This study examined the role of circulating tumor DNA (ctDNA) as a marker of early treatment response in patients with metastatic colorectal cancer (mCRC). A significant drop in ctDNA levels was observed after one cycle of chemotherapy and this drop predicted for subsequent tumor response, suggesting that ctDNA could be used for assessing early treatment failure in mCRC.
Keywords: circulating tumor DNA, metastatic colorectal cancer, biomarker, treatment response
Abstract
Background
Early indicators of treatment response in metastatic colorectal cancer (mCRC) could conceivably be used to optimize treatment. We explored early changes in circulating tumor DNA (ctDNA) levels as a marker of therapeutic efficacy.
Patients and methods
This prospective study involved 53 mCRC patients receiving standard first-line chemotherapy. Both ctDNA and CEA were assessed in plasma collected before treatment, 3 days after treatment and before cycle 2. Computed tomography (CT) scans were carried out at baseline and 8–10 weeks and were centrally assessed using RECIST v1.1 criteria. Tumors were sequenced using a panel of 15 genes frequently mutated in mCRC to identify candidate mutations for ctDNA analysis. For each patient, one tumor mutation was selected to assess the presence and the level of ctDNA in plasma samples using a digital genomic assay termed Safe-SeqS.
Results
Candidate mutations for ctDNA analysis were identified in 52 (98.1%) of the tumors. These patient-specific candidate tissue mutations were detectable in the cell-free DNA from the plasma of 48 of these 52 patients (concordance 92.3%). Significant reductions in ctDNA (median 5.7-fold; P < 0.001) levels were observed before cycle 2, which correlated with CT responses at 8–10 weeks (odds ratio = 5.25 with a 10-fold ctDNA reduction; P = 0.016). Major reductions (≥10-fold) versus lesser reductions in ctDNA precycle 2 were associated with a trend for increased progression-free survival (median 14.7 versus 8.1 months; HR = 1.87; P = 0.266).
Conclusions
ctDNA is detectable in a high proportion of treatment naïve mCRC patients. Early changes in ctDNA during first-line chemotherapy predict the later radiologic response.
introduction
Despite advances in screening and therapeutics, colorectal cancer continues to account for 694 000 deaths per year worldwide [1]. Ever-improving outcomes in patients with metastatic disease have resulted from an increasing number of active agents and biomarker-driven treatment selection. Important goals for ongoing biomarker studies include ensuring patients receive the benefit of being exposed to as many active therapies as possible while minimizing any treatment-related morbidity.
The current gold standard for assessing initial disease bulk, and for defining treatment response, is the image-based Response Evaluation Criteria in Solid Tumors (RECIST). RECIST limitations include poor inter- and intraobserver reproducibility and limited categorization [2]. Further, a minority of patients do not have ‘measurable disease’. Currently, blood biomarkers add little to imaging-based assessment, with CEA lacking sensitivity and specificity [3–5].
Central to the step-wise model of colorectal tumorigenesis is that mutations in multiple genes are required for cancer formation [6]. Digital approaches to DNA analysis can now enable rapid identification of somatic tumor mutations and subsequent quantification of circulating tumor DNA (ctDNA) [7–9]. While previous studies have demonstrated that ctDNA is detectable in a high proportion of patients with metastatic disease [10–12], the clinical utility of ctDNA as a cancer biomarker is just beginning to be evaluated. In this work, we describe the potential role of ctDNA as an early predictor of treatment response of patients with metastatic colorectal cancer (mCRC) undergoing chemotherapy and as a marker of disease bulk that could complement RECIST.
patients and methods
study design
This prospective multicenter study recruited patients from eight Australian hospitals. Eligible patients had RECIST measurable, chemotherapy naïve mCRC, and were to receive standard first-line combination chemotherapy (oxaliplatin- or irinotecan-based) with or without biological therapy. The study was approved by the human research ethics committees at each hospital and all patients provided written informed consent.
Serial blood samples were collected at three defined time-points: pretreatment (within 7 days before commencing chemotherapy), 3 days after starting chemotherapy and before cycle 2 (range 14–21 days after starting treatment). Computed tomography (CT) of the chest, abdomen and pelvis was carried out at baseline and at 8–10 weeks. These scans were centrally assessed by a single radiologist, and disease response evaluated according to RECIST version 1.1. Patients who died before the first follow-up imaging study were excluded from the tumor response analysis. CEA levels were measured by the diagnostic laboratory at each participating site.
Recent data suggest that a ≥20% reduction in the sum of largest tumor diameters (SLD) at first restaging is associated with progression-free (PFS) and overall survival (OS) in mCRC [13, 14]. Using this definition, we defined the ‘Response’ group as patients with at least a 20% reduction in SLD when assessed by CT 8–10 weeks after treatment initiation.
identification of somatic mutations in tumor tissue
Formalin-fixed paraffin-embedded tumor sections were macro-dissected under a dissecting microscope to ensure a neoplastic cellularity of >40%. DNA was purified with a Qiagen FFPE Kit (Qiagen cat #56494). PCR was used to amplify a panel of 15 genes as listed in supplementary Table S1, available at Annals of Oncology online. Primers were designed and sequencing results analyzed as previously described [8, 10, 15, 16].
circulating tumor DNA analysis
Blood samples were processed into plasma within 3 h of collection. Ten milliliter of plasma was collected from each patient. DNA from plasma was purified using QIAamp Circulating Nucleic Acid kit (Qiagen cat# 55114). One of the mutations identified in the tumor tissue was then assessed in the plasma by Safe-SeqS, a massively parallel sequencing (MPS)-based assay that permits the detection of low-frequency mutations [17]. In the MPS-based assay, plasma DNA was aliquotted into wells of a 96-well plate so that an average of 3 ng DNA was contained in each well. The DNA from each well was then amplified using barcodes that distinguished each template molecules. The DNA from all wells was pooled and subjected to MPS with an Illumina MiSeq instrument, as described [10]. ctDNA levels were quantified as the fraction of mutant alleles ×100; for example, if mutant alleles represented 5.1% of the total alleles (mutant plus normal alleles), then the ctDNA score was ‘5.1’. All ctDNA analyses were carried out by individuals blinded to the CEA levels, clinical status of the patients.
statistical analysis
Descriptive statistics and the Mann–Whitney U-test were used to assess the clinical and biochemical variables associated with baseline ctDNA levels. Correlations between the circulating biomarkers (ctDNA and CEA) and tumor burden (measured as per RECIST1.1) were assessed using Spearman's rank correlation.
The Wilcoxon-matched pairs signed-rank test was used to compare serial ctDNA and CEA levels. Receiver operating characteristic (ROC) curves were used to determine the most appropriate ctDNA index (pretreatment ctDNA, ctDNA before cycle 2 and fold reduction in ctDNA from baseline to cycle 2) and optimal cutoff for differentiating patients with ‘response’ and ‘no response’ at first restaging. Among the three ctDNA indices, fold change in ctDNA had the largest ROC area (supplementary Table S2, available at Annals of Oncology online). The optimal cutoff for the fold change in ctDNA was chosen based on the minimum ‘d’ value (the point on the ROC curve closest to (0,1)); d = √[(1 − Sn)2 + (1 − Sp)2], where Sn and Sp denote sensitivity and specificity, respectively. A 10-fold reduction in ctDNA was identified as the optimal cutoff for assessing early tumor response (supplementary Table S3, available at Annals of Oncology online). Comparisons of ROC curves were carried out using the DeLong method [18]. Kaplan–Meier survival plots were generated based on the 10-fold ctDNA reduction for assessment of PFS and OS. PFS and OS were measured from date of baseline blood collection to date of disease progression, death or censoring at last follow-up.
Statistical analysis was carried out using Stata version 12.1 (StataCorp LP, TX) and GraphPad Prism version 6.01 (GraphPad Software, Inc., CA), where P values of <0.05 were considered significant. Results are reported according to REMARK (Reporting Recommendations for Tumor Marker Prognostic Studies) criteria [19].
results
patient characteristics
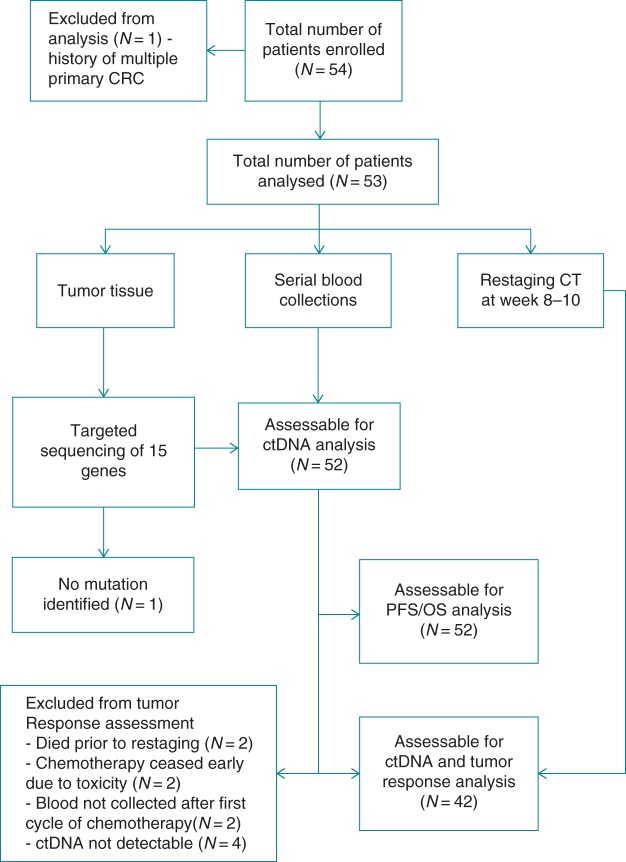
Between August 2011 and June 2013, 54 patients were enrolled, all of whom had at least one baseline blood draw. Figure 1 summarizes the flow of patients through the study, including reasons for exclusion from each stage of the analysis. At the time of analysis, 31 (60%) of the 52 patients evaluable for PFS had experienced disease progression and 20 (38%) patients had died, providing a median PFS of 8.2 months [interquartile range (IQR): 5.1–14.7 months] with the median OS being 17.4 months (25th percentile = 8.0, with no 75th percentile able to be estimated). Patient characteristics are described in Table 1.
Figure 1.
CONSORT diagram showing the flow of patients through the study, including the number of patients included in each of the analysis end points and reasons for exclusion.
Table 1.
Clinico-pathological characteristics and association with pretreatment ctDNA level (N = 52)
| Characteristic | N (%) | ctDNA level Median (IQR) |
P value |
|---|---|---|---|
| All patients | 52 | 16.2 (0.9–32.9) | |
| Age (years), median (IQR) | 64.6 (55.5–71.2) | 0.516 | |
| Age group | |||
| <65 | 27 (51.9) | 12.0 (2.4–30.0) | |
| 65+ | 25 (48.1) | 20.6 (0.6–34.5) | |
| Gender | |||
| Male | 32 (61.5) | 16.3 (0.6–36.2) | – |
| Female | 20 (38.5) | 16.2 (3.0–31.0) | 0.808 |
| ECOGa | |||
| 0 | 37 (72.6) | 9.8 (0.7–25.6) | – |
| 1 | 12 (23.5) | 32.8 (10.8)–42.4) | 0.193 |
| 2 | 2 (3.9) | 13.9 (0–27.9) | 0.865 |
| Primary tumor site | |||
| Right-sided colon | 19 (36.5) | 12.0 (0.5–26.5) | – |
| Left-sided colon | 28 (53.9) | 20.1 (2.5–42.0) | 0.277 |
| Rectum | 5 (9.6) | 12.2 (0.7–30.0) | 0.946 |
| Resected primary tumor | |||
| No | 34 (65.4) | 7.1 (0.6–29.4) | – |
| Yes | 18 (34.6) | 24.9 (13.6–34.5) | 0.409 |
| Number of metastatic sites | |||
| 1 | 22 (42.3) | 7.5 (0.7–32.7) | – |
| 2 | 15 (28.9) | 12.2 (6.9–26.5) | 0.849 |
| >2 | 15 (28.9) | 20.6 (0.1–46.9) | 0.527 |
| Baseline SLD, median (IQR) | 61.5 (35.5–103.5) | 0.008b | |
| Baseline serum CEA, median (IQR) | 26.6 (8.5–208.8) | 0.062b | |
| Chemotherapy regimena | |||
| Oxaliplatin-based | 47 (92.2) | 18.9 (1.4–33.0) | – |
| Irinotecan-based | 4 (7.8) | 13.8 (3.8–29.2) | 0.719 |
| Bevacizumab receiveda | |||
| Yes | 32 (64.0) | 16.3 (1.9–33.8) | – |
| Noc | 18 (36.0) | 16.2 (0.5–29.4) | 0.878 |
P value indicates a significance level of <0.05.
aOne missing observation each for ECOG and chemotherapy regimen; two missing observations for bevacizumab.
bCalculated using log-scale values of SLD and CEA.
cOne patient received cetuximab.
IQR, interquartile range; ECOG, Eastern Cooperative Oncology Group performance status; SLD, sum of longest diameters; CEA, carcinoembryonic antigen.
tissue mutation identification and ctDNA quantification
At least one mutation was identified in the tumor tissue of 52/53 (98.1%) cases. Of these 52 cases, the identical mutation was detectable in the baseline plasma of 48 patients, giving a sensitivity of 92.3%. The somatic mutations identified in tumor tissue and plasma, and the respective ctDNA levels are shown in supplementary Table S4, available at Annals of Oncology online. For one of the four patients in whom ctDNA was not detectable, both primary and metastatic tumor tissue contained the KRAS G13D mutation, indicating that intertumoral heterogeneity did not explain the absence of circulating mutant KRAS. Clinical characteristics of the patients with undetectable ctDNA are provided in the supplementary Table S5, available at Annals of Oncology online.
pretreatment ctDNA, CEA, clinical characteristics and tumor burden
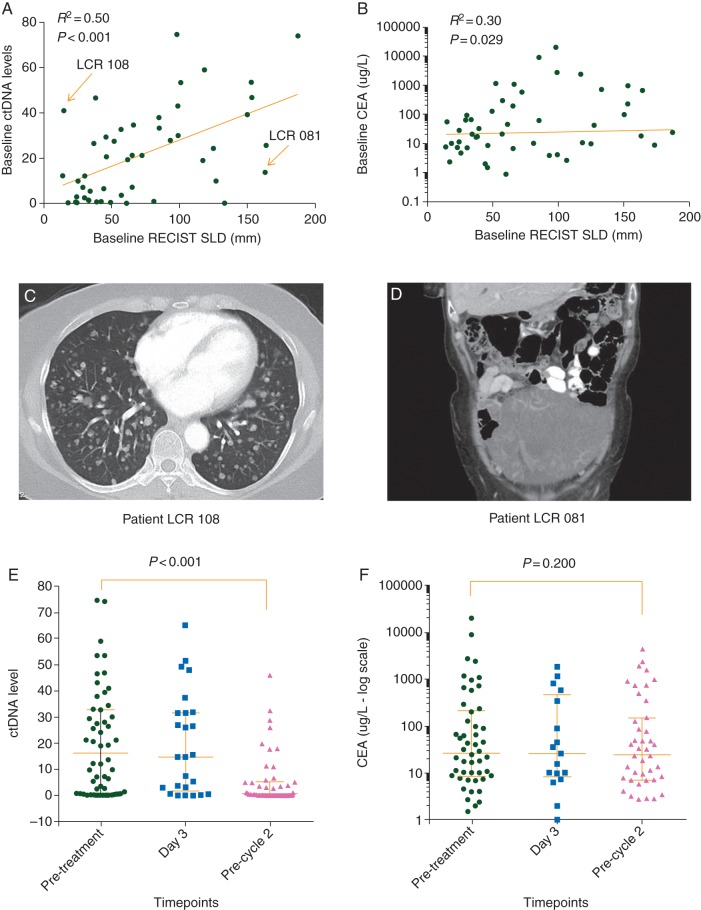
The median pretreatment ctDNA level was 16.2 [range 0–74.6; mean ± standard deviation (SD), 20.36 ± 20.34]. CEA was elevated at baseline in 42 of 50 patients (84%), with a median baseline level of 26.6 (range 0.9–19 850; mean ± SD, 817 ± 3064). Pretreatment ctDNA levels correlated more strongly with initial tumor burden as estimated from standard RECIST criteria (Spearman, r = 0.50, P < 0.001; Figure 2A) than pretreatment CEA (Spearman, r = 0.31, P = 0.029; Figure 2B). Figure 2C and D show the CT images from two outliers indicated by arrows in Figure 2A. LCR 108 had innumerable nonmeasurable small lung metastases with a commensurately high ctDNA, but a low bulk of disease as measured by RECIST. LCR 081 had a large cystic lesion, constituting bulky disease by RECIST criteria, but presumably containing only a modest amount of cancer cells as reflected in the relatively low ctDNA. The associations between pretreatment ctDNA and clinico-pathological variables are shown in Table 1.
Figure 2.
(A) Correlation between pretreatment ctDNA levels and the sum of longest tumor diameters by RECIST. Outliers emphasized in the text (LCR 108 and LCR 081) are indicated by arrows, with corresponding images shown in (C) and (D); (B) Correlation between pretreatment CEA levels and the sum of longest tumor diameters by RECIST; (C) CT images of a patient (LCR 108) with innumerable small lung metastases (majority are nonmeasurable by RECIST) and a high pretreatment ctDNA level (41); (D) CT images of a patient (LCR 081) with a large cystic pelvic mass but relatively low pretreatment ctDNA level (13.6); (E) Changes in ctDNA levels during cycle one of chemotherapy; (F) Changes in CEA levels during cycle one of chemotherapy. Day 3 = 3 days post-treatment, precycle 2 = before cycle 2 treatment.
changes in ctDNA and CEA during chemotherapy
No significant difference in ctDNA was observed between pretreatment and 3 days after chemotherapy (14.7 versus 16.4; P = 0.139). Intriguing results included four patients who had a spike in ctDNA 3 days after chemotherapy was initiated, which then rapidly declined (supplementary Figure S1, available at Annals of Oncology online). As three of these patients had an excellent response (≥20% reduction in tumor size at first restaging), this spike could reflect a rapid release of tumor DNA into the circulation from responsive tumors. Interestingly, two of these three patients also had a spike in their CEA levels at day 3.
Before cycle 2, ctDNA had decreased in 41 of the 48 patients, increased in three patients, and remained undetectable in four patients (supplementary Figure S1, available at Annals of Oncology online). The median ctDNA level before cycle 2 was significantly lower than the median pretreatment level (0.54 versus 16.24, P < 0.001; Figure 2E). Overall, no significant change in CEA levels was seen across the three time-points (median CEA: 3 days after therapy initiation versus pretreatment: 26.0 versus 26.6, P = 0.209; precycle 2 versus pretreatment: 24.0 versus 26.6, P = 0.198; Figure 2F).
ctDNA and tumor response as assessed by imaging
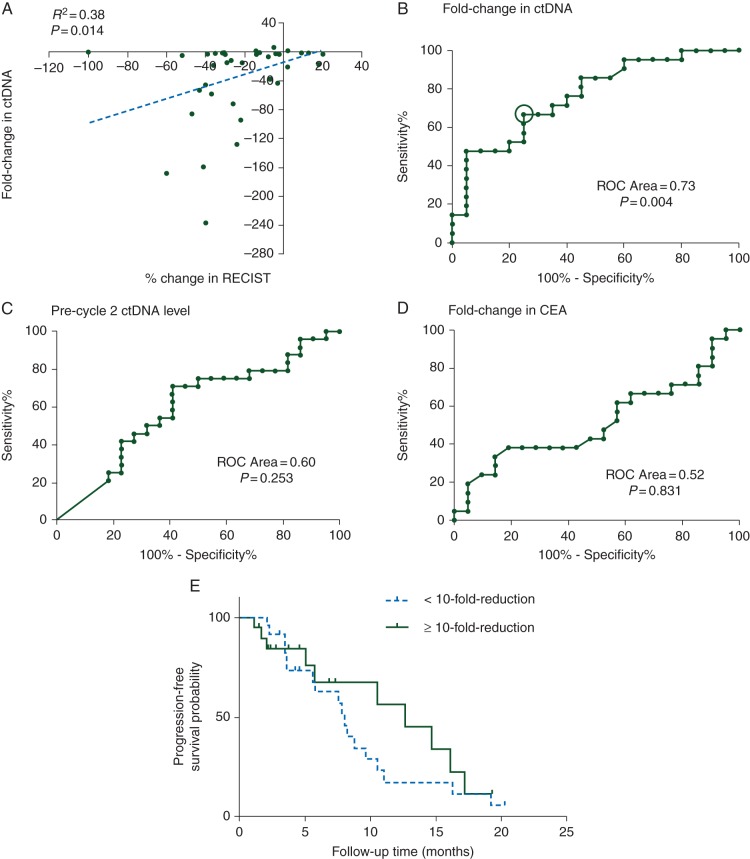
We next examined the correlation between ctDNA and disease status at first restaging in patients who had completed at least four cycles of chemotherapy. Of the 42 assessable patients, 22 (52%) responded (at least a 20% reduction in tumor size at first restaging imaging) and 20 (48%) did not respond (including a single patient with progressive disease). Figure 3A shows the positive correlation between percentage change in RECIST and fold change in ctDNA (Spearman r = 0.38, P = 0.014). The ROC plots in Figure 3B–D show that the fold reduction in ctDNA from pretreatment to before cycle 2 was a better predictor of radiologic response (ROC area = 0.73, P = 0.004) than the absolute level of ctDNA (ROC area = 0.6, P = 0.253). In contrast, the fold reduction in CEA levels at this early time point had no predictive value (ROC area = 0.52, P = 0.831) and was statistically inferior when compared with fold reduction in ctDNA (P = 0.017).
Figure 3.
(A) Correlation plot of percentage change in RECIST and fold change in ctDNA; (B–D) Receiver operating characteristic (ROC) analysis of fold change in ctDNA, ctDNA level after one cycle of chemotherapy and fold change in CEA, to predict tumor response; the circle in (B) indicates the location on the curve corresponding to a 10-fold change threshold (sensitivity 0.75, specificity 0.64); (E) Kaplan–Meier estimate for PFS in metastatic colorectal cancer patients with <10- and ≥10-fold reduction in ctDNA after one cycle of chemotherapy.
Overall, 14/19 (74%) patients who had a ≥10-fold reduction in ctDNA levels had a radiologic response measured at 8–10 weeks, while only 8/23 (35%) patients with lesser reductions in ctDNA levels responded [odds ratio = 5.25; 95% confidence interval (CI) 1.38–19.93; P = 0.016; Table 2]. The positive and negative predictive values of a ≥10-fold drop in ctDNA for response to chemotherapy were 65.2% and 73.7%, respectively. The changes in ctDNA displayed a wider dynamic range than changes in CEA with the median fold change in ctDNA before and after cycle one of chemotherapy being −5.7-fold (IQR: −1.8- to −48.9-fold) versus a median −1.1-fold for CEA (IQR: 1.2- to −1.3-fold).
Table 2.
Univariate association between tumor response at first imaging, CEA and ctDNA indices (N = 42)
| Variable | Tumor responsea |
P value | |
|---|---|---|---|
| No response | Response | ||
| Patients (N, %) | 20 (47.6) | 22 (52.4) | |
| CEA, pretreatmentb, median (IQR) | 46 (18–660) | 16.9 (10–89.5) | 0.171 |
| CEA after one cycle of chemotherapy, median (IQR) | 50.4 (19–520) | 11.3 (6.6–84.9) | 0.173 |
| Fold change in CEA, median (IQR) | 0.92 (0.78–1.11) | 0.91 (0.67–1.33) | 0.884 |
| ctDNA, pretreatment, median (IQR) | 12.9 (0.6–29.7) | 20.8 (2.7–34.5) | 0.339 |
| ctDNA after one cycle of chemotherapy, median (IQR) | 3.7 (0.3–6.6) | 0.4 (0.2–5.3) | 0.182 |
| Fold change in ctDNA (no. of patients, %) | 0.016 | ||
| <10-fold reduction | 15 (75.0) | 8 (36.4) | |
| ≥10-fold reduction | 5 (25.0) | 14 (63.6) | |
P value indicates a significance level of <0.05.
aDefined as ≥20% reduction in the sum of largest diameters according to standard RECIST criteria.
bCEA not collected before treatment in two patients.
prognostic significance of ctDNA for progression-free and overall survival
The optimal criterion for predicting response to therapy, as determined by the ROC plots, was ≥10-fold change in ctDNA after cycle 1 chemotherapy. Patients who met this criterion experienced a trend to longer PFS than patients with <10-fold drop in ctDNA (median PFS, 14.7 versus 8.1 months; HR = 1.87; 95% CI 0.62–5.61; Figure 3E). No significant relationship was found between fold change in ctDNA and OS.
discussion
Our study shows that ctDNA can be detected in a very high proportion of patients with untreated mCRC, with early changes in ctDNA during chemotherapy being associated with treatment response at first restaging. Serial ctDNA measurements could complement the information available from routine imaging-based assessments in evaluation of disease bulk and response to chemotherapeutic agents.
The evaluation of a modest panel of genes (15 in total) allowed us to identify mutations in 52 (98.1%) of the 53 cases examined. Matching mutations could be found in the plasma of 48 cases (90.6% of all 53 patients). We expect this level of detection might approach 100% as the technology continues to evolve and improve, including methods to identify somatic changes not identified by the current approach [10]. While CTCs are also a promising biomarker, they are only detectable in ∼50% of patients with mCRC [20, 21] and are found in some patients without cancer [22]. The narrow dynamic range of CTCs, with over 70% of patients having a count of <3 cells/7.5 ml blood [20, 21], also restricts the value of serial sampling over time.
Logically, the most informative way of using ctDNA in assessing treatment response is through serial assessments of this ratiometric measure, just as CEA or imaging is currently used. That very early changes in ctDNA levels (3 days after chemotherapy) were not predictive of response was not unexpected, given that reduction of tumor burden is typically a slow process. Notably, the change in ctDNA after completion of cycle 1 of chemotherapy was predictive of the response to treatment.
There are several limitations to our study. We evaluated only three time-points; additional later samples may have added value. Further, the thresholds for positivity that we selected based on ROC analyses need to be validated in an independent study. Additionally, given the modest sample size, our inferences from associations with P values marginally <0.05 should be viewed cautiously. However, our major finding, which changes in ctDNA measured 14–21 days after therapy was initiated, predict later tumor response as seen on imaging, seems robust (odds ratio = 5.25; 95% CI 1.38–19.93; P = 0.016).
New studies are demonstrating anticancer activity in patients that have already received three or more lines of therapy, but patients in routine practice often do not live long enough to enjoy the potential benefits of additional types of therapy [23]. If a patient's nonresponse to each treatment could be reliably assessed earlier, such as with serial ctDNA analysis, an earlier switch to an alternate therapy may be of benefit, minimizing the side-effects of the ineffective therapy and providing the opportunity for a more effective one.
Our data also suggest how ctDNA analysis could complement RECIST assessment. The notable outliers emphasized in Figure 2C and D illustrate two of the well-known limitations of RECIST measurements. A third case (supplementary Figure S2, available at Annals of Oncology online), with substantial necrosis evident on repeat imaging, and a commensurate large fall (>20-fold reduction) in ctDNA but stable disease by RECIST criteria, suggests another instance where ctDNA analysis may be superior to RECIST. However, the greatest impact of serial ctDNA measurement may be in patients with nonmeasurable disease by RECIST criteria, where a reliable measure of treatment response would greatly assist clinical decision making.
In summary, the most important findings of our study are that early changes in ctDNA are associated with later tumor responses as assessed by imaging, and that serial ctDNA measurement has significant potential to complement standard RECIST-based disease assessment. While these findings do not have immediate clinical impact, this study is an essential first step towards achieving this long-term goal. Serial ctDNA analysis should be incorporated into future clinical trials to provide a more robust assessment of this promising biomarker.
funding
This work was supported by Victorian Cancer Agency Translational Research Grant; Hilton Ludwig Cancer Prevention Initiative funded by the Conrad N. Hilton Foundation and the Ludwig Institute for Cancer Research; National Institutes of Health (grant numbers CA152753, CA57345 and CA62924); and Victorian Cancer Agency Clinical Research Fellowship (to JT).
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
We thank the Victorian Cancer Biobank for processing some of the plasma samples for this study.
references
- 1.Ferlay J, Soerjomataram I, Ervik M et al. GLOBOCAN 2012 v1.0—cancer incidence and mortality worldwide: IARC CancerBase No. 11. [Internet] Lyon, France: International Agency for Research on Cancer, 2013; http://globocan.iarc.fr (17 March 2015, date last accessed). [Google Scholar]
- 2.Sharma MR, Maitland ML, Ratain MJ. RECIST: no longer the sharpest tool in the oncology clinical trials toolbox. Cancer Res 2012; 72: 5145–5149. [DOI] [PubMed] [Google Scholar]
- 3.Bast RC Jr, Ravdin P, Hayes DF et al. 2000 update of recommendations for the use of tumor markers in breast and colorectal cancer: clinical practice guidelines of the American Society of Clinical Oncology. J Clin Oncol 2001; 19: 1865–1878. [DOI] [PubMed] [Google Scholar]
- 4.Sorbye H, Dahl O. Carcinoembryonic antigen surge in metastatic colorectal cancer patients responding to oxaliplatin combination chemotherapy: implications for tumor marker monitoring and guidelines. J Clin Oncol 2003; 21: 4466–4467. [DOI] [PubMed] [Google Scholar]
- 5.Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest 2005; 23: 338–351. [DOI] [PubMed] [Google Scholar]
- 6.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61: 759–767. [DOI] [PubMed] [Google Scholar]
- 7.Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci USA 1999; 96: 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diehl F, Schmidt K, Choti MA et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008; 14: 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dressman D, Yan H, Traverso G et al. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc Natl Acad Sci USA 2003; 100: 8817–8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bettegowda C, Sausen M, Leary RJ et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014; 6: 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SJ, Tsui DW, Murtaza M et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013; 368: 1199–1209. [DOI] [PubMed] [Google Scholar]
- 12.Newman AM, Bratman SV, To J et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014; 20: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki C, Blomqvist L, Sundin A et al. The initial change in tumor size predicts response and survival in patients with metastatic colorectal cancer treated with combination chemotherapy. Ann Oncol 2012; 23: 948–954. [DOI] [PubMed] [Google Scholar]
- 14.Mansmann UR, Laubender RP, Giessen CA et al. Validating the prognostic relevance of initial change in tumor size using a series of therapeutic regimens for patients with metastatic colorectal cancer (mCRC). J Clin Oncol 2012;30(suppl 4); abstr 580. [Google Scholar]
- 15.Diaz LA Jr, Williams RT, Wu J et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 2012; 486: 537–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinde I, Bettegowda C, Wang Y et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013; 5: 167ra164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinde I, Wu J, Papadopoulos N et al. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci USA 2011; 108: 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44: 837–845. [PubMed] [Google Scholar]
- 19.McShane LM, Hayes DF. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol 2012; 30: 4223–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen SJ, Punt CJ, Iannotti N et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol 2008; 26: 3213–3221. [DOI] [PubMed] [Google Scholar]
- 21.Tol J, Koopman M, Miller MC et al. Circulating tumour cells early predict progression-free and overall survival in advanced colorectal cancer patients treated with chemotherapy and targeted agents. Ann Oncol 2010; 21: 1006–1012. [DOI] [PubMed] [Google Scholar]
- 22.Allard WJ, Matera J, Miller MC et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004; 10: 6897–6904. [DOI] [PubMed] [Google Scholar]
- 23.Abrams TA, Meyer G, Moloney J et al. Patterns of chemotherapy (CT) use in a U.S.-wide population-based cohort of patients (pts) with metastatic colorectal cancer (mCRC). J Clin Oncol 2012; 30(suppl); abstr 3537. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.