Abstract
Background
Human Herpesvirus-6 (HHV-6) latently infects a majority of adults. In about 1% of the population HHV-6 exists in a chromosomally integrated form (ciHHV-6) that resides in every somatic and germ cell and can be transmitted through the germ line. Patients with ciHHV-6 have been misdiagnosed and unnecessarily treated for active HHV-6 infection, sometimes with significant side effects, based on results from quantitative molecular HHV-6 tests.
Methods
A droplet digital PCR (ddPCR) assay was developed to identify ciHHV-6 in cellular patient specimens by precisely determining the ratio of HHV-6 to cellular DNA. We validated the assay on confirmed ciHHV-6 patient specimens and a cell line derived from a ciHHV-6 patient, and analyzed hematopoietic stem cell transplant patients suspected of ciHHV-6. We additionally evaluated whether the assay could be applied to stored plasma samples from a study of clinical correlates of HHV-6.
Results
The ddPCR assay accurately identified ciHHV-6 in cellular samples (buffy coat, PBMCs), giving a ratio very close to 1 HHV-6/cell (1.02 ± .03 in FISH-confirmed samples). In stored plasma samples, the assay performance was set by design to have 100% sensitivity, which resulted in 82% specificity for ciHHV-6.
Conclusions
The possibility of ciHHV-6 is often overlooked in patients with detectable HHV-6 viral loads by quantitative PCR. Our ddPCR test provides rapid and accurate laboratory identification of ciHHV-6 from easily obtained cellular specimens. In addition, the assay provides excellent sensitivity and specificity using stored plasma samples, facilitating retrospective analysis of the clinical significance of ciHHV-6.
Keywords: Human Herpesvirus 6, ciHHV-6, digital PCR
Introduction
Human Herpesvirus 6 (HHV-6) latently infects more than 90% of adults,(1) and reactivates in 30–50% of transplant recipients.(1) HHV-6 has two species, HHV-6A and HHV-6B, and both are capable of chromosomal integration.(2, 3) Recent studies demonstrate that about one percent of the population has HHV-6 stably integrated into the chromosome telomere regions of all somatic and germ cells,(4–6) and this chromosomally integrated form of the virus (ciHHV-6) can be passed from parent to child through the germline (7)
The presence of ciHHV-6 in all cells of an individual complicates interpretation of HHV-6 real-time PCR testing on plasma, serum or whole blood. Because typical plasma PCR assays for HHV-6 also detect integrated virus within any cellular DNA present in the specimen, i.e. from lysed cells, patients with ciHHV-6 may be falsely diagnosed with active HHV-6 infection. Misdiagnosis is detrimental because antiviral treatment for HHV-6 involves drugs including ganciclovir, foscarnet, and cidofovir, which are costly and carry significant side effects.
Patients with HHV-6 viral loads greater than 1×106 copies/ml in whole blood or 1×104 copies/ml in plasma are currently presumed to have ciHHV-6.(8, 9) However, a recent study demonstrated that qualitative or quantitative HHV-6 PCR of plasma is not sufficient to distinguish active viral replication from the chromosomally integrated form of HHV-6.(10) A review of 21 case reports of confirmed ciHHV-6 patients determined that antiviral therapy was mistakenly administered to five asymptomatic patients presumed to have active HHV-6 infection due to high HHV-6 DNA levels by standard real-time PCR testing.(11) Conversely, patients with symptomatic active HHV-6 infection can have very high levels of HHV-6 consistent with ciHHV-6, which may confound or delay the diagnosis of active infection.
Currently, identification of ciHHV-6 requires fluorescence in situ hybridization (FISH), a lengthy procedure with limited availability, or HHV-6 PCR testing of hair follicle cells(2), an atypical sample type for many molecular diagnostics labs. Here we utilize an emerging molecular quantitation method called droplet digital PCR (ddPCR) to perform a ratio-based assay that rapidly and accurately detects ciHHV-6 from cellular specimens, typically buffy coat collected from whole blood.
Methods
Droplet Digital PCR (ddPCR)
Droplet digital PCR uses TaqMan chemistry like real-time PCR, but partitions the reaction into thousands of individual droplets, which are each read as positive or negative for DNA template allowing absolute quantification of DNA copies without the use of a standard curve (12–14). Development and validation of our ciHHV-6 assay was performed in accordance with the MIQE guidelines for digital PCR (15). The HHV-6 primer and probe set amplify a 150 bp region of U67 as previously described(16) and are as follows: 5R(A) 5’-GTTAGGATATACCGA TGTGCGTGAT-3’, 5R(B) 5’-TACAGATACGGAGGCAATAGATTTG-3’, 5R probe, 5R(P), 5’-FAM-TCCGAAACAACTGTCTGACTGGCAAAA-BHQ1-3’. These sequences target the U67 gene in a region that is conserved between HHV-6A and B. Because both HHV-6A and B integrate, this assay does not discriminate. In our laboratory, typing is performed with a secondary PCR reaction designed to target a non-conserved region(17). RPP30 is a ribonuclease reference gene for cell count. The RPP30 primer and probe set amplify a 60 bp region, were provided by Bio-Rad Laboratories and have the following sequences: RPP30for 5’-GATTTGGACC TGCGAGCG-3’, RPP30rev 5’GCGGCTGTC TCCACAAGT-3’, RPP30probe 5’-HEX-TCTGACCTGAAGGCTCTGCG CG-BHQ1-3’.
The ddPCR reaction mixture consisted of 12·5µl of 2× ddPCR Supermix for Probes (Bio-Rad, catalogue no. 1863010), 1·25µl of each 20× primer-probe mix (18µM each PCR primer, 5µM probe), and 10µl of template DNA in a final volume of 25µl. If derived from a cellular sample, the template DNA was digested with restriction enzyme HindIII (New England BioLabs, Ipswich, MA) before adding it to the ddPCR reaction. The digestion reaction used 5 µl template DNA, 1 µl HindIII, 1µl NEB buffer 4, 3 µl water, digested at 37°C for 1 hour and diluted 1:5 with the addition of 40 µl water. Dilute NEB buffer 4 did not inhibit these reactions, but inhibitory effects of any reaction additives on dPCR should be determined empirically (18). DNA from plasma samples was used undigested. 20µl of each reaction mixture was loaded onto a disposable plastic cartridge (Bio-Rad) with 70µl of droplet generation oil (Bio-Rad) and placed in the Droplet Generator (Bio-Rad). The droplets generated from each sample were transferred to a 96-well PCR plate and PCR amplification was performed on a 2720 Thermal Cycler (Applied Biosystems, Carlsbad, CA) with the conditions: 94°C for 10 minutes, 40 cycles of 94°C for 30 seconds and 60°C for 1 minute, followed by 98°C for 10 minutes and ending at 4°C. After amplification, the plate was loaded onto the Droplet Reader (Bio-Rad) and the droplets from each well of the plate were automatically read at a rate of 32 wells/hour. Data were analyzed with QuantaSoft analysis software (V1.3.2.0) and quantification of target molecules presented as copies/µl of PCR reaction. Around 15,000 droplets (0.89nl/droplet) were analyzed per well (19). Data from any wells with less than 10,000 droplets analyzed were discarded. The result of HHV-6 copies/cell was obtained using the formula: HHV-6 copies/ (RPP30 copies/2) because two copies of RPP30 are present in a diploid genome.
Cell lines and patient samples
The Hector-2 cell line (Bioworld Consulting Laboratories, Mt. Airy, MD) was derived from B-lymphocytes of a donor with FISH-confirmed ciHHV-6 (7) and contains one HHV-6 integration per cell (in chromosome 18). Hector-2 cells and whole blood samples from an IRB-approved ciHHV-6 registry (HHV-6 Foundation, Santa Barbara, CA) were utilized to validate and calibrate the ratiometric ddPCR assay. DNA was extracted from cells previously frozen at −80°C and the duplex ddPCR assay for HHV-6 and RPP30 performed.
Patient buffy coat, plasma, and tissue samples saved frozen at −20 to −80°C from routine clinical HHV-6 and ciHHV-6 real-time PCR testing were evaluated for ciHHV-6 by digital PCR. Plasma and tissue samples from patients with or without real-time PCR-confirmed HHV-6 reactivation, and patients with suspected ciHHV-6 (determined a priori as increasing HHV-6 plasma DNA levels during the first 2 weeks after transplantation and persistent levels ≥ 100 copies per/mL in ≥ 80% of subsequent plasma samples), were selected from a hematopoietic stem cell transplantation study population.(20, 21) Study subjects with HHV-6 reactivation were selected based on the following criteria: 1) negative sample within the first ten days post-transplant 2) two consecutive positive samples 3) negative sample collected seven to ten days after the last positive. Patients without HHV-6 reactivation had three consecutive negative samples collected within the same range of days post-transplant as those with reactivation as well as a negative sample collected seven to ten days after the last of the three consecutive negative samples. Samples were run blind by a single technician.
DNA extractions of cultured cells and tissue specimens were performed on a Maxwell 16 (Promega, Madison, WI) utilizing the total viral nucleic acid extraction kit with varying volumes of cell or tissue sample extracted to 50–100 µl in water. Plasma samples were extracted on a MagnaPure LC (Roche, Basel, Switzerland) utilizing the DNA Isolation Kit I with a volume of 200µl plasma extracted to 100µl DNA in elution buffer. DNA was stored frozen at −20°C until use. DNA from plasma specimens was run in triplicate ddPCR reactions while DNA from cellular specimens was run as a single ddPCR reaction. Use of all patient specimens was approved by the University of Washington Institutional Review Board.
Statistical analysis of plasma results
The ratio of HHV-6 to cellular DNA in plasma samples from patients without ciHHV-6 depends on the number of lysed cells in the sample and the amount of infectious virus circulating in the blood. These could by chance have a ratio of one even in active infection. To distinguish active HHV-6 from ciHHV-6, we analyzed the plasma HHV-6/cell ratios of known ciHHV-6 positive and negative samples. To aid analysis, we transformed these ratios to the absolute log scale, as ratios near one approach zero on this scale while ratios larger (or smaller) than one will be larger than zero. Then we set as a cutoff for differentiation the maximum observed absolute log ratio in confirmed ciHHV-6 participants and used this cutoff to define the non-transformed ratio range indicative of ciHHV-6. Specificity of this cutoff was estimated from participants with real-time PCR-confirmed HHV-6 reactivation as described above. Confidence intervals for sensitivity and specificity were computed using the normal approximation to the binomial distribution. Positive and negative predictive values were computed from sensitivity, specificity and prevalence using Bayes’ rule.
Results
Analytical Performance of ddPCR for ciHHV-6
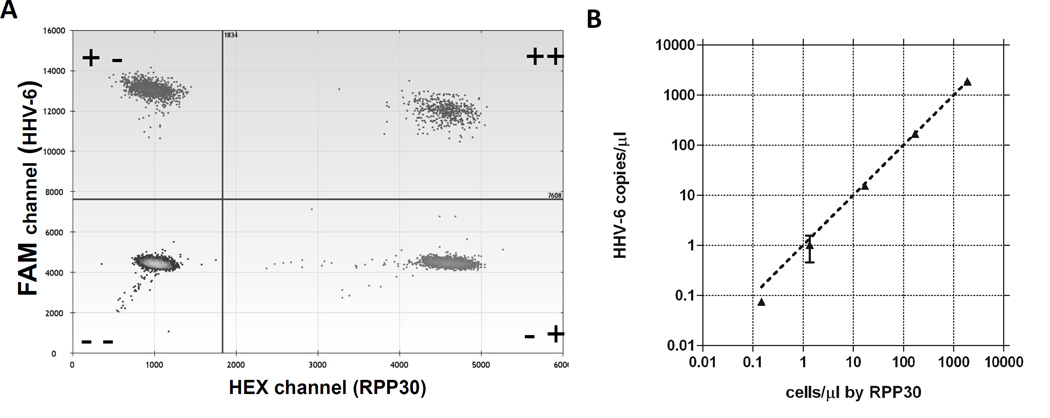
A duplex ddPCR assay for HHV-6 and RPP30 was optimized on the QX100 Droplet Digital PCR system (Bio-Rad Laboratories, Hercules, CA). To evaluate the performance of ddPCR for the quantitation of HHV-6 vs human genome copies in cellular material, we used the Hector-2 cell line, which contains one HHV-6 integration per cell. The duplexed assay showed consistent, wide separation of positive and negative droplets in the FAM and HEX channels (Figure 1A) and could detect as few as 0.1 HHV-6 copies/µl or 2 copies/reaction (Figure 1B). The false positive background of the assay was minimal. Of twenty negative control wells, each run on a separate day, 3 had a single positive droplet for HHV-6 and none were positive for RPP30. Therefore, the cutoff for detection would conservatively be placed at 2 positive droplets/reaction, corresponding to 4 copies/reaction.
Figure 1.
A. Droplet plot of positive and negative droplets for HHV-6/RPP30 duplex ddPCR assay using template DNA from Hector-2 ciHHV-6 positive control cells. Upper left dots are positive for the HHV-6 FAM assay, lower right dots are positive for the RPP30 HEX assay, upper right dots are positive for both assays and lower left dots are negative for both assays. B. Concordance of HHV-6 copy number with cell number in ciHHV-6 positive control Hector-2 cells assayed by the duplexed digital PCR assay for HHV-6 and RPP30 run in duplicate.
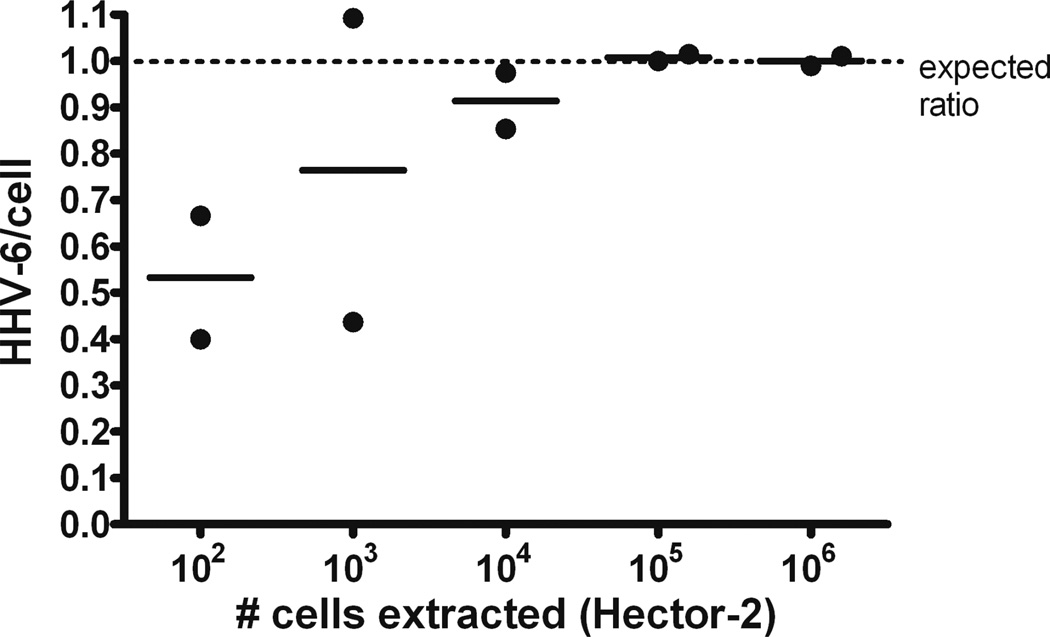
The assay provided a precise ratio of HHV-6/cell close to one with as few as 104 cells (15 cells/µl ddPCR reaction as counted by RPP30) (Figure 2). A typical whole blood sample from a healthy individual contains 4–7 × 106 leukocytes/ml of blood,(9) so this cell input is well within the range applicable to buffy coat samples taken from two to five ml of whole blood. Moreover, the interassay precision is high. Results for five independent runs using Hector-2 template DNA gave a mean and standard deviation of 0.96 ± 0.03 with a coefficient of variation of 3%.
Figure 2.
Dilution series 10-fold) of Hector-2 ciHHV-6 cell line indicates that the ddPCR assay provides a precise ratio of HHV-6/cell with as few as 104 cells.
Assay performance on FISH-confirmed clinical specimens
To confirm the performance of this assay on patient specimens, we obtained samples from two patients with previously identified, FISH-confirmed ciHHV-6. FISH results and patient background were provided for both of these patients in previous publications (patient one is sibling three(7) and patient B(22), and patient two is the father of sibling three(7)). Only two buffy coat samples were available given the low number of confirmed samples available worldwide. Buffy coat specimens from the two patients resulted in ratios near 1 HHV-6/cell by the ddPCR assay (patient one =1.01 and patient two =1.05). The striking precision of this assay on buffy coat specimens underscores its utility as a rapid diagnostic tool for identification of ciHHV-6 from blood samples.
ciHHV-6 status of hematopoietic stem cell transplant study patients
We analyzed samples from four hematopoietic stem cell transplant (HCT) patients who were candidates for a study of HHV-6 reactivation, but were excluded from that study for suspected ciHHV-6.(20, 21) Three of the four patients (patient 30, 199, 329) had white blood cell samples that gave a ratio of HHV-6/cell very close to one (Table 1). In the cases of patients 199 and 329, donor white blood cell samples were available to confirm that ciHHV-6 was transferred from donor to recipient through stem cell transplantation (Table 1). Patient 30 (Table 1) was particularly interesting, with a PBSC (peripheral blood stem cell) HHV-6/cell ratio of 0.83 on a PBSC sample taken 22 days post transplant. This patient received a non-myeloablative transplant and had bone marrow chimerism of 99.8% donor six days later, at 28 days post transplant. Thus, the ratio of HHV-6/cell in the tested PBSC sample most likely corresponds to the ratio of donor and recipient stem cells present at day 22. The fourth patient (patient 98) was suspected of being ciHHV-6 positive and receiving a transplant from a ciHHV-6 negative donor. Consistent with this, post-transplant PBMC showed a very low HHV-6/cell ratio (Table 1). Although pre-transplant PBMC were not available, we were able to obtain two post-transplant formalin-fixed, paraffin embedded (FFPE) skin tissue samples. Since skin is not derived from the donor hematopoietic cells, these samples should reflect the recipient's pre-transplant ciHHV-6 status. Although the cell numbers available from these specimens were not adequate to calculate an accurate HHV-6/cell ratio, the samples were positive for HHV-6, supporting the hypothesis of recipient ciHHV-6.
Table 1.
Suspected ciHHV-6 positive patient cell and plasma samples from hematopoietic stem cell transplant study
| Patient | Sample type* | Days post-transplant | HHV-6/cell |
|---|---|---|---|
| Patients with ciHHV-6 donor | |||
| 30 | PBSC | 22 | 0.83 |
| 30 | plasma | 81 | 0.97 |
| 199 | Pre-transplant PBMC | NA | 0.01 |
| 199-donor | PBMC | NA | 0.99 |
| 199 | PBMC | 95 | 0.99 |
| 199 | plasma | 92 | 0.57 |
| 329-donor | PBSC | NA | 1.04 |
| 329 | PBMC | 84 | 0.98 |
| 329 | plasma | 1334 | 1.32 |
| ciHHV-6 patient with non ciHHV-6 donor | |||
| 98 | PBMC | 145 | 0.002 |
| 98 | plasma | 838 | 0.18 |
PBSC indicates peripheral blood stem cell
PBMC indicates peripheral blood mononuclear cell
samples are post-transplant unless otherwise noted
NA= not applicable
It is worth noting that these four patients were singled out as possible carriers of ciHHV-6 on the basis of HHV-6 plasma viral load levels that were being monitored regularly over time in the context of a research study. However, in a typical clinical setting, HHV-6 viral load levels are not routinely interrogated over time, and without a high index of suspicion and a rapid molecular assay available, ciHHV-6 status could easily be unrecognized.
Identification of ciHHV-6 in residual clinical cellular samples
Further analyses utilizing the ddPCR assay were performed on cellular samples left over from routine clinical HHV-6 testing from eight patients. While five of the patients were clearly negative for ciHHV-6, three patients had specimens that indicated possible ciHHV-6 by ddPCR testing (Table 2). Patient A had a buffy coat sample with an HHV-6/cell ratio that indicates two chromosomal integrations, a rare but documented condition.(23) Patient B had HHV-6 viremia with HHV-6-associated hepatitis, and a liver biopsy sample was tested for ciHHV-6 by ddPCR. The analysis demonstrated 1.2 HHV-6/cell, but the liver biopsy was a poor sample with only one to two cells/µl, prompting testing of additional samples from multiple sites (see Table 2). These specimens all had ratios close to 0, which ruled out ciHHV-6 and suggested that the ratio near 1 in the liver tissue was likely due to an active HHV-6 infection. An in-depth report of this patient's presentation and clinical course has been accepted for publication (24). Patient C had a buffy coat sample with a ratio precisely indicating a single chromosomal integration.
Table 2.
Residual clinical cellular samples tested for ciHHV-6
| Patient | Specimen | Sample type | HHV-6/cell |
|---|---|---|---|
| A | 1 | Buffy coat | 2.07 |
| B | 1 | Pre-transplant PBMC | 0 |
| B | 2 | Donor PBMC* | 0 |
| B | 3 | Buffy coat* | 0.03 |
| B | 4 | Bone marrow* | 0.03 |
| B | 5 | Liver biopsy | 1.2 |
| C | 1 | Buffy coat | 0.99 |
indicates donor derived specimen
Suitability of plasma samples for ddPCR ciHHV-6 analysis
Identification of ciHHV-6 status from plasma samples is of interest to researchers working from sample registries, many of which only save plasma. Until now the field has assumed that ciHHV-6 status cannot be determined from plasma because the integrated form of HHV-6 resides in cells. However, cellular DNA is known to be released into plasma due to cell lysis during sample processing and handling.(6) We therefore asked whether the ddPCR assay might be applicable to plasma samples.
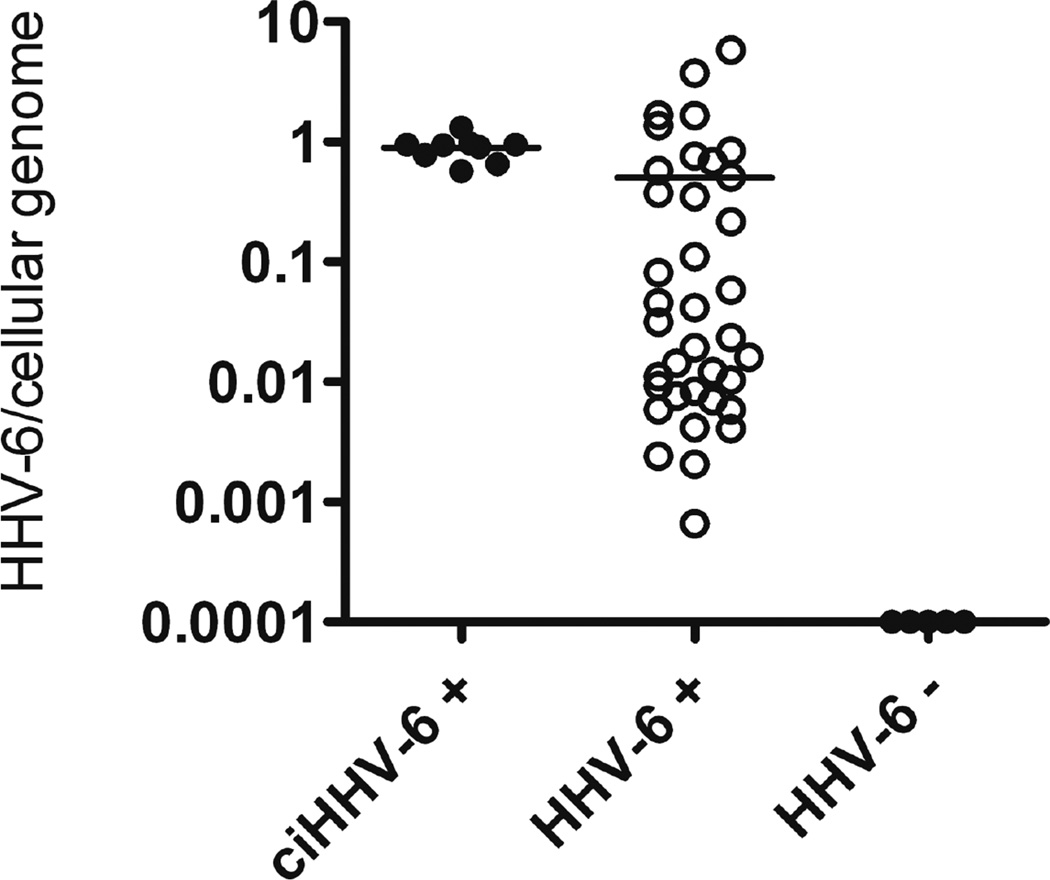
To evaluate the performance of the assay in plasma, we determined the HHV-6/cell ratios in plasma samples from the 9 available ciHHV-6 patients (6 from the ciHHV-6 registry, 3 from the HCT study). The two FISH-confirmed ciHHV-6 patients who contributed buffy coat samples described earlier also contributed plasma specimens. Both of these plasma specimens resulted in ratios close to one (0·72, 0·95), but as predicted results were not as precise as with the cellular samples. It is worth noting that the real-time PCR results in plasma from the FISH-confirmed ciHHV-6 patients were in the 1,000–2,000 copies/ml range. Such levels can be observed in active HHV-6 infection without ciHHV-6. Thus, if a physician were presented with a plasma viral load at this level without prior knowledge of the patient’s ciHHV-6 status, the patient could be administered unnecessary treatment for active HHV-6 infection. Plasma specimens from the three ciHHV-6 positive HCT patients also resulted in ratios close to 1 HHV-6/cell (Table 1, plasma samples, Figure 4). Based on this data, absolute log10 ratios from 0–0·25, corresponding to 0·56–1·78 HHV-6/cell, were identified as the range in which ciHHV-6 would be suspected from tests on plasma samples (Figure 3), chosen in order to achieve 100% sensitivity among these persons.
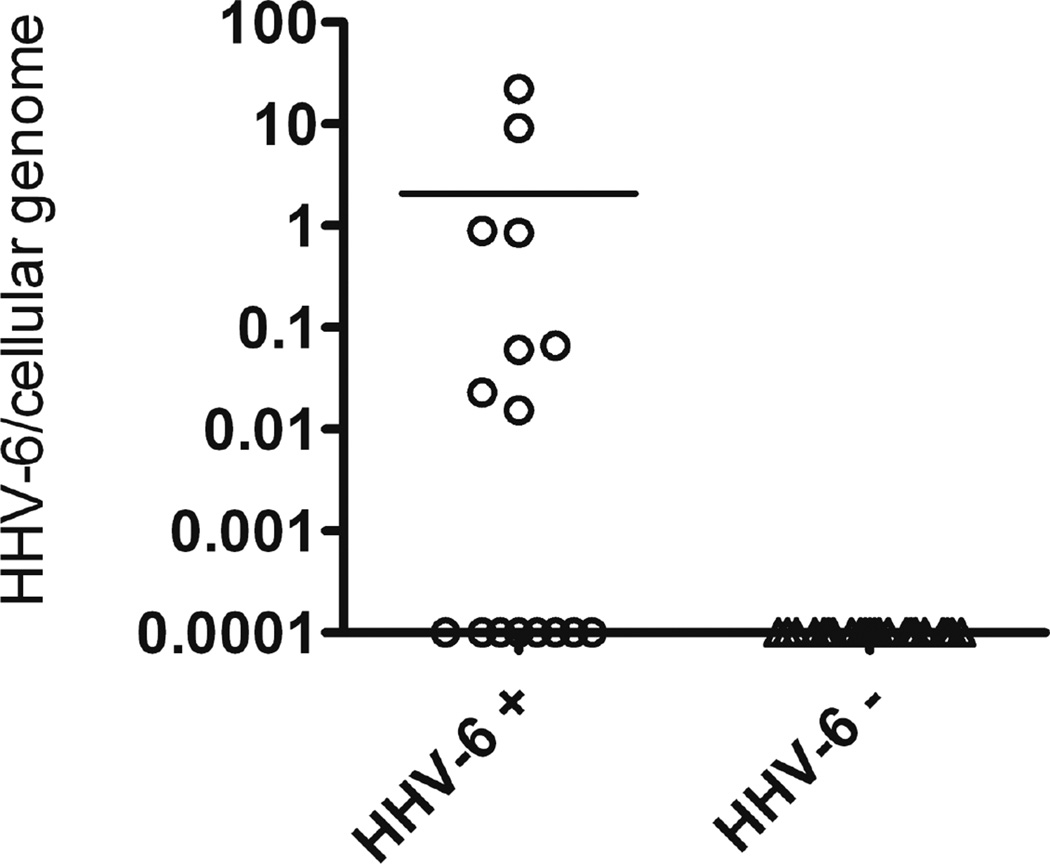
Figure 4.
HHV-6/cell ddPCR results for plasma samples from residual clinical specimens that were leftover from routine qPCR testing for HHV-6 viral load and were positive by qPCR (HHV-6+). Negative control samples (HHV-6 −) are leftover samples from routine qPCR testing for Human Cytomegalovirus (CMV).
Figure 3.
HHV-6/cell ddPCR results for plasma samples from hematopoietic stem cell transplant study patients with ciHHV-6 (ciHHV-6 +), reactivated HHV-6 infection (HHV-6 +), or no HHV-6 reactivation (HHV-6−).
To investigate the specificity of the assay on plasma, we used control patients with active HHV-6 infection and without ciHHV-6 (Figure 3). The control patients were selected from the population of patients in an HCT study(20, 21) who reactivated HHV-6 but were negative for ciHHV-6 as determined by subsequent negative results on longitudinal real-time PCR monitoring, or who were presumed ciHHV-6 negative because they never had positive HHV-6 PCR results on longitudinal testing. From the cohort of 38 available reactivation patients, 7 had ratios outside our cutoff of 0·56–1·78. Additionally, 5 specimens from 5 non-reactivators were tested and negative. Based on this, we calculate a specificity of the plasma assay for ciHHV-6 of 82% ± 12%.
We further investigated the use of the ddPCR assay on plasma by testing sixteen residual plasma specimens that previously tested positive for HHV-6 by routine clinical real-time PCR testing but whose ciHHV6 status was unconfirmed. Fourteen of these patients were ciHHV-6 negative according to our cutoff range of 0·56–1·78 (Figure 4). Based on the reported population frequency of ciHHV-6 (one percent(4–6)) we would expect none of the 16 patients to be positive for ciHHV-6. Using this assumption, the specificity of the assay in plasma would be 88%, which falls within the range of our previous results.
Discussion
We have developed a rapid, precise assay for identifying patients with a chromosomally integrated and heritable form of HHV-6. The assay utilizes droplet digital PCR technology (Figure 1A) to identify integrations from cellular samples by precisely assaying the ratio between HHV-6 and cellular genomic DNA (Figure 1B). A similar method for detecting ciHHV-6 by real-time PCR was described by Ward and colleagues, which relied on quantitation of HHV-6 in tandem with quantitation of β-globin to determine cell number (or genome equivalent copy number).(2) However, ratiometric assays utilizing real-time PCR are problematic because of inherent assay imprecision(14), which results in a broad distribution of ratios. As demonstrated here, the extreme precision of ddPCR provides highly precise HHV-6/cell ratios, and thus avoids this problem.
Our assay has been validated on specimens confirmed for chromosomal integration by fluorescence in situ hybridization, the current diagnostic gold standard for ciHHV-6 (Figure 2). Notably, in these FISH-confirmed cases, plasma HHV-6 viral loads by real-time PCR were lower than the levels that have been suggested for suspicion of ciHHV-6 in acellular fluids (>3·5 log10)(9), calling attention to the fact that an HHV-6 real-time PCR result alone cannot identify ciHHV-6. The need for a more robust, rapid clinical assay for ciHHV-6 is met by utilizing ddPCR, which provides a precise ratio of HHV-6/cellular DNA. For clinical use, this ddPCR assay should be performed on patient buffy coat samples obtained from whole blood to identify patients with ciHHV-6.
The clinical significance of ciHHV-6 is an area of active research. Several recent reports have suggested links between ciHHV-6 and long-term sequelae;(22, 25–29) however, no larger studies have systematically examined how often clinical disease occurs in persons with ciHHV-6. In addition to preventing misdiagnosis of active HHV-6 infection, our ddPCR assay should also help identify individuals with ciHHV-6 to aid in determining how this condition may contribute to disease states. The assay could also prove useful in the transplant setting, as data from the HCT study population presented here highlights the need for screening patients and potential donors for ciHHV-6.
Our ddPCR assay is a powerful tool for rapid distinction of active infection from ciHHV-6 in cellular samples, but caution must be taken with certain samples. Specimens with low genomic DNA content, such as the liver biopsy sample described in Table 2, may yield imprecise ratios that make distinguishing ciHHV-6 from active infection difficult. In cases such as these, it is advisable to obtain additional tissue or cellular samples to determine the likelihood of ciHHV-6. Additionally, in rare cases a patient may have multiple HHV-6 genomic integrations, such as in Patient A (Table 2). Although only one case has been published,(23) with a high quality cellular sample a ratio of 2 HHV6/cell is likely indicative of ciHHV-6.
Beyond its clear clinical utility when using cellular samples, we have also shown our ddPCR assay is suitable for plasma samples when cellular specimens are not available, such as for large retrospective studies, where plasma is often the only specimen stored long term. Based on our estimates of sensitivity (100%), specificity (82±12%) and the prevalence in the overall population (1%), the negative predictive value (NPV) of the plasma test (using an absolute log10 ratio <0.25) is estimated at 100%, and thus the test is ideal for ruling out ciHHV-6. In contrast, the positive predictive value (PPV), the proportion of positive results that are true positives would only be 5–8% in the general population (although it would be substantially higher in populations selected for a higher pre-test probability of ciHHV-6); thus, the plasma test would be best suited for screening specimens for additional follow up. We should note that the number of cellular genomes in an acellular specimen such as plasma varies according to length of time between specimen collection and fractionation, so the ratio cutoffs described here may not be directly transferable to other laboratories. Finally, for clinical testing, we would reemphasize that the optimal specimen type is buffy coat cells from whole blood, which permit the definitive identification of ciHHV-6. This assay is the first clinically available viral diagnostic test that utilizes ddPCR and paves the path for additional genetic diagnostic assays based on digital PCR technology.
Summary.
Chromosomally integrated Human Herpesvirus 6 (ciHHV-6), present in about 1% of the general population, complicates diagnosis of active HHV-6 infection by standard molecular quantitative methods. We describe the design and application of a digital PCR assay for rapid clinical identification of ciHHV-6. The assay is validated for utilizing cellular specimens that previously tested positive for ciHHV-6 by the current gold-standard FISH assay. In addition, the assay provides excellent sensitivity and specificity using stored plasma samples. A rapid molecular test for ciHHV-6 may be particularly useful in the transplant setting and will facilitate retrospective analysis of the clinical significance of ciHHV-6.
Acknowledgements
We thank David Myerson for providing FFPE patient samples. We thank Kristin Loomis and the HHV-6 Foundation for orchestrating collaborations and generously providing patient specimens from their ciHHV-6 registry. Bio-Rad Laboratories provided funding and reagents for this work. This work was supported by the NIH under award numbers R01AI57639, U19 AI096111, and CA18029. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Zerr DM. Human herpesvirus 6 (hhv-6) disease in the setting of transplantation. Curr Opin Infect Dis. 2012;25:438–444. doi: 10.1097/QCO.0b013e3283553362. [DOI] [PubMed] [Google Scholar]
- 2.Ward KN, Leong HN, Nacheva EP, Howard J, Atkinson CE, Davies NW, et al. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral dna in blood, sera, and hair follicles. J Clin Microbiol. 2006;44:1571–1574. doi: 10.1128/JCM.44.4.1571-1574.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Razonable RR, Lautenschlager I. Impact of human herpes virus 6 in liver transplantation. World J Hepatol. 2010;2:345–353. doi: 10.4254/wjh.v2.i9.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward KN. The natural history and laboratory diagnosis of human herpesviruses-6 and -7 infections in the immunocompetent. J Clin Virol. 2005;32:183–193. doi: 10.1016/j.jcv.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Leong HN, Tuke PW, Tedder RS, Khanom AB, Eglin RP, Atkinson CE, et al. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of uk blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 6.Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, et al. Chromosomally integrated human herpesvirus 6: Questions and answers. Rev Med Virol. 2012;22:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, et al. The latent human herpesvirus-6a genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci U S A. 2010;107:5563–5568. doi: 10.1073/pnas.0913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flamand L, Komaroff AL, Arbuckle JH, Medveczky PG, Ablashi DV. Review, part 1: Human herpesvirus-6-basic biology, diagnostic testing, and antiviral efficacy. J Med Virol. 2010;82:1560–1568. doi: 10.1002/jmv.21839. [DOI] [PubMed] [Google Scholar]
- 9.Potenza L, Barozzi P, Torelli G, Luppi M. Translational challenges of human herpesvirus 6 chromosomal integration. Future Microbiol. 2010;5:993–995. doi: 10.2217/fmb.10.74. [DOI] [PubMed] [Google Scholar]
- 10.Caserta MT, Hall CB, Schnabel K, Lofthus G, Marino A, Shelley L, et al. Diagnostic assays for active infection with human herpesvirus 6 (hhv-6) J Clin Virol. 2010;48:55–57. doi: 10.1016/j.jcv.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SO, Brown RA, Razonable RR. Chromosomally integrated human herpesvirus-6 in transplant recipients. Transpl Infect Dis. 2012;14:346–354. doi: 10.1111/j.1399-3062.2011.00715.x. [DOI] [PubMed] [Google Scholar]
- 12.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital pcr system for absolute quantitation of dna copy number. Anal Chem. 2011;83:8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sedlak RH, Jerome KR. Viral diagnostics in the era of digital polymerase chain reaction. Diagn Microbiol Infect Dis. 2013;75:1–4. doi: 10.1016/j.diagmicrobio.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, et al. Absolute quantification by droplet digital pcr versus analog real-time pcr. Nat Methods. 2013 doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huggett JF, Foy CA, Benes V, Emslie K, Garson JA, Haynes R, et al. The digital miqe guidelines: Minimum information for publication of quantitative digital pcr experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 16.Zerr DM, Gooley TA, Yeung L, Huang ML, Carpenter P, Wade JC, et al. Human herpesvirus 6 reactivation and encephalitis in allogeneic bone marrow transplant recipients. Clin Infect Dis. 2001;33:763–771. doi: 10.1086/322642. [DOI] [PubMed] [Google Scholar]
- 17.Zerr DM, Gupta D, Huang ML, Carter R, Corey L. Effect of antivirals on human herpesvirus 6 replication in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2002;34:309–317. doi: 10.1086/338044. [DOI] [PubMed] [Google Scholar]
- 18.Dingle TC, Sedlak RH, Cook L, Jerome KR. Tolerance of droplet-digital pcr vs real-time quantitative pcr to inhibitory substances. Clin Chem. 2013;59:1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinheiro LB, Coleman VA, Hindson CM, Herrmann J, Hindson BJ, Bhat S, Emslie KR. Evaluation of a droplet digital polymerase chain reaction format for dna copy number quantification. Anal Chem. 2012;84:1003–1011. doi: 10.1021/ac202578x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zerr DM, Fann JR, Breiger D, Boeckh M, Adler AL, Xie H, et al. Hhv-6 reactivation and its effect on delirium and cognitive functioning in hematopoietic cell transplantation recipients. Blood. 2011;117:5243–5249. doi: 10.1182/blood-2010-10-316083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerr DM, Boeckh M, Delaney C, Martin PJ, Xie H, Adler AL, et al. Hhv-6 reactivation and associated sequelae after hematopoietic cell transplantation. Biol Blood Marrow Transplant. 2012;18:1700–1708. doi: 10.1016/j.bbmt.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montoya JG, Neely MN, Gupta S, Lunn MR, Loomis KS, Pritchett JC, et al. Antiviral therapy of two patients with chromosomally-integrated human herpesvirus-6a presenting with cognitive dysfunction. J Clin Virol. 2012;55:40–45. doi: 10.1016/j.jcv.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 23.Daibata M, Taguchi T, Nemoto Y, Taguchi H, Miyoshi I. Inheritance of chromosomally integrated human herpesvirus 6 dna. Blood. 1999;94:1545–1549. [PubMed] [Google Scholar]
- 24.Hill JA, Myerson D, Sedlak RH, Jerome KR, Zerr DM. Hepatitis due to human herpesvirus 6b after hematopoietic stem cell transplantation and a review of the literature. Vol. Transplant Infectious Disease. doi: 10.1111/tid.12208. Accepted 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amirian ES, Scheurer ME. Chromosomally-integrated human herpesvirus 6 in familial glioma etiology. Med Hypotheses. 2012;79:193–196. doi: 10.1016/j.mehy.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Troy SB, Blackburn BG, Yeom K, Caulfield AK, Bhangoo MS, Montoya JG. Severe encephalomyelitis in an immunocompetent adult with chromosomally integrated human herpesvirus 6 and clinical response to treatment with foscarnet plus ganciclovir. Clin Infect Dis. 2008;47:e93–e96. doi: 10.1086/593315. [DOI] [PubMed] [Google Scholar]
- 27.Wittekindt B, Berger A, Porto L, Vlaho S, Grüttner HP, Becker M, Lehrnbecher T. Human herpes virus-6 dna in cerebrospinal fluid of children undergoing therapy for acute leukaemia. Br J Haematol. 2009;145:542–545. doi: 10.1111/j.1365-2141.2009.07641.x. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi D, Kogawa K, Imai K, Tanaka T, Hiroi S, Satoh H, et al. Quantitation of human herpesvirus-6 (hhv-6) dna in a cord blood transplant recipient with chromosomal integration of hhv-6. Transpl Infect Dis. 2011;13:650–653. doi: 10.1111/j.1399-3062.2011.00693.x. [DOI] [PubMed] [Google Scholar]
- 29.Pantry S, Medveczky M, Arbuckle J, Luka J, Montoya J, Hu J, et al. Persistent human herpesvirus-6 infection in patients with an inherited form of the virus. Vol. Journal of Medical Virology. doi: 10.1002/jmv.23685. [DOI] [PMC free article] [PubMed] [Google Scholar]