Abstract
Background
Traditional diagnostic technology with tumor biomarkers is inefficient, expensive and requires a large number of serum samples. The purpose of this study was to construct human lung cancer protein chips with new lung cancer biomarkers screened by the T7-phage display library, and improve the early diagnosis rate of lung cancer.
Methods
A T7-phage cDNA display library was constructed of fresh samples from 30 lung cancer patients. With biopanning and high-throughput screening, we gained the immunogenic phage clones from the cDNA library. The insert of selected phage was blasted at GeneBank for alignment to find the exact or the most similar known genes. Protein chips were then constructed and used to assay their expression level in lung cancer serum from 217 cases of lung cancer groups:80 cases of benign lung disease and 220 healthy controls.
Results
After four rounds of Biopanning and two rounds of enzyme-linked immunosorbent assay, 12 phage monoclonal samples were selected from 2880 phage monoclonal samples. After blasting at GeneBank, six similar genes were used to construct diagnostic protein chips. The protein chips were then used to assay expression level in lung cancer serum. The expression level of six genes in lung cancer groups was significantly higher than those in the other two groups (P < 0.05).
Conclusions
In this study, we successfully constructed diagnostic protein chips with biomarkers selected from the lung cancer T7-phage cDNA library, which can be used for the early screening of lung cancer patients.
Keywords: Biomarkers, lung neoplasms, protein microarray, T7 phage
Introduction
Lung cancer has recently become the leading cause of cancer-related mortality worldwide.1 Clinical studies have shown that the survival rate is related to stage.2 Unfortunately, because of the trait of rapid metastasis, most patients are only diagnosed at advanced stage.3,4 Despite improved modern diagnosis and treatment strategies, the five-year survival rate has not correspondingly improved.5 Therefore, finding a way to screen lung cancer patients from healthy groups is of great significance. Identifying tumor biomarkers is useful and convenient for earlier detection of lung cancer.6 Many lung cancer tumor biomarkers have been reported, including carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA 21-1), and neuron-specific enolase (NSE). However, because of their limited sensitivity and specificity, and false-positive rate in normal populations, we cannot screen cancer patients with one kind of tumor biomarker.7 At present, several kinds of biomarkers are frequently used in conjunction to screen cancer patients, but false-positive cases remain. Moreover, the traditional test method with tumor biomarkers is inefficient, requiring a large number of serum samples. Thus, it is necessary to find new biomarkers and a minimally invasive, simple, and efficient method, which can be used for lung cancer screening of the high-risk population.
Protein microarray is becoming a vital tool for large scale and high-throughput miniaturized protein analysis and represents great potential in many aspects, including the identification of protein-protein interactions, protein-phospholipid interactions, small molecule targets, and substrates of protein kinases.8,9 Compared with the traditional test method of tumor markers, protein microarrays have more advantages, such as requiring low samples and miniaturization. In our study, we aimed to determine new markers in order to construct protein chips with high sensitivity and specificity that may be applied in clinics.
Patients and methods
Patients
Lung cancer samples were obtained from 30 patients who were not treated with chemotherapy and/or radiotherapy before initial surgical treatment at the Department of Respiratory and Thoracic Tumor Research Institute of the Affiliated Hospital of Qingdao University from March 2013 through March 2014. Pathohistological examination showed no evidence of remaining tumor. Clinical stage was classified using the Union for International Cancer Control Tumor Node Metastasis Atlas. Parts of the surgically resected tumors were immediately shock-frozen and stored at −70°C until used for lung-cancer T7 phage cDNA library construction. A total of 517 serum samples, including 217 lung cancer patients were collected: 80 patients with benign lung disease and 220 healthy controls. Participants had been diagnosed by pathohistological examination, did not have any other malignant or autoimmune disease, and were matched for age, gender, and smoking history. All patients gave informed consent. Two millilitres of limosis blood was collected and stored at 4°C within an hour. The blood was then centrifuged for 20 minutes at 4000 rpm, distributed into 100 μl aliquots, and stored at −80°C until used. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the institutional review board of Qingdao University.
Ribonucleic acid extraction and construction of T7 phage display cDNA library
Total ribonucleic acid (RNA) was extracted from 30 fresh samples from lung cancer patients using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Messenger (m)RNA was purified with the Oligotex mRNA purification kit (Qiagen, Mainz, Germany). We generated an orientation-directed cDNA library using a random priming method. The cDNA library was amplified by frame-shift polymerase chain reaction (PCR) for 30 cycles with three forward and three reverse primers (Forward primers: 5′-AACTAGATAAGAATAGCGGCCGCGCGCCGCGACC-3′, 5′-AACTAGATAAGAATAGCGGCCGCAGCGCCGCGACC-3′, 5′-AACTAGATAAGAATAGCGGCCGCAAGCGCCGCGACC-3′; Reverse primers: 5′-AATGATGTCGAGTAGGCCGGCCTCC-3′, 5′-AATGATCTCGAGTGGCCGGCCTCC-3′, 5′-AATGATCTCGAGTAGGCCGGCCTCC-3′). The PCR product between 300/bp-1.5 kb was purified on 2% agarose gel, digested with NotI and XhoI, and ligated into T7Bio phage vector10 at NotI and XhoI. After packaging with T7 phage package extract (Novagen, Merck, KGaA, Germany), the original phage library titer was ∼2.0 × 108 pfu. The library was amplified once on BLT5615 bacterial plates, bound to streptavidin-coated paramagnetic beads (Promega, Madison, WI, USA), and eluted with 3C protease (10 units in 400 μl) at 4°C for 16 hours to generate phage libraries. All procedures for T7 phage display have been described previously.10,11
Phage display biopanning and high-throughput screening by enzyme-linked immunosorbent assay
The T7 phage display (1200 μl, diluted with 10% bovine serum albumin [BSA] 1:40) was added to the healthy control serum (20 μl), incubated for two hours at 4°C, and centrifuged for three minutes at 1000 rpm/minute. The supernatant was added to the lung-cancer group serum (30 μl), incubated at 4°C overnight, and centrifuged for three minutes at 1000 rpm/min. The supernatant was discarded. The precipitation was added to 1% DSD (100 μl), incubated for 10 minutes at room temperature, and centrifuged for 10 minutes at 500 rpm/minute. From the supernatant, we extracted the phage clones that bind specifically to serum antibodies associated with lung cancer. These phages were eluted, amplified in Escherichia coli BLT5615, and were input for the next round of selection. The titer of the phages was calculated after each round of selection. After four rounds of selection, the inserts of the selected phages were amplified by PCR. The insert fragments >200bp were then submitted for high-throughput screening by enzyme-linked immunosorbent assay (ELISA).
ELISA plates were coated with T7 tailer antibody (diluted by phosphate buffered saline [PBS] [pH 7.4] 1:1000) at 4°C overnight. After washing three times with PBS Triton (T), 200 μl of blocking solution (2% BSA in PBS) was added, and the plates were incubated for two hours at room temperature and subsequently washed four times with PBST. Enriched phages, at 100 μl per well (diluted by 2% BSA 1:5), were then added to two wells. The plates were incubated for 90 minutes and then washed with PBST. Then, 100 μl per well of lung cancer group serum (diluted by 1% BSA) and 100 μl per of healthy control serum (diluted by 1% BSA) were added to the two wells as previously described. The plates were incubated for one hour and then washed four times with PBST. Horseradish peroxidase goat anti-human immunoglobulin G (IgG) was added, and the plates were incubated for one hour at room temperature. After washing with PBST four times, the chromogenic reagent was added. ELISA was ceased with a stop buffer (30 μl) after incubation for 20 minutes at 37°C. A negative control was given to each plate. Absorbances were measured at 450 nm wavelengths. The positive samples were screened and used as input for next round of ELISA. The twice-positive samples were selected and the insert of these phages was amplified by PCR and then sequenced. The gene alignment was then blasted at GeneBank to find exact or similar genes. Products of selected genes were used to construct protein chips.
Diagnostic chip design
The cDNA fragments of selected phages were inserted into the XhoI-KpnI sites of a green fluorescent protein (GFP) pET-14b plasmid to express the antigens on BLT5615 bacterial plates.10 Antigen-protein level and quality was assessed by Lowry protein assay and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Antigen-protein was diluted to 25.6 mol/mL (0.796-1.87 mg/mL) in PBS. Each antigen-protein solution was printed on FAST slides (Truelab, Shanghai, China) as an array of seven lines, each with five 200 μm spots of 10 μL using an array spotter SPBIO2000 (Hitachi Solution, Tokyo, Japan). The designed chips were incubated overnight in the dark at room temperature and then washed three times with PBS. Protein chips were stored in PBS at 4°C in the dark and used immediately or within six months.
Antigen-bound chips were washed with PBS for five minutes, placed in a 24-well plate, and washed with tris-buffered saline Tween 20 (TBS-T, 0.5 mL TBS, 0.1% Tween-20) for 10 minutes. Chips were transferred to PCR tubes containing 200 μL of SDS/2-mercaptoethanol (2ME) sample buffer (2% SDS in 50 mM Tris-HCl [pH 6.8] with 5% 2-mercaptoethanol, 10% glycerol, and 0.0012% Bromophenol Blue), heated at 95°C for five minutes, washed five times in TBS-T, and incubated for one hour in blocking solution (TBS-T containing 5% BSA). Chips were incubated in serum prepared previously (217 lung cancer patients, 80 benign lung disease patients, 220 healthy controls) diluted five-fold in 50 μl blocking solution for 1.5 hours of gentle shocking, and then washed five times in TBS-T. Chips incubated for 30 minutes with 1:500 Alexa Flour 633 (Alexa633)-conjugated anti-human IgG antibody (Invitrogen, Life Technologies, Carlsbad, CA, USA), followed by a further 30 minutes, adding the same volume of 1:500 Alexa Fluor 555 (Alexa555)-conjugated anti-mouse IgG antibody (Invitrogen). The chips were then washed five times with TBS-T and once with PBS. All chips were placed on a slide holder, mounted with PBS. The chips were then observed by BZ-9000 fluorescent microscope (Keyence, Osaka, Japan) at 90% laser power for both Alexa555 and Alexa633 fluorescence using Cy3 and Cy5 detection channels, respectively. The Alexa633/555 ratio calculation of each spot was used as positive criteria.
Statistical analysis
Statistical analysis was performed with SPSS 10.0 statistical software(SSPS Inc., Chicago, IL, USA). All of the experimental data was analyzed by chi-square test or variance analysis. Significance was determined when P < 0.05.
Results
Screening of lung-cancer-associated phages
The titer of the original phage library was 2.0 × 108.After four rounds of biopanning, the titer of the phage library was 4.2 × 109, 4.0 × 1010, 2.0 × 1011, and 4.6 × 1012, respectively; an improvement on the original library. The results showed evidence that phages associated with lung cancer had been enriched. After four rounds of biopanning, we selected 2880 phages whose insert fragments were >200 bp. These phages reacted with antibodies in the serum of lung cancer patients but did not react in the serum of healthy controls. Therefore, we believe these phages could express lung cancer associated antigens. To ensure exact tumor biomarkers, these phages were submitted for two rounds of ELISA high-throughput screening. The index value of each phage tested by ELISA was defined by the following formula: index = OD of sample group/OD of negative control. The results were considered positive if the index ≥ 2.0. If the OD of the negative control < 0.05, the OD of the negative control = 0.05; if not, it is the actual measured value. The phage was selected if it was positive in the lung cancer group and negative in the healthy group. If the phage was positive after two rounds of ELISA, we determined that the antigens expressed in these phages also had a high express level in lung cancer tissue. After two rounds of ELISA, we selected 12 positive phages (Table 1). Inserts of these 12 phages were sequenced and then blasted at GeneBank. Finally, we found that these 12 phages contained six exact or similar genes: olfactomedin 1 (OLFM1), A-kinase anchor protein-4 (AKAP-4), transthyretins (TTRs), vacuolar inhibitor of fructosidase 1 (VIF-1), cytokeratin-18 (CK-18), and squalene epoxidase (SQLE). Previous studies have shown that these genes are related to cancer.12–16
Table 1.
The selected phages by twice-positive ELISA
| Group | OD value | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 290 | 302 | 315 | 376 | 815 | 897 | 1012 | 1206 | 1365 | 1524 | 1874 | 1942 | |
| A: Positive phages of the first round of ELISA | ||||||||||||
| Lung cancer group | 0.176 | 0.115 | 0.142 | 0.115 | 0.165 | 0.198 | 0.220 | 0.154 | 0.165 | 0.203 | 0.146 | 0.157 |
| Healthy control | 0.086 | 0.074 | 0.054 | 0.049 | 0.075 | 0.095 | 0.102 | 0.083 | 0.079 | 0.101 | 0.081 | 0.079 |
| Twice negative value | 0.120 | 0.100 | 0.100 | 0.100 | 0.100 | 0.100 | 0.133 | 0.100 | 0.108 | 0.132 | 0.100 | 0.100 |
| B: Positive phages of the second round of ELISA | ||||||||||||
| Lung cancer group | 0.180 | 0.106 | 0.153 | 0.119 | 0.167 | 0.185 | 0.197 | 0.152 | 0.171 | 0.186 | 0.148 | 0.149 |
| Healthy control | 0.092 | 0.075 | 0.052 | 0.054 | 0.072 | 0.091 | 0.102 | 0.083 | 0.081 | 0.093 | 0.085 | 0.073 |
| Twice negative value | 0.114 | 0.100 | 0.102 | 0.102 | 0.100 | 0.105 | 0.108 | 0.100 | 0.100 | 0.100 | 0.100 | 0.100 |
ELISA, enzyme-linked immunosorbent assay.
Detection of protein chips among three groups
All proteins were constructed to contain a GFP. A GFP construct with no fusion partner (31 kd) was used as a control. The expression of recombinant GFP-tagged antigenic proteins was confirmed by SDS-PAGE after purification using Ni-affinity gel (Fig 1). The protein amount in each spot is essential for quantification of antibodies that specifically bind to the immobilized antigenic proteins. The amount of antigenic proteins on each chip was verified by measuring GFP fluorescence with a fluorescent microscope (Fig 2). Denaturing antigenic proteins on the chip by heating in SDS/2ME solution eliminated this aggregation. To check the effect of the heat-denaturing step in this protocol, GFP fluorescence was checked at each step by fluorescent microscopy. GFP fluorescence disappeared after treatment with SDS/2ME. In addition, Alexa555 fluorescence from the GFP tag increased for all protein spots. The Alexa633/555 ratios of the healthy controls and benign lung disease groups were below 0.8. However, the value of the lung cancer group was very high, above 0.8. Thus, the Alexa633/555 ratio >0.8 is the positive criteria of diagnostic chips.
Figure 1.
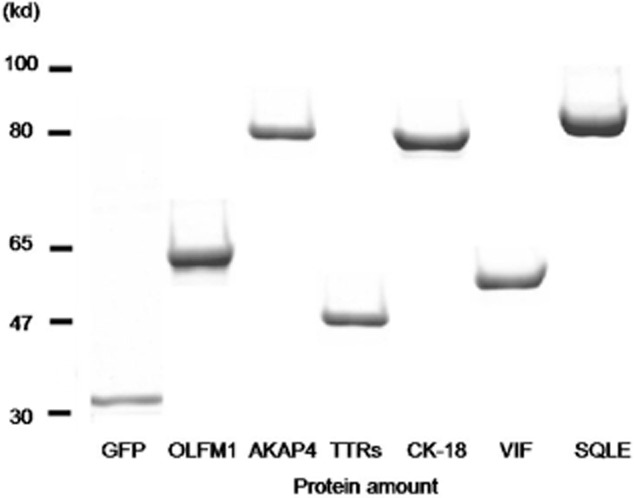
The six recombinant proteins confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Figure 2.
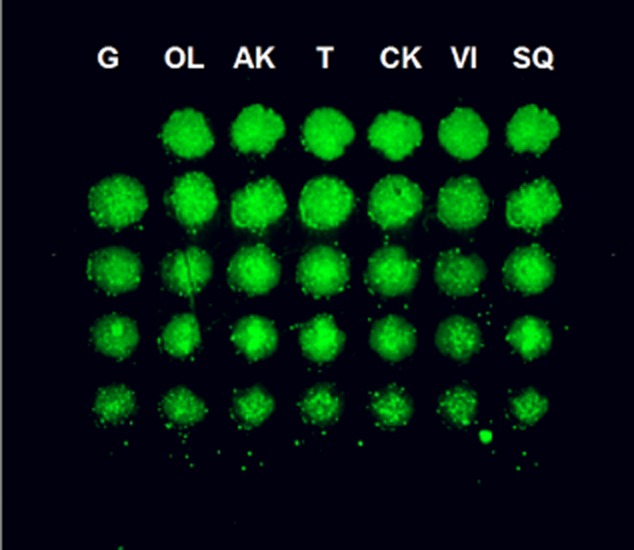
Green fluorescent protein from the protein chip as detected by fluorescence microscopy.
The detection result of three groups
The results show that the positive rate of the lung cancer group is significantly higher than the benign lung disease and healthy groups (χ2 = 61.29, P < 0.05; χ2 = 135.68, P < 0.05, respectively) (Table 2). The positive rate of each marker among the three groups shows evidence that the protein chip can screen lung cancer patients from high-risk or healthy groups (Table 3). Protein chips have a great potential in the early diagnosis of lung cancer. These six markers are likely to become new lung cancer markers.
Table 2.
Positive rate of protein chip in each group
| Group | N | Positive case | Negative case | Positive rate |
|---|---|---|---|---|
| Lung cancer | 217 | 158 | 59 | 72.81% |
| Benign lung disease | 80 | 18 | 62 | 22.50% |
| Healthy | 220 | 37 | 183 | 16.82% |
Comparison of lung cancer and healthy groups, χ2 = 135.68, P < 0.05; comparison of lung cancer and benign lung disease groups, χ2 = 61.29, P < 0.05; comparison of benign lung disease and healthy groups, χ2 = 1.26, P > 0.10.
Table 3.
The positive rate of six markers among three groups (%)
| Group | OLFM1 | AKAP4 | TTRs | CK-18 | VIF-1 | SQLE |
|---|---|---|---|---|---|---|
| Lung cancer | 15.74** | 10.19** | 21.30* | 33.33** | 7.41* | 24.07** |
| Benign lung disease | 14.58 | 6.05 | 8.33▴ | 25.00 | 8.33 | 14.58 |
| Healthy | 2.04 | 0.69 | 3.45 | 22.07 | 0.69 | 1.38 |
Comparison of lung cancer and healthy
P < 0.05
P < 0.01; comparison of lung cancer and benign lung disease groups
P < 0.05. AKAP-4, A-kinase anchor protein-4
CK-18, cytokeratin-18
OLFM1, olfactomedin 1
SQLE, squalene epoxidase
TTRs, transthyretins
VIF-1, vacuolar inhibitor of fructosidase 1.
Discussion
Multi-biomarkers have the potential to improve the early diagnosis rate of lung cancer. However, because of the drawback of traditional biomarkers and diagnostic methods, they are not an ideal diagnostic tool in clinical practice. Therefore, we need to find new biomarkers and simple and efficient test methods.17 As it requires a smaller sample and miniaturization, protein microarray technology may have great potential in this respect. Protein microarray technology has made enormous progress in the last decade, increasingly becoming an important research tool for the study and detection of proteins.8,9
In our study, we successfully constructed a T7 phage display library with cDNA of fresh sections from 30 lung cancer patients. These patients contained various pathologic classifications, were diagnosed for the first time, and had not been treated with radiotherapy or chemotherapy. The phage display library contained a large number of non-specific phages, which were not used to construct protein chips for lung cancer diagnosis. Studies show that antibodies against the surface antigens of tumor cells have been generated in sera in early stage patients. Using the antigen-antibody reaction, the T7 phage display library was screened with serum antibodies of lung cancer patients and healthy controls. In each round of biopanning, we selected phages that reacted to the serum of lung cancer patients but did not react to the serum of healthy controls. After four rounds biopanning, 2880 phages related to lung cancer were purified and enriched. In order to find tumor markers with the potential for clinical lung cancer diagnosis, 2880 selected phages were submitted twice for ELISA high-throughput analysis. Finally, 12 phages that combined with serum antibodies in lung cancer patients, but did not combine in the healthy controls, were selected. According to the ELISA results, we determined that the surface antigens of phages might also exist on the surface of lung cancer cells, leading to the production of serum antibodies.
All of the inserts of the 12 selected phages were amplified by PCR and sequenced. Compared with known sequence genes at GeneBank, we found six exact or similar genes: OLFM1, AKAP-4, TTRs, VIF-1, CK-18, and SQLE. These genes have been located in chromosomes. Some studies have shown evidence that OLFM1, AKAP-4, CK-18, and TTRs are closely related to the occurrence of tumors.12–15 AKAP-4, as a new biomarker, has been verified to have a high expression level in lung cancer.16 All of these results show that the T7 phage display library constructed from 30 fresh sections from lung cancer patients is reliable. Therefore, it is warranted to use these antigens in the design of protein chips for lung cancer diagnosis.
In the last stage of our study, we successfully constructed protein chips with the six selected antigens. The protein chips were then used to test the serum of 217 cases in the lung cancer group,80 cases of benign lung disease, and 220 healthy controls. We found that the Alexa633/555 ratio of healthy control chips was below 0.8, but the value of the lung cancer group chips was above 0.8. Therefore, we determined that this value (0.8) could be used as diagnostic value. Comparing the positive rate among groups, we found that the positive rate of the lung cancer group was significantly higher than in the benign lung disease (P < 0.05) and healthy control groups (P < 0.05). However, there was no significant difference in the positive rate between the benign lung disease and healthy control groups (P > 0.10). The protein chip designed with six selected antigens is effective and may have great future potential in a clinical setting.
Conclusion
In summary, we successfully identified six lung cancer related biomarkers and constructed lung cancer diagnostic protein chips. The protein chip can screen lung cancer patients from a high-risk population. However, as false-positive samples remained, the protein chips need to be improved. Therefore, follow-up research upon the relationship between these markers and pathological type and stage is required to verify the effect on the occurrence of lung cancer.
Acknowledgments
This study was supported by the Public Science and Technology Support Program of the Qingdao Technology Bureau (NO. 09-1-1-9-NSH).
Disclosure
No authors report any conflict of interest.
References
- 1.Ferlay J. 2011. GLOBOCAN 2009, Cancer incidence and mortality worldwide IARC Scientific Publications No. 10.
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. (Published erratum appears in 2011; 61: 133–4.) [DOI] [PubMed] [Google Scholar]
- 3.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: new biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 4.Owonikoko TK, Ragin CC, Belani CP, et al. Lung cancer in elderly patients: an analysis of the surveillance, epidemiology, and end results database. J Clin Oncol. 2007;25:5570–5577. doi: 10.1200/JCO.2007.12.5435. [DOI] [PubMed] [Google Scholar]
- 5.International Early Lung Cancer Action Program Investigators. Henschke CI, Yankelevitz DF, et al. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;335:1763–1771. doi: 10.1056/NEJMoa060476. (Published errata appear in 2008; 359: 877; 2008; 358: 1862; 2008; 358: 1875.) [DOI] [PubMed] [Google Scholar]
- 6.Risch A, Plass C. Lung cancer epigenetics and genetics. Int J Cancer. 2008;123:1–7. doi: 10.1002/ijc.23605. [DOI] [PubMed] [Google Scholar]
- 7.Hur J, Lee HJ, Nam JE, et al. Additional diagnostic value of tumor markers in cytological fluid for diagnosis of non-small-cell lung cancer. BMC Cancer. 2009;12:392. doi: 10.1186/1471-2407-12-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Curr Opin Chem Biol. 2012;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinrich D, Jonkheijm P, Niemeyer CM, Waldmann H. Applications of protein biochips in biomedical and biotechnological research. Angew Chem Int Ed Engl. 2009;48:7744–7751. doi: 10.1002/anie.200901480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Davis JL, Li W. Identification of tribbles homolog 2 as an autoantigen in autoimmune uveitis by phage display. Mol Immunol. 2005;42:1275–1281. doi: 10.1016/j.molimm.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Caberoy NB, Zhou Y, Li W. Can phage display be used as a tool to functionally identify endogenous eat-me signals in phagocytosis? J Biomol Screen. 2009;14:653–661. doi: 10.1177/1087057109335679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scott LC, Evans TR, Cassidy J, et al. Cytokeratin 18 in plasma of patients with gastrointestinal adenocarcinoma as a biomarker of tumour response. Br J Cancer. 2009;101:410–417. doi: 10.1038/sj.bjc.6605175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiriva-Internati M, Yu Y, Mirandola L, et al. Identification of AKAP-4 as a new cancer/testis antigen for detection and immunotherapy of prostate cancer. Prostate. 2012;72:12–23. doi: 10.1002/pros.21400. [DOI] [PubMed] [Google Scholar]
- 14.Nakaya N, Lee HS, Takada Y, Tzchori I, Tomarev SI. Zebrafish olfactomedin 1 regulates retinal axon elongation in vivo and is a modulator of Wnt signaling pathway. J Neurosci. 2008;28:7900–7910. doi: 10.1523/JNEUROSCI.0617-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gericke B, Raila J, Sehouli J, et al. Microheterogeneity of transthyretin in serum and ascitic fluid of ovarian cancer patients. BMC Cancer. 2005;5:133. doi: 10.1186/1471-2407-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radhi S, Alalawi R, Mirandola L, Yu Y, et al. Novel tumor markers in non-small cell lung cancer detected both in serology and tissue. J Clin Oncol. 2012;30(Suppl):Abstract c21105. 2012 ASCO Annual Meeting Proceedings. [Google Scholar]
- 17.Sutandy FX, Qian J, Chen CS, Zhu H. Overview of protein microarrays. Curr Protoc Protein Sci. 2013 doi: 10.1002/0471140864.ps2701s72. Chapter 27: Unit 27.1. [DOI] [PMC free article] [PubMed] [Google Scholar]


