Variations of genes involved in dormancy predict response, recurrence patterns and clinical outcome in patients with colorectal liver metastases undergoing perioperative bevacizumab-based chemotherapy and curative liver resection. The findings of this study may help identifying predictive and/or prognostic biomarkers and novel therapy targets.
Keywords: bevacizumab, colorectal liver metastases, dormancy, neoadjuvant chemotherapy, single-nucleotide polymorphisms
Abstract
Background
Tumor dormancy has been described as a state of hibernation. Dormancy can be switched to proliferation by different pathways, which may play a critical role in tumor recurrence. In this study, we investigated genetic variations within genes involved in tumor dormancy and their association with recurrence and outcome in patients with colorectal liver metastases (CLM) who underwent neoadjuvant bevacizumab-based chemotherapy.
Patients and methods
Genomic DNA was extracted from resected CLM (FFPE) from 149 patients. Single-nucleotide polymorphisms (SNPs) in 14 genes associated with dormancy were analyzed by direct Sanger DNA sequencing and evaluated for response, recurrence-free survival (RFS), overall survival (OS) and recurrence patterns.
Results
NME1 rs34214448 C>A was significantly associated with RFS in univariable analysis (P = 0.039) and with intrahepatic recurrence (P = 0.014). NOTCH3 rs1044009 T>C and CD44 rs8193 C>T showed a significant difference in 3-year OS rates (P = 0.004 and P = 0.042, respectively). With respect to radiological response, CD44 rs8193 C>T variant genotypes were associated with a significantly higher response rate (P = 0.033). Recursive partitioning analyses revealed that Dll4 rs12441495 C>G, NME1 rs34214448 C>A and NOTCH3 rs1044009 T>C were the dominant SNPs predicting histological response, RFS and OS, respectively.
Conclusion
Our data suggest that gene variations within genes involved in tumor dormancy pathways are associated with response and outcome in patients with resected CLM. These data may lead to new and more effective treatment strategies targeting tumor dormancy.
introduction
Dormancy is a state of hibernation, in which disseminated tumor cells (DTCs) remain until they are switched on to start growing, sometimes years or even decades after apparent cure [1, 2]. It is generally assumed that dormant cancer stem cells (CSCs) cause these late recurrences [3]. The microenvironment of CSCs appears to play a crucial role in the maintenance of dormancy. Three categories have been defined: (i) cellular dormancy of single cells caused by intrinsic and extrinsic factors, (ii) angiogenic dormancy of tumors maintained by a balance of cell proliferation and death due to insufficient blood vessels, and (iii) immune-mediated dormancy maintained by cytotoxic immunosurveillance [4].
Dormancy is playing a critical role in cancer recurrence because dormant cells are known to be refractory to adjuvant chemotherapy [5]. Recent data suggest that CSCs are disseminated very early, in contrast to the traditional doctrine that considers it to be a late step in cancer progression [6]. This may explain that recurrence may occur widespread in some distant organs (e.g. liver and lung), while these cells remain in a dormant state in other organs (e.g. bone marrow) [7].
Patients who underwent curative resection for liver-limited metastases are an interesting cohort to test whether genes involved in dormancy pathways are associated with clinical outcome since only 25% of them are cured [8]. Our hypothesis is that undetected dormant DTCs left behind may account for recurrences. After liver resection, liver regeneration with activation of multiple different pathways has been accused to be responsible for switching on dormant cancer cells left in the liver [9]. Understanding the molecular pathway leading to awakening of these dormant cells will be critical to identify new targets and development of novel treatment strategies.
We tested whether variations in a comprehensive panel of genes involved in dormancy pathways will predict clinical outcome in patients with colorectal liver metastases (CLM) treated with neoadjuvant bevacizumab-based chemotherapy. This panel included CSC- and angiogenesis-related genes involved in regulating cell proliferation through integrin signaling via the extracellular matrix (ITGA5, ITGB1), genes regulating integrin receptor activation (PLAU, PLAUR, EGF, EGFR), genes regulating angiogenic dormancy (DLL4, NOTCH3), genes suppressing metastasis-associated aspects, such as invasion and survival (NME1), and genes associated with CSCs (TCF7L2, AXIN2, CD44, LGR5, ALDH1A1) [10–14]. A better understanding of genes and pathways relevant for dormancy may help to identify predictive and/or prognostic biomarkers and potential targets for future therapies.
patients and methods
One hundred forty-nine consecutive patients [87 (58.4%) male, median age 62 years (range 30–80 years)] were enrolled (Table 1). Median follow-up was 3.9 years (range 0.02–7.7 years). Patients received 3 months of neoadjuvant and adjuvant fluoropyrimidine-based combination chemotherapy [oxaliplatin (N = 124), irinotecan (N = 18), both (N = 7)] including bevacizumab and curative liver resection (2005–2011). Last dose of bevacizumab was administered 5 weeks before surgery. Clinical data were provided from a prospectively maintained database. The study was approved by the local ethics committee.
Table 1.
Baseline characteristics (N = 149)
| N | % | |
|---|---|---|
| Age | ||
| Median (range) | 62 (30–80) | |
| <65 years | 90 | 60.4 |
| ≥65 years | 59 | 39.6 |
| Sex | ||
| Male | 87 | 58.4 |
| Female | 62 | 41.6 |
| Timing of metastases | ||
| Metachronous | 55 | 36.9 |
| Synchronous | 94 | 63.1 |
| Number of metastasesa | ||
| 1–2 | 88 | 59.5 |
| >2 | 60 | 40.5 |
| Size of metastasesa | ||
| 1–50 mm | 127 | 85.8 |
| >50 mm | 21 | 14.2 |
| Distribution of metastases | ||
| Unilobar | 76 | 51.0 |
| Bilobar | 73 | 49.0 |
| Primary tumor sitea | ||
| Right colon | 39 | 26.4 |
| Left colon | 60 | 40.5 |
| Rectum | 49 | 33.1 |
| KRAS status | ||
| Wild-type | 97 | 66.9 |
| Codons 12, 13 or 61 mutant | 48 | 33.1 |
aOne patient had missing information.
Patients with progressive disease (<5%) under neoadjuvant chemotherapy according to Response Evaluation Criteria In Solid Tumours (RECIST 1.1) did not undergo liver resection and were not enrolled [15].
According to the classification by Rubbia-Brandt et al., tumor regression grade (TRG) 1 and 2 were categorized as major histological response, TRG 3 as partial histological response and TRG 4 and 5 as no histological response [16].
To detect disease recurrence, clinical examination, computed tomography scans of thorax, liver and abdomen and blood tests including carcinoembryonic antigen were carried out every 3 months for 3 years, and then every 6 months.
single-nucleotide polymorphisms selection and genotyping
Genes (DLL4, NOTCH3, PLAU, PLAUR, ITGA5, ITGB1, EGF, EGFR, TCF7L2, AXIN2, CD44, LGR5, ALDH1A1, NME1) were selected because of their association with dormancy (supplementary Table S1, available at Annals of Oncology online) [10–14]. Candidate single-nucleotide polymorphisms (SNPs) were selected for analyses when having a minor allele frequency of ≥10% in Caucasians according to the Ensembl database and when having functional relevance for transcriptional regulation, protein coding or splicing according to Queen's University F-SNP and National Institute of Environmental Health Sciences SNP Function Prediction. Polymorphisms described in previous studies were also included [17–19].
The QIAamp DNAeasy Kit (Qiagen, Hilden, Germany) was used to extract genomic DNA from formalin-fixed paraffin-embedded CLM. DNA analyses were carried out by direct Sanger sequencing. Supplementary Table S1, available at Annals of Oncology online, lists primers used for polymerase chain reaction (PCR). To test primers for quality control, PCRs were carried out on known test DNA samples extracted from whole blood. The ABI Sequencing Scanner v1.0 (Applied Biosystems, Foster City, CA) was used to analyze DNA sequences. Investigators performing DNA analyses were blinded to patients' clinical data.
statistical analysis
The primary objective was to explore the predictive value of SNPs of 14 genes involved in the dormancy pathways. The primary end point was recurrence-free survival (RFS), and secondary end points were overall survival (OS) and radiological and histological responses. RFS was calculated as the interval from the day of liver resection to the first day of documented disease recurrence or death. In patients without recurrence and who were alive at the last follow-up, RFS was censored on the day of the last CT scan. OS was calculated as the time from liver resection to the date of death or censored on the date of the last follow-up. A 1-degree-freedom χ2 test was used to test if the allelic distribution of each dormancy polymorphism was within Hardy–Weinberg equilibrium. The associations between polymorphisms and RFS and OS were investigated using Kaplan–Meier estimation and log-rank tests in univariable analysis. A Cox proportional hazards regression model was used in multivariable analysis. The following baseline characteristics were adjusted in the multivariable analysis: age (<65 versus ≥65 years), number (1–2 versus >2), distribution (unilobar versus bilobar) and timing of resectable liver metastases (metachronous versus synchronous) and KRAS mutation status because they were associated with RFS or OS at a 0.10 significance level. To assess relationships between polymorphisms and response, χ2 tests were used in univariable analyses and logistic regression model controlling for potential predictive variables were used in multivariable analyses. Recursive partitioning analyses were used to explore and identify profiles of polymorphism associated with RFS, OS and radiological and histological responses.
A mixture cure model was used to assess the cure rate by individual polymorphisms [20]. The recurrence patterns (intrahepatic only or extrahepatic) were assessed using cumulative incidence of recurrence in the competing risks model.
Leave-one-out cross validation, an internal validation method, was carried out to assess the process of polymorphism selection to predict clinical outcomes.
All analyses were carried out using SAS/STAT 12.3 (SAS Institute, Cary, NC), a SAS macro (%pspmcm), along with rpart and cmprsk functions in R package. All analyses were carried out with two-sided tests at a significance level of 0.05. P values were not adjusted for multiple hypothesis testing.
results
Hardy–Weinberg equilibrium
All SNPs were in Hardy–Weinberg equilibrium except PLAUR rs2302524 A>G and ITGB5 rs7306692 C>T.
gene variants and recurrence-free survival, overall survival and response
The A/A genotype of NME1 rs34214448 C>A was associated with significantly shorter RFS compared with other genotypes [any C 12.8, A/A 8.3 months; hazard ratio (HR) 1.72 (1.02, 2.91), P = 0.039]. This difference did not remain significant in multivariable analysis (HR 1.58 (0.84, 2.99), P = 0.16) (supplementary Figure S1, available at Annals of Oncology online). For NOTCH3 rs1044009 T>C, variant genotypes were associated with a significantly lower 3-year OS rate (T/T 73%, any C 55%; univariable HR 2.22 (1.19, 4.16), P = 0.010; multivariable HR 2.62 (1.37, 5.04), P = 0.004) (supplementary Figure S2, available at Annals of Oncology online). For CD44 rs8193 C>T, variant genotypes were associated with nonsignificantly higher 3-year OS rate in univariable analysis [C/C 60%, any T 74%; HR 0.63 (0.35, 1.14); P = 0.12], which became significant in multivariable analysis [HR 0.53 (0.29, 0.98); P = 0.042] (supplementary Figure S3, available at Annals of Oncology online). Data on RFS and OS are presented in supplementary Table S2, available at Annals of Oncology online. In keeping with the OS data, CD44 rs8193 C>T variant genotypes were associated with a significantly higher radiological response rate compared with the C/C genotype (C/C 72%, any T 88%, P = 0.033). None of the SNPs was associated with histological response (supplementary Table S3, available at Annals of Oncology online).
gene variants and recurrence patterns
Patients with an A/A genotype of NME1 rs34214448 C>A had a significantly higher 2-year intrahepatic recurrence rate than those harboring any C (27% (±11%) versus 8% (±3%), P = 0.014). Other polymorphisms were not associated with intra- or extrahepatic recurrence (data not shown).
recursive partitioning for recurrence-free survival, overall survival and histological response
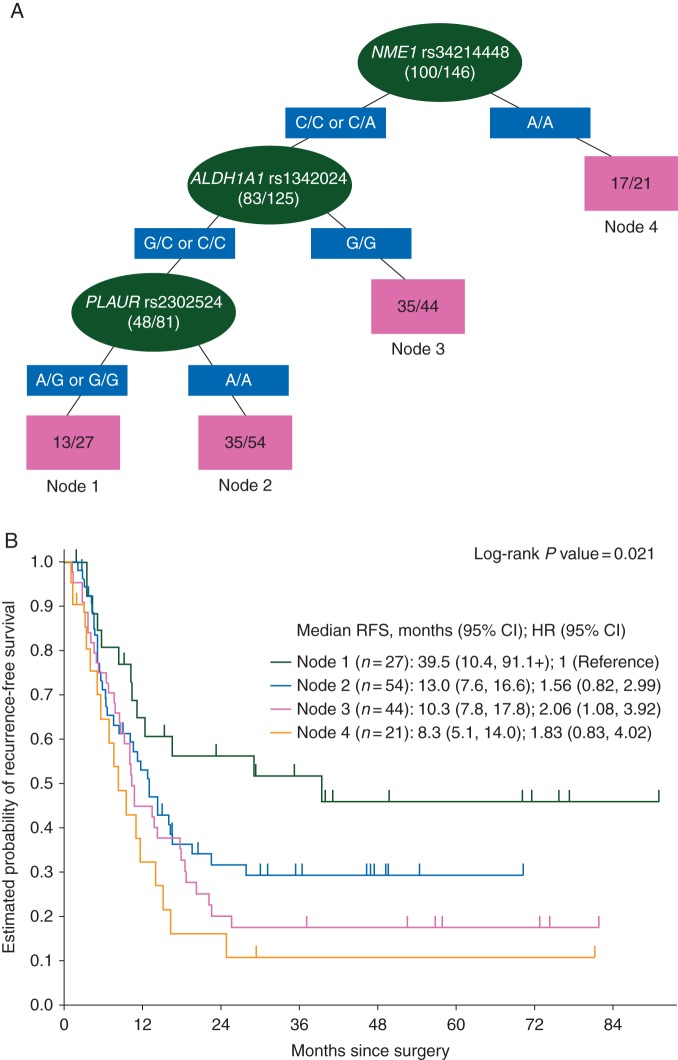
NME1 rs34214448 C>A was the dominant SNP to predict RFS. ALDH1A1 rs1342024 G>C and PLAUR rs2302524 A>G were predictive in subgroups (Figure 1). NOTCH3 rs1044099 T>C was the dominant SNP to predict OS and PLAU rs4065 A>G, CD44 rs8193 C>T and ITGA5 rs7306692 C>T in subgroups (supplementary Figure S4, available at Annals of Oncology online). DLL4 rs12441495 C>G was the dominant SNP to predict histological response, and EGF rs4444903 T>C, DLL4 rs4923888 G>C and ITGA5 rs7306692 C>T in subgroups (supplementary Figure S5, available at Annals of Oncology online). None of the SNPs was associated with radiological response.
Figure 1.
(A) Recursive partitioning tree for RFS. Light grey (green online) ovals represent intermediate subgroups; light grey (pink online) squares represent terminal nodes. Dark grey (blue online) rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who relapsed/total patients with that node. Node 1 represents low-risk patients. Node 2 and 3 represent intermediate risk patients. Node 4 represents high-risk patients. (B) Kaplan–Meier curves for terminal nodes.
internal validation
Among 149 sets, all P values were <0.05 for both univariable and multivariable OS analyses for NOTCH3 rs1044099 T>C. For CD44 rs8193 C>T, in none of 149 sets, P value was <0.05 in univariable OS analysis; in 122 of 149 sets, P value was <0.05 in multivariable OS analysis. For NME1 rs34214448 C>A, in 139 of 149 datasets, P value was <0.05 in univariable RFS analysis, but in none of them in multivariable analysis. For CD44 rs8193 C>T, in 136 of 149 datasets, P value was <0.05 for radiological response. For NME1 rs34214448 C>A, 144 of 149 LOOCV datasets showed a P value of <0.05 for intrahepatic recurrence.
discussion
The goal of liver resection in patients with CLM is cure; however, the majority of patients develop disease recurrence within the liver. We tested whether variations of dormancy-related genes in a unique cohort of patients who underwent neoadjuvant bevacizumab-based chemotherapy and curative liver resection will predict tumor recurrence.
Our data showed for the first time that SNPs in cellular and angiogenic dormancy-related (NOTCH3 and NME1) and CSC-related genes (CD44) are associated with response, recurrence patterns and clinical outcome in patients with CLM. These findings suggest that SNPs in these genes may be predictive biomarkers to identify patients who benefit most from these resections. These findings also identify potential novel targets and hopefully lead to new drug development strategies, which might increase cure rates.
NME1 rs34214448 C>A was associated with RFS and intrahepatic recurrence. Although the exact mechanism of action of the metastasis suppressor NME1 (NME/NM23 nucleoside diphosphate kinase 1) to promote dormancy remains elusive, various roles have been described. NME1 may induce dormancy through inhibition of Ras/Raf/MAPK (mitogen-activated protein kinase) signaling via phosphorylation and thereby inactivation of KSR1 (kinase suppressor of Ras 1), a scaffold protein in this pathway [12]. Furthermore, NME1 negatively regulates LPAR1 (lysophosphatidic acid receptor 1), which is an activator of the MAPK ERK (extracellular signal regulated kinase) 1/2 [21]. This suppression of LPAR1 leads to a reduction ERK1/2 signaling and to activation of stress-activated kinase p38 signaling, which has been described to facilitate dormancy [22]. Another mechanism of ERK/p38 signaling is modulation of angiogenic dormancy via proangiogenic vascular endothelial growth factor (VEGF) and antiangiogenic thrombospondin [23]. These data suggest that mechanisms of cellular and angiogenic dormancy are overlapping and that targeting one pathway may have effects on multiple categories of dormancy. The finding that NME1 rs34214448 C>A was associated with only intrahepatic recurrence is of special interest as these patients may be candidates for repeat resection. However, one has to be aware of the fact that statistical significance for RFS was lost in multivariable analysis.
In addition, NOTCH3 rs1044009 T>C was associated with OS. NOTCH3 and its ligand DLL4 (delta-like 4), who are important regulators of CSCs, have been shown to be involved in escape to dormancy by upregulation of DLL4 on endothelial cells [11, 24]. This escape was facilitated by activation of antiapoptotic and survival-promoting nuclear factor kappa-light-chain-enhancer of activated B cells signaling downstream of NOTCH3 [11, 25]. These data demonstrate that the vasculature is not only mediating angiogenic dormancy, but also modulating cellular dormancy via direct NOTCH3/DLL4-mediated contact of endothelial cells and cancer cells. Interestingly, NOTCH3 signaling may be influenced by the anti-VEGF treatment in our patient population as DLL4 expression is upregulated by VEGF-A [26]. These data suggest that bevacizumab may also modulate dormancy through DLL4/NOTCH4 signaling. In summary, the DLL4/NOTCH3 axis appears to be an attractive target to block escape from dormancy and potentially prevent recurrence, and NOTCH inhibitors are in early clinical trials.
Our data also demonstrated that CD44 rs8193 C>T C/C was associated with inferior radiological response, OS and a trend toward shorter RFS. These data are consistent with a previous study that demonstrated an association of CD44 rs8193 C/C genotype with shorter time-to-recurrence in stage II and III patients receiving adjuvant treatment [14]. CD44 has been described as CSC marker in colorectal cancer (CRC) that drives cell proliferation via phosphatidylinositol-4,5-bisphosphate 3-kinase/AKT (protein kinase B) signaling and is a target gene for the Wnt pathway [27]. CSCs show abilities of self-renewal, chemoresistance and are considered the true tumor dormancy. The C/C genotype of CD44 rs8193 C>T may be associated with higher transcriptional activity of this gene as predicted by F-SNP [18] consistent with the poor prognosis in these patients.
The identification of variations in genes involved in promoting dormancy provides further insights into the facilitation of dormancy in the clinical impact. Thereby, novel treatment targets may be identified to develop new treatment strategies. A potential treatment strategy could be keeping cancer cells in a dormant state to avoid regrowth of metastases after curative resection [28]. Some authors favor this concept as the concept of awakening dormant cells to make them susceptible to cytotoxic treatment harbors potential risks as they may activate alternative pathways to increase chemoresistance [4, 29].
Recursive partitioning showed which genes were relevant in subgroups and that the resulting genetic profile was associated with substantial differences in survival. These analyses revealed that different SNPs predict histological response, RFS and OS. The observation that some biomarkers may be predictive for one clinical end point, while others may be predictive for other end points may be owed to the different underlying biological mechanisms.
A limitation of this study is the absence of a validation cohort as the investigated cohort is very unique; however, the results appear to be valid due to their consistency in various clinical end points. Another limitation is the lack of a preclinical rationale of investigating dormancy in CRC as dormancy data come predominantly from breast and prostate cancers. However, the fact that dormancy is closely linked to CSCs that facilitate recurrence make it worthwhile to investigate these pathways in resected CLM.
In summary, this study shows that variations in genes and pathways involved in the regulation of dormancy are associated with treatment efficacy and clinical outcome in patients with CLM. These insights may help to identify predictive and/or prognostic biomarkers to guide treatment strategies. These biomarkers could help to exclude patients who do not benefit from this multidisciplinary treatment approach, which is complex and bears potential complications. Moreover, a deeper knowledge of the dormancy implications in cancer recurrence may contribute to find new dormancy-related treatment targets to improve the therapy of CRC.
funding
This work was supported by the National Institute of Health (P30CA014089 to H-JL); and the Gloria Borges Wunderglo Foundation. SS is a recipient of an Erwin Schrödinger fellowship of the Austrian Science Fund (J3501-B13). AS is a recipient of a Rio Hortega Research Grant from the Insituto de Salud Carlos III (CM11/00102). SS is supported by a postdoctoral fellowship from the German Cancer Aid (Mildred-Scheel Foundation).
disclosure
TG receives financial support by Roche, Merck-Serono, Sanofi-Aventis, Bayer and Amgen, and is member of the advisory boards of Roche, Merck-Serono and Sanofi-Aventis. H-JL is member of the advisory board of Genentech, Merck KG and BMS. All remaining authors have declared no conflict of interest.
Supplementary Material
references
- 1.Paez D, Labonte MJ, Bohanes P et al. Cancer dormancy: a model of early dissemination and late cancer recurrence. Clin Cancer Res 2012; 18: 645–653. [DOI] [PubMed] [Google Scholar]
- 2.Karrison TG, Ferguson DJ, Meier P. Dormancy of mammary carcinoma after mastectomy. J Natl Cancer Inst 1999; 91: 80–85. [DOI] [PubMed] [Google Scholar]
- 3.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell 2013; 155: 750–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer 2014; 14: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naumov GN, Townson JL, MacDonald IC et al. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat 2003; 82: 199–206. [DOI] [PubMed] [Google Scholar]
- 6.Husemann Y, Geigl JB, Schubert F et al. Systemic spread is an early step in breast cancer. Cancer Cell 2008; 13: 58–68. [DOI] [PubMed] [Google Scholar]
- 7.Heiss MM, Simon EH, Beyer BC et al. Minimal residual disease in gastric cancer: evidence of an independent prognostic relevance of urokinase receptor expression by disseminated tumor cells in the bone marrow. J Clin Oncol 2002; 20: 2005–2016. [DOI] [PubMed] [Google Scholar]
- 8.Nordlinger B, Sorbye H, Glimelius B et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet 2008; 371: 1007–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panis Y, Ribeiro J, Chretien Y, Nordlinger B. Dormant liver metastases: an experimental study. Br J Surg 1992; 79: 221–223. [DOI] [PubMed] [Google Scholar]
- 10.Aguirre Ghiso JA, Kovalski K, Ossowski L. Tumor dormancy induced by downregulation of urokinase receptor in human carcinoma involves integrin and MAPK signaling. J Cell Biol 1999; 147: 89–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Indraccolo S, Minuzzo S, Masiero M et al. Cross-talk between tumor and endothelial cells involving the Notch3-Dll4 interaction marks escape from tumor dormancy. Cancer Res 2009; 69: 1314–1323. [DOI] [PubMed] [Google Scholar]
- 12.Hartsough MT, Morrison DK, Salerno M et al. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem 2002; 277: 32389–32399. [DOI] [PubMed] [Google Scholar]
- 13.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature 2005; 434: 843–850. [DOI] [PubMed] [Google Scholar]
- 14.Gerger A, Zhang W, Yang D et al. Common cancer stem cell gene variants predict colon cancer recurrence. Clin Cancer Res 2011; 17: 6934–6943. [DOI] [PubMed] [Google Scholar]
- 15.Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009; 45: 228–247. [DOI] [PubMed] [Google Scholar]
- 16.Rubbia-Brandt L, Giostra E, Brezault C et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol 2007; 18: 299–304. [DOI] [PubMed] [Google Scholar]
- 17.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 2009; 37: W600–W605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res 2008; 36: D820–D824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P, Amode MR, Barrell D et al. Ensembl 2014. Nucleic Acids Res 2014; 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corbiere F, Joly P. A SAS macro for parametric and semiparametric mixture cure models. Comput Methods Programs Biomed 2007; 85: 173–180. [DOI] [PubMed] [Google Scholar]
- 21.Horak CE, Mendoza A, Vega-Valle E et al. Nm23-H1 suppresses metastasis by inhibiting expression of the lysophosphatidic acid receptor EDG2. Cancer Res 2007; 67: 11751–11759. [DOI] [PubMed] [Google Scholar]
- 22.Marshall JC, Collins JW, Nakayama J et al. Effect of inhibition of the lysophosphatidic acid receptor 1 on metastasis and metastatic dormancy in breast cancer. J Natl Cancer Inst 2012; 104: 1306–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007; 7: 834–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashley N. Regulation of intestinal cancer stem cells. Cancer Lett 2013; 338: 120–126. [DOI] [PubMed] [Google Scholar]
- 25.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature 2006; 441: 431–436. [DOI] [PubMed] [Google Scholar]
- 26.Ridgway J, Zhang G, Wu Y et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature 2006; 444: 1083–1087. [DOI] [PubMed] [Google Scholar]
- 27.Du L, Wang H, He L et al. CD44 is of functional importance for colorectal cancer stem cells. Clin Cancer Res 2008; 14: 6751–6760. [DOI] [PubMed] [Google Scholar]
- 28.Sosa MS, Avivar-Valderas A, Bragado P et al. ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clin Cancer Res 2011; 17: 5850–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Essers MA, Trumpp A. Targeting leukemic stem cells by breaking their dormancy. Mol Oncol 2010; 4: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.