Abstract
Background
Lung cancer is the leading cause of morbidity and death worldwide. Although the available lung cancer animal models have been informative and further propel our understanding of human lung cancer, they still do not fully recapitulate the complexities of human lung cancer. The pathogenesis of lung cancer remains highly elusive because of its aggressive biologic nature and considerable heterogeneity, compared to other cancers. The association of Cul4A amplification with aggressive tumor growth and poor prognosis has been suggested. Our previous study suggested that Cul4A is oncogenic in vitro, but its oncogenic role in vivo has not been studied.
Methods
Viral delivery approaches have been used extensively to model cancer in mouse models. In our experiments, we used Cre-recombinase induced overexpression of the Cul4A gene in transgenic mice to study the role of Cul4A on lung tumor initiation and progression and have developed a new model of lung tumor development in mice harboring a conditionally expressed allele of Cul4A.
Results
Here we show that the use of a recombinant adenovirus expressing Cre-recombinase (“AdenoCre”) to induce Cul4A overexpression in the lungs of mice allows controls of the timing and multiplicity of tumor initiation. Following our mouse models, we are able to study the potential role of Cul4A in the development and progression in pulmonary adenocarcinoma as well.
Conclusion
Our findings indicate that Cul4A is oncogenic in vivo, and this mouse model is a tool in understanding the mechanisms of Cul4A in human cancers and for testing experimental therapies targeting Cul4A.
Keywords: AdenoCre, Cre, Cul4A, lung cancer, mouse models
Introduction
Lung cancer is the leading cause of cancer death worldwide.1 This high mortality rate is because of the fact that most cases of lung cancer are diagnosed at an advanced stage of development. Although morphological differences between the categories are well recognized, the underlying molecular alterations are poorly understood. Perhaps because of an inability to recognize premalignant lesions, physicians are seldom able to diagnose lung cancer before it has reached an advanced stage.2
The cullin 4A/B (CRL4)-based ubiquitin ligase complex has recently become the subject of intense investigation after publication of the crystal structure of its core components and identification of some of its substrates.3,4 The central role of CRL4Cdt2 in regulating the expression of various proteins that regulate the cell cycle, and its role in determining the integrity of DNA replication and repair, suggest that various components of CRL4Cdt2 could be involved in tumorigenesis. We previously found that Cul4A is an oncogene in mesothelioma cells.5 However, the relationship between Cul4A and lung tumor occurrence has not been reported, and the potential oncogenic role of the gene in vivo is yet to be determined. In our experiments, we used Cre-recombinase-induced overexpression of the Cul4A gene in transgenic mice to study the role of Cul4A on lung tumor initiation and progression and to discover a new cell type contributing to the development of pulmonary adenocarcinoma.
To date, most mouse models generate lung adenocarcinomas with various stages of malignancy. A frequently used method to induce lung tumorigenesis is through conditional or spontaneously activated Kirsten rat sarcoma viral oncogene homolog (K-ras) mutations.6 However, it is obvious that more complex mouse models are needed to further our understanding of highly complex lung cancer genetics. Simulating more complex lung tumor genetics in human lung cancer requires an expanded approach with other conditional genetic systems. Cre-mediated recombination can be achieved through the administration of an engineered adenovirus expressing Cre-recombinase (AdenoCre) via nasal or tracheal instillation.7 In an effort to improve mouse lung tumor modeling, we developed a Lox-Stop-Lox Cul4A conditional mouse strain (referred to as LSL-Cul4A), in which overexpression of Cul4A is activated by the removal of a transcriptional termination stop element, achieved through exposure to AdenoCre.8 An advantage of this method is that a limited amount of adult mouse lung cells can be targeted in a localized and controlled fashion. Infection of adult mouse lungs with AdenoCre virus rapidly resulted in the onset of pre-invasive lesions at 16 weeks, followed by the development of adenocarcinoma at 40 weeks. Through synchronized tumor initiation, we were able to characterize the early stages of lung tumor progression. Another important aspect of this model was that lung tumor multiplicity could be controlled, because application of AdenoCre infects only a subset of lung epithelial cells. This, together with control of the time-points of AdenoCre application, ensured a precise mimicking of the sporadic character of human lung cancer development and progression.
Methods
AdenoCre induction in Cul4A transgenic mice
Mice were infected with AdenoCre or Ade-green fluorescent protein (GFP; as control group) at 6–10 weeks of age. The University of California, San Francisco (UCSF) Institutional Animal Care and Use Committee approved the study (Approval No. AN085516-03C). The intranasal administration of the adenoviruses was performed as previously described.9 The AdenoCre (Ad5CMVCre) and Ade-GFP (Ad5CMVeGFP) were purchased from the Gene Transfer Vector Core of the University of Iowa, and have been used in several studies. To generate a mouse model that would conditionally overexpress Cul4A protein, we used the pCALL2 vector.10 Transgenic mice were mated with FVB/N mice to produce transgenic offspring. The pCALL2-Cul4A transgene was genotyped by polymerase chain reaction (PCR) using genomic DNA isolated from the tail.11 Transgenic mice were anesthetized with 2.5% Avertin via intraperitoneal injection, after which half of the mice inhaled approximately 106 or 107 particles of AdenoCre introduced directly into the lungs. The other half inhaled Ade-GFP without AdenoCre. Eight weeks later, the mice were killed and the lungs dissected and sections analyzed.
Histological analysis and immunohistochemistry
Mice were killed at the post-infection times indicated and subjected to full necropsy for histological and immunohistochemical analyses. Endogenous peroxidase was quenched for 15 minutes at room temperature with 3% H2O2 in methanol. Sections were blocked with 4% normal goat serum in phosphate buffered saline (PBS) with 0.2% Triton for two hours at room temperature before incubating overnight at 4°C with the properly diluted antibodies: anti-Cul4A1:500 (ab34897, Abcam, Cambridge, UK); anti-CCSP:2000 (07-623, Millipore, Billerica, MA, USA); anti-proS-PC1:4000 (AB3786, Millipore).
For immunofluorescence, sections were blocked at room temperature for two hours in PBS containing 0.2% Triton X-100 and normal horse serum. They were then incubated at 4°C overnight with dilutions of the appropriate antibodies in blocking buffer. After incubation, sections were washed three times for five minutes with 0.2% Triton X-100 in PBS. Sections were then incubated for 30 minutes at room temperature with GFP-conjugated anti-goat at a 1:200 dilution, followed by a 30-minute incubation with fluorescein isothiocyanate-conjugated anti-rabbit at a 1:1000 dilution. Sections were then counterstained for five minutes with 4′,6-diamidino-2-phenylindole.
Immunohistochemical and statistical analyses
Three independent researchers blindly scored positivity, and the data represent the samples that were scored positive by all three individuals. The following scoring system was used: −, no stain; +, weak staining (10% or above stained cellularity considered as positive); ++, moderate staining (30% or above stained cellularity considered as positive); and +++, strong staining (50% or above stained cellularity considered as positive). All scoring was performed under magnification (20×) with a Zeiss Axioskop 2 microscope (Carl Zeiss Inc., Dusseldorf, Germany) and photomicrographs were obtained with a Carl Zeiss AxioCam MrC5 camera.
Correlation analyses were performed using SPSS 13.0 (SPSS Inc., Chicago, IL, USA). Zero, 1, 2, and 3 were used to represent −, +, ++, and +++, respectively.
Results
Overexpression of Cul4A in mouse lungs elicits tumor formation
Infected LSL-Cul4A mice (hereafter referred to as Lox-Cul4A) were killed at eight, 12, 16, 20, and 24 weeks after infection with Ade-GFP and AdenoCre. On examination, the lungs from the wild-type mice induced with Ade-GFP and those induced with AdenoCre appeared grossly normal and uniform at 12 weeks post-infection (Fig 1a, b), whereas small, scattered hyperplasia lesions were observed histologically at eight and 12 weeks post-infection (Fig 1f, g). The lungs of the Lox-Cul4A mice had numerous bumpy lesions at 16 weeks post-infection, and histological sections showed a single lesion (Fig 1c, h). At 20 weeks post-infection with AdenoCre, the surface of Lox-Cul4A lungs had a cobblestone appearance, and histological sections showing a single lesion (Fig 1d, i). This may more closely mimic human lung cancer, in which patients typically develop a single tumor that progresses over time to metastatic disease. At 24 weeks post-infection with AdenoCre, numerous adenocarcinoma-like lesions were clearly visible on the surface of the Lox-Cul4A mouse lungs (Fig 1e, j).
Figure 1.
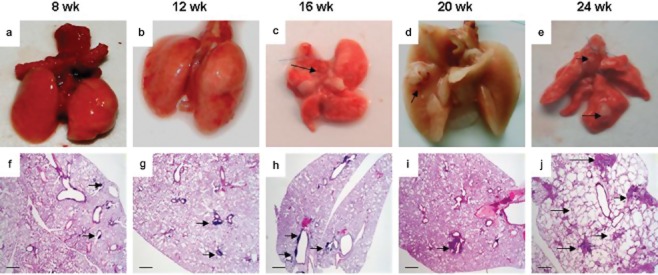
Tumor pathology in mouse lungs with Cul4A overexpression. (a) Wild-type mouse lungs eight weeks post-infection with Ade-green fluorescent protein. The surface of the lungs is smooth and uniform. (b) Lox-Cul4A mouse lungs 12 weeks post-infection with adenovirus expressing Cre-recombinase (AdenoCre). The surface of the lungs is also smooth and uniform. (c) Lox-Cul4A mouse lungs 16 weeks post-infection with AdenoCre. The surface of the lungs have a bumpy appearance, arrows indicate the lesions. (d) Lox-Cul4A mouse lungs 20 weeks post-infection with AdenoCre. An isolated cobblestone-like nodule on the surface of the lungs is visible; the arrow indicates the lesion. (e) Lox-Cul4A mouse lungs 24 weeks post-infection with AdenoCre. Several nodules on the surface of the lungs have appeared; arrows indicate the lesions. (f) Histological sections of wild-type mouse lungs eight weeks post-infection with AdenoCre. (g) Histological sections of Lox-Cul4A mouse lungs 12 weeks post-infection with AdenoCre; the arrow shows a single lesion. (h) Histological sections of Lox-Cul4A mouse lungs 16 weeks post-infection with AdenoCre; arrows indicate isolated lesions. (i) Histological sections of Lox-Cul4A mouse lungs 20 weeks post-infection with AdenoCre; arrow shows a single adenocarcinoma-like lesion. (j) Histological sections of Lox-Cul4A mouse lungs 24 weeks post-infection with AdenoCre; arrows show diffuse severe adenocarcinoma-like lesions. Scale bar indicates 200 μm. Wk, weeks.
Distinct types of preneoplastic and adenocarcinomous lesions allow progression to be followed over time
Three types of pulmonary preneoplastic lesions have been recognized in Lox-Cul4A mouse lungs at eight, 12, and 16 weeks post-infection with AdenoCre, according to the recent World Health Organization (WHO) classification system.
Immunohistochemical analysis reveals that hyperplasias and adenocarcinomas are related to both Clara cells and alveolar type II pneumocytes
We detected positive cells lining the bronchioles by staining for Clara cell secretory protein (CCSP), but found that Cul4A-induced lesions were largely CCSP negative (Fig 2a, c, g). In contrast, most cells within Cul4A-induced lesions expressed surfactant protein-C (SP-C), suggesting that they have properties reminiscent of type II pneumocytes (Fig 2b, d, h). Serial section analysis indicated that Cul4A induced adenomas often arose near terminal bronchioles (Fig 2e, f).
Figure 2.
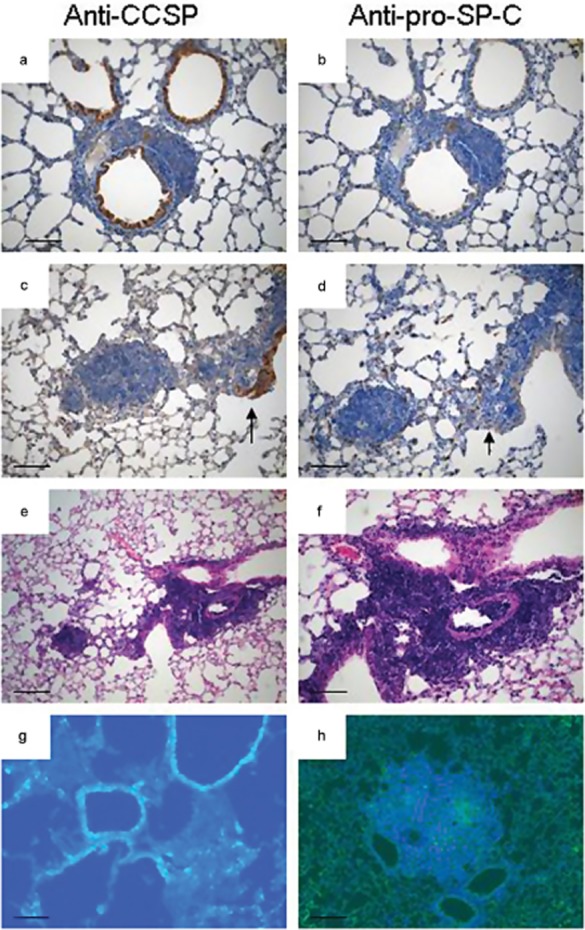
Immunophenotype of lesions in Lox-Cul4A mice. (a) Clara cell secretory protein (CCSP)-positive in nonrespiratory bronchiole epithelial cells and negative in atypical adenomatous hyperplasias. (b) Surfactant protein-C (SP-C) positive adenoma and SP-C negative bronchiole epithelial cells in the same lesion of (a). (c) CCSP positive papillary structures in epithelial hyperplasia (EH) continuous with an adenoma. (d) SP-C-positive papillary structures and adenoma in the same continuous lesion in the adjacent serial section. Arrows indicate the same papillary structure in (c) and (d) that is positive for both CCSP and SP-C. (e) Histological section of EH continuous with an adenoma. (f) Higher magnification of papillary structures in which there are double-positive SP-C/CCSP cells. (g) CCSP immunofluorescence alone in the adenocarcinomatous lesion 24 weeks post-infection with adenovirus expressing Cre-recombinase (AdenoCre). (h) SP-C immunofluorescence alone in the adenocarcinomatous lesion 24 weeks post-infection with AdenoCre. Scale bar indicates 100 μm in A–F, and 50 μm in G–H.
Discussion
Our study demonstrates that Cul4A is oncogenic in vivo and that the development of lung tumors can be induced by Cul4A overexpression. The very reproducible tumor latency obviously depends on strain susceptibility and/or the application of carcinogen-induction protocols. Mouse lung cells may be especially sensitive to the proliferative effects of Cul4A, such that a tumor will more readily arise when the Cre-recombination event occurs in these cell types. A similar situation happens in K-ras and B-Raf mutations, which are both associated with tumor progression in mouse models.12 By 20 weeks post-infection, we observed no adenocarcinomas or metastases in any of the models with overexpression of Cul4A, most probably because by itself, the overexpression of Cul4A is not enough to allow the adenocarcinoma to progress to a later state of malignancy. However, these straightforward experiments elucidate the important role of Cul4A in mouse lung tumor initiation and progression.
To test that the lesions were associated with excision of the transcriptional stop element, genomic DNA was prepared from portions of the infected lungs, and PCR confirmed the presence of the Cul4A allele. To verify that excision of the stop element resulted in the expression of Cul4A protein, immunostainings of Cul4A in mouse lung sections post-infection were analyzed at the indicated time points. The rearranged Cul4A allele was not detected in any of the other tissues from the infected mice, indicating that infection and Cul4A overexpression were limited to the lungs (data not shown; see Methods).11 The lungs of mice inducted with AdenoCre were strongly positive for Cul4A, whereas the lungs of mice without Cre-excision were negative (Fig 3). Interestingly, we detected overexpressions of Cul4A in some hyperplasia and adenocarcinomous lesions in the mouse lungs after AdenoCre inhalation. Possibly, this is a result of the oncogenic functions of the Cul4A protein.
Figure 3.

Immunostaining of anti-Cul4A in mouse lung sections. (a) Eight weeks with Ade-green fluorescent protein (GFP); (b) eight weeks with adenovirus expressing Cre recombinase (AdenoCre); (c) 12 weeks with Ade-GFP; (d) 12 weeks with AdenoCre; (e) 16 weeks with Ade-GFP; (f) 16 weeks with AdenoCr; (g) 20 weeks with Ade-GFP; (h) 20 weeks with AdenoCre.
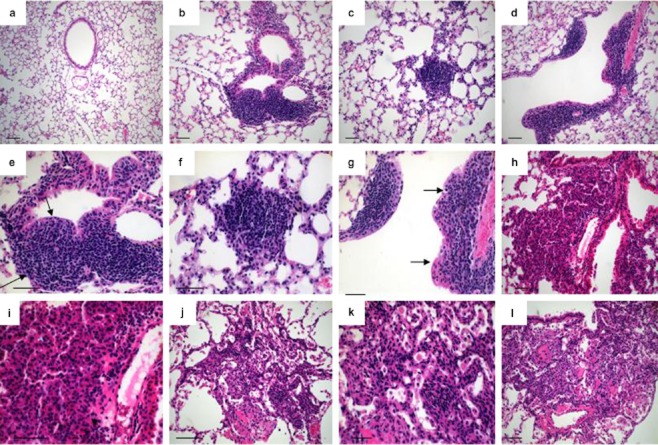
The first preneoplastic lesion is atypical adenomatous hyperplasia (AAH), a proliferation of atypical epithelial cells growing along the alveolar septae, which do not disrupt underlying lung architecture. In the 1999 and 2004 WHO classifications, AAH was recognized as a pre-invasive lesion for lung adenocarcinoma.13 The AAH present in Lox-Cul4A mice closely resembles human AAH, a dysplastic lesion proposed to be a precursor of pulmonary adenocarcinoma.1 Lesions of the second type are epithelial hyperplasia (EH), the papillary proliferations of epithelial cells lining the bronchioles, for which analogous lesions have not been identified in humans. Lesions of the third type are adenomas, which are neoplasms with papillary, solid or mixed architecture that distort or obliterate the alveolar septae. Adenomas of the lung are reported infrequently in humans, but this may reflect the fact that lung cancer patients typically present in an advanced stage of the disease.
Early on in our study (around 8 weeks post-infection), mice with Cul4A overexpression had AAH arising within the terminal bronchioles and within the central lung parenchyma (n = 5). The lesions appeared to arise in alveolar ducts and expand outward and around bronchioles. This hyperplastic epithelium displayed papillary excrescences (Fig 4b, e), but at early time points (4–8 weeks), nodules had not formed (Fig 1a, b). At 12 weeks post-infection with AdenoCre (n = 7), the mouse lungs contained a spectrum of lesions ranging from AAH to small adenomas. We detected changes in the papillary adenomas where they appeared to undergo alterations at their periphery, appearing to take on a more solid architecture (Fig 4c, f). EH of the bronchioles also persisted at this time-point, and a subset of EH appeared to be continuous with AAH, as before, as well as with adenomas. It is striking that the Cul4A-induced lung adenomas formed rapidly (8–12 weeks). At 16 weeks post-infection with AdenoCre, we observed the continued presence of AAH (n = 5), but these lesions were outnumbered by adenomas. The adenomas were significantly larger than those seen at eight weeks. There was no invasive component. In two slides at 16 weeks, AAH lesions were rare, but moderate EH was more prevalent, suggesting that these lesions are indeed precursor lesions that progress over time, eventually becoming tumors of more advanced stages (Fig 4d, g). At 20 weeks (n = 7) post-infection with AdenoCre, AAH lesions were no longer seen, and overt minimally invasive adenocarcinoma (MIA) was present. MIA is usually nonmucinous but may rarely be mucinous (Fig 4h, i). It is a small, solitary adenocarcinoma, with a predominantly lepidic growth pattern, with tumor cells infiltrating myofibroblastic stroma. These findings provide strong evidence for a progression from AAH lesion to adenoma, and then to adenocarcinoma, and an overexpression of Cul4A appears sufficient to rapidly induce the hyperplastic lesions that progress to adenoma. At 24 weeks (n = 6), the multiple adenocarcinomas showed numerous mitoses and nuclear pleomorphisms, including enlarged nuclei with prominent nucleoli (Fig 4j, k, l). Major histologic predominant patterns of invasive adenocarcinoma consist of lepidic and acinar growth. The lepidic pattern consists of a proliferation of type II pneumocytes and Clara cells along the surface alveolar walls. Acinar adenocarcinoma consists of round to oval-shaped malignant glands invading a fibrous stroma.
Figure 4.
Histological analysis of distinct types of lesions and stages of tumor progression in Lox-Cul4A mice. (a) Eight weeks post-infection with Ade-green fluorescent protein (GFP. (b) Slightly atypical adenomatous hyperplasia of terminal (bottom) and epithelial hyperplasia (EH) of a respiratory (top) bronchiole in Lox-Cul4A lungs eight weeks post-infection; (c) Small papillary adenoma in Lox-Cul4A lungs 12 weeks post-infection. (d) Moderate EH of a terminal bronchiole 16 weeks post-infection; arrow indicates the destroyed basal membrane. (e) Higher magnification of the lesion in B; arrows indicate the lesions. (f) Higher magnification of the lesion in (c). (g) Higher magnification of the lesion in (d). (h) Nonmucinous minimally invasive adenocarcinoma (MIA). This subpleural adenocarcinoma tumor consists primarily of lepidic growth area of invasion 20 weeks post-infection. (i) Higher magnification of the lesion in (h). (j) Mucinous MIA 24 weeks post-infection. This consists of a tumor showing lepidic growth. (k) Higher magnification of the lesion in (j). The tumor cells grow mostly in a lepidic pattern along the surface of alveolar walls. (l) The tumor invades the areas of stromal fibrosis in an acinar pattern. Scale bar indicates 100 μm.
In our observations, the short latency and high penetrance of AAH formation was particularly striking, and suggested that overexpression of Cul4A is sufficient for the induction of hyperplastic lesions. The time-course study performed here indicated that AAH lesions became progressively larger and more solid, until it became impossible to make a distinction between advanced AAH and adenoma. Depending on the extensiveness of the search, AAH may be multiple in up to 7% of resected lung adenocarcinomas.14 Multiple subtypes with different growth patterns show that these mouse lung tumors are bilologically heterogeneous and contain genotypically and phenotypically diverse subpopulations of tumor cells, each of which has the potential to complete some but not all of the steps in the metastatic process. Meanwhile, we found that most lesions were located subpleurally (Fig 1j), which is consistent with the ability of metastatic cells to grow at a foreign site, and suggests that metastasis favors the survival and growth of a few subpopulations of cells that preexist within the parent neoplasm.
The cellular origins of human lung adenocarcinoma and mouse lung adenomas are unclear.15 To assess the properties of mouse lung lesions brought about by the overexpression of Cul4A, we used immunohistochemical and immunofluorescence analysis to detect CCSP/CC10 and SP-C, a surface marker of type II pneumocytes (AT2 cells).16 Non-small cell lung cancers frequently express markers of Clara cells or alveolar type II pneumocytes (AT2 cells). We discovered that the areas of alveolar hyperplasia, AAH, adenomas, and adenocarcinomas observed in the alveolar septa following Cul4A activation induced by the AdenoCre virus were found to be negative for CCSP but strongly positive for SP-C.
The earliest studies focused on AT2 cells and a rare population found within the bronchioalveolar duct junction (BADJ) termed bronchioalveolar stem cells (BASCs).17 Based on our staining of alveolar type II pneumocytes and Clara cell-specific markers in Cul4A-induced lesions, we discovered that the hyperplasia and adenocarcinomous lesions were mostly located at the bifurcation between bronchiolar and respiratory epithelium. We therefore hypothesize that BASCs constitute a stem or progenitor cell population that maintains Clara cells and alveolar type II pneumocyte cells in mouse lungs.
The cell of origin for most lung cancers remains elusive, but several studies have pointed to cells residing in the BADJ region.18 As with these other studies, we observed an increased incidence of tumor formation in this region, especially in the Kras activation model. Although it was difficult to explain why this also occurred in our Cul4A overexpression mouse model, our results suggest that the origin of these tumors is either BASC-like cells or Clara cells within the bronchiolar epithelium. In fact, lesions present at early post-infection time points appeared to be continuations of bronchiolar hyperplasia and alveolar hyperplasia arising in terminal bronchioles. To explore this hypothesis, we will need to isolate and further characterize BASCs.
Conclusion
The role Cul4A plays in the initiation and development of lung tumors in our mouse model has implications for understanding cancer biology and introduces new avenues for therapeutic development targeting Cul4A. Identifying the cells of origin of lung cancer may lead to new therapeutic strategies. This new Cul4A mouse model of lung cancer has many advantages over existing models. A better understanding of Cul4A signaling pathways and mechanisms and their effects on the growth of lung tumors in our mouse model could provide new insights, thus contributing to future experimental therapies.
Acknowledgments
This study was supported by a NIH grant R01 CA140654-01A1. We are grateful for support from the Kazan, McClain, Abrams, Fernandez, Lyons, Greenwood, Harley & Oberman Foundation, Inc; the Estate of Robert Griffiths; the Jeffrey and Karen Peterson Family Foundation; Paul and Michelle Zygielbaum; the Estate of Norman Mancini; and the Barbara Isackson Lung Cancer Research Fund. We thank Lorretta Chan in the UCSF Cancer Center Tissue Core for her help. We also thank Pamela Derish in the UCSF Department of Surgery for editorial assistance with the manuscript.
Disclosure
No authors report any conflict of interest.
References
- 1.Kerr KM. Pulmonary preinvasive neoplasia. J Clin Pathol. 2001;54:257–271. doi: 10.1136/jcp.54.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuveson DA, Jacks T. Modeling human lung cancer in mice: Similarities and shortcomings. Oncogene. 1999;18:5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- 3.Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 2006;443:590–593. doi: 10.1038/nature05175. [DOI] [PubMed] [Google Scholar]
- 4.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 5.Hung MS, Mao JH, Xu Z, et al. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2011;15:350–358. doi: 10.1111/j.1582-4934.2009.00971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- 7.Jackson EL, Willis N, Mercer K, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li T, Hung MS, Wang Y, et al. Transgenic mice for cre-inducible overexpression of the Cul4A gene. Genesis. 2011;49:134–141. doi: 10.1002/dvg.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 11.Li T, Hung MS, Wang Y, et al. Transgenic mice for cre-inducible overexpression of the Cul4A gene. Genesis. 2011;49:134–141. doi: 10.1002/dvg.20708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson L, Mercer K, Greenbaum D, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 13.Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeshima AM, Tochigi N, Yoshida A, Asamura H, Tsuta K, Tsuda H. Clinicopathologic analysis of multiple (five or more) atypical adenomatous hyperplasias (AAHs) of the lung: Evidence for the AAH-adenocarcinoma sequence. J Thorac Oncol. 2010;5:466–471. doi: 10.1097/JTO.0b013e3181ce3b73. [DOI] [PubMed] [Google Scholar]
- 15.Mainardi S, Mijimolle N, Francoz S, Vicente-Dueñas C, Sánchez-García I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proc Natl Acad Sci. 2014;111:255–260. doi: 10.1073/pnas.1320383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawlins EL, Okubo T, Xue Y, et al. The role of Scgb1a1 + Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim CF, Jackson EL, Woolfenden AE, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 18.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci. 2014;111:4952–4957. doi: 10.1073/pnas.1319963111. [DOI] [PMC free article] [PubMed] [Google Scholar]