dr. J. Avruch for the Mob-pT12 antibody, and the Drosophila Genomics Resource Center for the mats cDNA clone. We also thank Dr. D. Tomchick for assistance with the refinement of the pMst2–Mob1 structure. Use of the Argonne National Laboratory Structural Biology Center beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy under contract DE-AC02-06CH11357. The DD2 console of the 600-MHz NMR spectrometer was supported by a grant from the National Institutes of Health (S10RR026461-01). This work was supported in part by grants from the National Institutes of Health (GM085004 to X.L. and EY015708 to D.P.). D.P. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.264929.115.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. 2012. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev Biol 23: 770–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badouel C, Garg A, McNeill H. 2009. Herding Hippos: regulating growth in flies and man. Curr Opin Cell Biol 21: 837–843. [DOI] [PubMed] [Google Scholar]
- Barry ER, Camargo FD. 2013. The Hippo superhighway: signaling crossroads converging on the Hippo/Yap pathway in stem cells and development. Curr Opin Cell Biol 25: 247–253. [DOI] [PubMed] [Google Scholar]
- Bichsel SJ, Tamaskovic R, Stegert MR, Hemmings BA. 2004. Mechanism of activation of NDR (nuclear Dbf2-related) protein kinase by the hMOB1 protein. J Biol Chem 279: 35228–35235. [DOI] [PubMed] [Google Scholar]
- Boggiano JC, Fehon RG. 2012. Growth control by committee: intercellular junctions, cell polarity, and the cytoskeleton regulate Hippo signaling. Dev Cell 22: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos J, Tuttle RL, Ottey M, Luca FC, Halazonetis TD. 2005. Human LATS1 is a mitotic exit network kinase. Cancer Res 65: 6568–6575. [DOI] [PubMed] [Google Scholar]
- Chan EH, Nousiainen M, Chalamalasetty RB, Schafer A, Nigg EA, Sillje HH. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene 24: 2076–2086. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA. 2006. From cell structure to transcription: Hippo forges a new path. Cell 124: 267–273. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle L, McNeill H. 2013. Hippo gains weight: added insights and complexity to pathway control. Sci Signal 6: re7. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. 2011. Hippo signaling: growth control and beyond. Development 138: 9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X. 2008. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. 2007. The Salvador–Warts–Hippo pathway—an emerging tumour-suppressor network. Nat Rev Cancer 7: 182–191. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. 2003. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114: 457–467. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. 2013. The Hippo pathway and human cancer. Nat Rev Cancer 13: 246–257. [DOI] [PubMed] [Google Scholar]
- Hergovich A. 2011. MOB control: reviewing a conserved family of kinase regulators. Cell Signal 23: 1433–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A. 2013. Regulation and functions of mammalian LATS/NDR kinases: looking beyond canonical Hippo signalling. Cell Biosci 3: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergovich A, Hemmings BA. 2009. Mammalian NDR/LATS protein kinases in hippo tumor suppressor signaling. Biofactors 35: 338–345. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Schmitz D, Hemmings BA. 2006. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun 345: 50–58. [DOI] [PubMed] [Google Scholar]
- Hong W, Guan KL. 2012. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol 23: 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434. [DOI] [PubMed] [Google Scholar]
- Hwang E, Ryu KS, Paakkonen K, Guntert P, Cheong HK, Lim DS, Lee JO, Jeon YH, Cheong C. 2007. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci 104: 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, Zhang L. 2012. Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J Biol Chem 287: 5784–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. 1994. NMRView: a computer program for visualization and analysis of NMR data. J Biomol NMR 4: 603–614. [DOI] [PubMed] [Google Scholar]
- Johnson R, Halder G. 2014. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov 13: 63–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. 1998. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol Cell Biol 18: 2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X. 2010. Snapshots of a hybrid transcription factor in the Hippo pathway. Protein Cell 1: 811–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrkobrada S, Boucher L, Ceccarelli DF, Tyers M, Sicheri F. 2006. Structural and functional analysis of Saccharomyces cerevisiae Mob1. J Mol Biol 362: 430–440. [DOI] [PubMed] [Google Scholar]
- Ni L, Li S, Yu J, Min J, Brautigam CA, Tomchick DR, Pan D, Luo X. 2013. Structural basis for autoactivation of human Mst2 kinase and its regulation by RASSF5. Structure 21: 1757–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Hamada K, Kawahara K, Sasaki M, Noguchi F, Chiba S, Mizuno K, Suzuki SO, Dong Y, Tokuda M, et al. 2012. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J Clin Invest 122: 4505–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pan D. 2010. The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. 2003. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol 5: 921–927. [DOI] [PubMed] [Google Scholar]
- Praskova M, Xia F, Avruch J. 2008. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol 18: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro PS, Josue F, Wepf A, Wehr MC, Rinner O, Kelly G, Tapon N, Gstaiger M. 2010. Combined functional genomic and proteomic approaches identify a PP2A complex as a negative regulator of Hippo signaling. Mol Cell 39: 521–534. [DOI] [PubMed] [Google Scholar]
- Rock JM, Lim D, Stach L, Ogrodowicz RW, Keck JM, Jones MH, Wong CC, Yates JR III, Winey M, Smerdon SJ, et al. 2013. Activation of the yeast Hippo pathway by phosphorylation-dependent assembly of signaling complexes. Science 340: 871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Zheng Y, Ruiz-Trillo I, Pan D. 2012. Premetazoan origin of the hippo signaling pathway. Cell Rep 1: 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley BK, Irvine KD. 2012. Hippo signaling in Drosophila: recent advances and insights. Dev Dyn 241: 3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Shields DC, Farooq A. 2012. Structures of YAP protein domains reveal promising targets for development of new cancer drugs. Semin Cell Dev Biol 23: 827–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. 2002. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478. [DOI] [PubMed] [Google Scholar]
- Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. 1993. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol 229: 105–124. [DOI] [PubMed] [Google Scholar]
- Wei X, Shimizu T, Lai ZC. 2007. Mob as tumor suppressor is activated by Hippo kinase for growth inhibition in Drosophila. EMBO J 26: 1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114: 445–456. [DOI] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. 2013. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell 154: 1342–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. 2010. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lei QY, Guan KL. 2008a. The Hippo–YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol 20: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008b. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. 2010a. The Hippo–YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24: 862–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010b. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 24: 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.