Abstract
We consider the problem of using high dimensional data residing on graphs to predict a low-dimensional outcome variable, such as disease status. Examples of data include time series and genetic data measured on linear graphs and imaging data measured on triangulated graphs (or lattices), among many others. Many of these data have two key features including spatial smoothness and intrinsically low dimensional structure. We propose a simple solution based on a general statistical framework, called spatially weighted principal component regression (SWPCR). In SWPCR, we introduce two sets of weights including importance score weights for the selection of individual features at each node and spatial weights for the incorporation of the neighboring pattern on the graph. We integrate the importance score weights with the spatial weights in order to recover the low dimensional structure of high dimensional data. We demonstrate the utility of our methods through extensive simulations and a real data analysis based on Alzheimer’s disease neuroimaging initiative data.
Keywords: Graph, Principal component analysis, Regression, Spatial, Supervise, Weight
1 Introduction
Our problem of interest is to predict a set of response variables Y by using high-dimensional data x = {xg : g ∈
 } measured on a graph ζ = (
} measured on a graph ζ = (
 ,
,
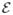 ), where
), where
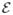 is the edge set of ζ and
is the edge set of ζ and
 = {g1, …, gm} is a set of vertexes, in which m is the total number of vertexes in
= {g1, …, gm} is a set of vertexes, in which m is the total number of vertexes in
 . The response Y may include cognitive outcome, disease status, and the early onset of disease, among others. Standard graphs including both directed and undirected graphs have been widely used to build complex patterns [10]. Examples of graphs are linear graphs, tree graphs, triangulated graphs, and 2-dimensional (2D) (or 3-dimensional (3D)) lattices, among many others (Figure 1). Examples of x on the graph ζ = (
. The response Y may include cognitive outcome, disease status, and the early onset of disease, among others. Standard graphs including both directed and undirected graphs have been widely used to build complex patterns [10]. Examples of graphs are linear graphs, tree graphs, triangulated graphs, and 2-dimensional (2D) (or 3-dimensional (3D)) lattices, among many others (Figure 1). Examples of x on the graph ζ = (
 ,
,
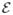 ) include time series and genetic data measured on linear graphs and imaging data measured on triangulated graphs (or lattices). Particularly, various structural and functional neuroimaging data are frequently measured in a 3D lattice for the understanding of brain structure and function and their association with neuropsychiatric and neurodegenerative disorders [9].
) include time series and genetic data measured on linear graphs and imaging data measured on triangulated graphs (or lattices). Particularly, various structural and functional neuroimaging data are frequently measured in a 3D lattice for the understanding of brain structure and function and their association with neuropsychiatric and neurodegenerative disorders [9].
Fig. 1.
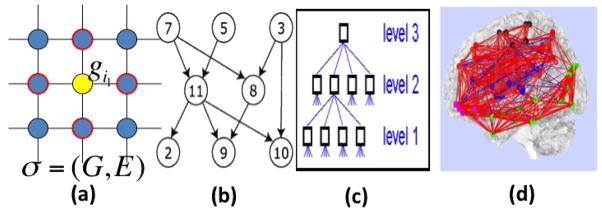
Illustration of graph data structure ζ = (
 ,
,
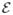 ): (a) two-dimensional lattice; (b) acyclic directed graph; (c) tree; (d) undirected graph.
): (a) two-dimensional lattice; (b) acyclic directed graph; (c) tree; (d) undirected graph.
The aim of this paper is to develop a new framework of spatially weighted principal component regression (SWPCR) to use x on graph ζ = {
 ,
,
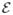 } to predict Y. Four major challenges arising from such development include ultra-high dimensionality, low sample size, spatially correlation, and spatial smoothness. SWPCR is developed to address these four challenges when high-dimensional data on graphs ζ share two important features including spatial smoothness and intrinsically low dimensional structure. Compared with the existing literature, we make several major contributions as follows:
} to predict Y. Four major challenges arising from such development include ultra-high dimensionality, low sample size, spatially correlation, and spatial smoothness. SWPCR is developed to address these four challenges when high-dimensional data on graphs ζ share two important features including spatial smoothness and intrinsically low dimensional structure. Compared with the existing literature, we make several major contributions as follows:
(i) SWPCR is designed to efficiently capture the two important features by using some recent advances in smoothing methods, dimensional reduction methods, and sparse methods.
(ii) SWPCR provides a powerful dimension reduction framework for integrating feature selection, smoothing, and feature extraction.
(iii) SWPCR significantly outperforms the competing methods by simulation studies and the real data analysis.
2 Spatially Weighted Principal Component Regression
In this section, we first describe the graph data that are considered in this paper. We formally describe the general framework of SWPCR.
2.1 Graph Data
Consider data from n independent subjects. For each subject, we observe a q × 1 vector of discrete or continuous responses, denoted by yi = (yi,1, …, yi,q)T, and a m × 1 vector of high dimensional data xi = {xi,g : g ∈
 } for i = 1, …, n. In many cases, q is relatively small compared with n, whereas m is much larger than n. For instance, in many neuroimaging studies, it is common to use ultra-high dimensional imaging data to classify a binary class variable. In this case, q = 1, whereas m can be several million number of features. In many applications,
} for i = 1, …, n. In many cases, q is relatively small compared with n, whereas m is much larger than n. For instance, in many neuroimaging studies, it is common to use ultra-high dimensional imaging data to classify a binary class variable. In this case, q = 1, whereas m can be several million number of features. In many applications,
 = {g1, …, gm} is a set of prefixed vertexes, such as voxels in 2D or 3D lattices, whereas the edge set
= {g1, …, gm} is a set of prefixed vertexes, such as voxels in 2D or 3D lattices, whereas the edge set
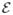 may be either prefixed or determined by xi (or other data).
may be either prefixed or determined by xi (or other data).
2.2 SWPCR
We introduce a three-stage algorithm for SWPCR to use high-dimensional data x to predict a set of response variables Y. The key stages of SWPCR can be described as follows.
Stage 1. Build an importance score vector (or function) WI:
 → R+ and the spatial weight matrix (or function) WE:
→ R+ and the spatial weight matrix (or function) WE:
 ×
×
 → R.
→ R.Stage 2. Build a sequence of scale vectors {s0 = (sE,0, sI,0), ···, sL = (sE,L, sI,L)} ranging from the smallest scale vector s0 to the largest scale vector sL. At each scale vector sℓ, use generalized principal component analysis (GPCA) to compute the first few principal components of an n × m matrix X = (x1 ··· xn)T, denoted by A(sℓ), based on WE(·, ·) and WI(·) for ℓ = 0, …, L.
Stage 3. Select the optimal 0 ≤ ℓ* ≤ L and build a prediction model (e.g., high-dimensional linear model) based on the extracted principal components A(sℓ*) and the responses Y.
We slightly elaborate on these stages. In Stage 1, the important scores wI,g play an important feature screening role in SWPCR. Examples of wI,g = WI(g) in the literature can be generated based on some statistics (e.g., Pearson correlation or distance correlation) between xg and Y at each vertex g. For instance, let p(g) be the Pearson correlation at each vertex g and then define
| (1) |
In Stage 1, without loss of generality, we focus on the symmetric matrix WE = (wE,gg′) ∈ Rp×p throughout the paper. The element wE,gg′ is usually calculated by using various similarity criteria, such as Gaussian similarity from Euclidean distance, local neighborhood relationship, correlation, and prior information obtained from other data [21]. In Section 2.3, we will discuss how to determine WE and WI while explicitly accounting for the complex spatial structure among different vertexes.
In Stage 2, at each scale vector sℓ = (sE,ℓ, sI,ℓ), we construct two matrices, denoted by QE,ℓ and QI,ℓ based on WE and WI as follows:
| (2) |
where F1 : Rp×p × R+ → Rp×p and F2 : Rp × R+ → Rp are two known functions. For instance, let 1(·) be an indicator function, we may set
| (3) |
to extract ‘significant’ vertexes. There are various ways of constructing QE,ℓ. For instance, one may set QE,ℓ as
where sE,ℓ = (sE,ℓ;1, sE,ℓ;2)T and D(g, g′) is a graph-based distance between vertexes g and g′. The value of sE,ℓ;2 controls the number of vertexes in {g′ ∈
 : D(g, g′) ≤ sE,ℓ;2}, which is a patch set at vertex g [18], whereas sE,ℓ;1 is used to shrink small |wE,gg′|s into zero.
: D(g, g′) ≤ sE,ℓ;2}, which is a patch set at vertex g [18], whereas sE,ℓ;1 is used to shrink small |wE,gg′|s into zero.
After determining QE,ℓ and QI,ℓ, we set and Σr = In for independent subjects. Let X̃ be the centered matrix of X. Then we can extract K principal components through minimize the following objective function given by
| (4) |
If we consider correlated observations from multiple subjects, we may use Σr to explicitly model their correlation structure. The solution (Uℓ, Dℓ, Vℓ) of the objective function (4) at sℓ is the SVD of X̃R,ℓ = X̃QE,ℓQI,ℓ. The we can use a GPCA algorithm to simultaneously calculate all components of (Uℓ, Dℓ, Vℓ) for a fixed K as follows. In practice, a simple criterion for determining K is to include all components up to some arbitrary proportion of the total variance, say 85%.
For ultra-high dimensional data, we consider a regularized GPCA to generate (Uℓ, Dℓ, Vℓ) by minimizing the following objective function
| (5) |
subject to and for all k, where uk,ℓ and vk,ℓ are respectively the k-th column of Uℓ and Vℓ. We use adaptive Lasso penalties for P1(·) and P2(·) and then iteratively solve (5) [1]. For each k0, we define and minimize
| (6) |
subject to and . By using the sparse method in [12], we can calculate the solution of (6), denoted by (d̂k0,ℓ, ûk0,ℓ, v̂k0,ℓ). In this way, we can sequentially compute (d̂k,ℓ, ûk,ℓ, v̂k,ℓ) for k = 1, …;, K.
In Stage 3, select ℓ* as the minimum point of the objective function (5) or (6). let and then K principal components A(sℓ*) = XQF,ℓ*. Moreover, K is usually much smaller than min(n, m). Then, we build a regression model with yi as responses and Ai (the i-th row of A(sℓ*)) as covariates, denoted by R(yi, Ai; θ), where θ is a vector of unknown (finite-dimensional or nonparametric) parameters. Specifically, based on {(yi, Ai)}i≥1, we use an estimation method to estimate θ as follows:
where ρ(·, ·, ·) is a loss function, which depends on both the regression model and the data, and P3(·) is a penalty function, such as Lasso. This leads to a prediction model R(yi, Ai; θ). For instance, for binary response yi = 1 or 0, we may consider a sparse logistic model given by for R(yi, Ai; θ).
Given a test feature vector x*, we can do predictions from our prediction model as follows:
Center each component of x* by calculating x̃* = x* − μ̂x, in which μ̂x is the mean and learnt from the training data;
Optimize an objective function based on R(y, x̃*TQF,ℓ*; θ̂) to calculate an estimate of y, denoted by ŷ*.
Our prediction model is applicable to various regression settings for continuous and discrete responses and multivariate and univariate responses, such as survival data and classification problems.
2.3 Importance Score Weights and Spatial Weights
There are two sets of weights in SWPCR including (i) importance score weights enabling a selective treatment for individual features, and (ii) spatial weights accommodating the underlying spatial dependence among features across neighboring vertexes on graph. Below, we propose the strategy of determining both importance score weights and spatial weights.
Importance Score Weights
As discussed in Section 2.3, at each vertex g, wI,g, such as the Pearson correlation in (1), is calculated based on a statistical model between xg and Y in order to perform feature selection according to each feature’s discriminative importance. Statistically, most existing methods use a marginal (or vertex-wise) model by assuming
where β = (β(g): g ∈
 ) and β(g) is introduced to quantify the association between yi and xi,g at each vertex g ∈
) and β(g) is introduced to quantify the association between yi and xi,g at each vertex g ∈
 . At the g–th vertex, wI,g is a statistic based on the marginal model
. However, those wI,gs largely ignore complex spatial structure, such as homogenous patches defined below, across all vertexes on graph.
. At the g–th vertex, wI,g is a statistic based on the marginal model
. However, those wI,gs largely ignore complex spatial structure, such as homogenous patches defined below, across all vertexes on graph.
For a graph ζ = (
 ,
,
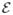 ), it is common to assume that β(g) across all vertexes are naturally clustered into P homogeneous patches, denoted by {
), it is common to assume that β(g) across all vertexes are naturally clustered into P homogeneous patches, denoted by {
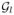 : l = 1, …, P}, such that P ≪ m,
, and β(g) varies smoothly in each
: l = 1, …, P}, such that P ≪ m,
, and β(g) varies smoothly in each
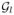 . Note that a patch
. Note that a patch
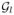 consists of a set of vertexes that are completely connected through edges in
consists of a set of vertexes that are completely connected through edges in
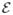 . That is, if g, g′ ∈
. That is, if g, g′ ∈
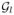 , then there is a sequence of vertexes g0 = g, ···, gM = g′ in
, then there is a sequence of vertexes g0 = g, ···, gM = g′ in
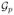 such that (gj−1, gj) ∈
such that (gj−1, gj) ∈
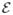 for all j = 1, …, M. It has been shown that for graph data, algorithms based on patch information have led to state-of-the art techniques for classification and denoising. See for example, [18] for overviews of imaging patches.
for all j = 1, …, M. It has been shown that for graph data, algorithms based on patch information have led to state-of-the art techniques for classification and denoising. See for example, [18] for overviews of imaging patches.
We propose the strategy to jointly model xi and yi and simultaneously calculate wI,g across all vertexes, while learning the homogenous patches
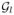 . The strategy is to model the conditional distribution of xi given yi, denoted by p(xi|yi, β). Then we can learn the patches
. The strategy is to model the conditional distribution of xi given yi, denoted by p(xi|yi, β). Then we can learn the patches
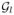 in
in
 from the estimated β.
from the estimated β.
Here we consider a set of vertexes
 with unknown edge information
with unknown edge information
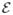 . It is important to learn the homogeneous patches
. It is important to learn the homogeneous patches
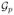 and then form the edge set
and then form the edge set
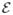 . Let
. Let
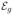 (h) be an edge set at scale h at each vertex g. We consider a sequence of nested edge sets across multiscales hs such that h0 = 0 ≤ h1 ≤ ··· ≤ hS and
(h) be an edge set at scale h at each vertex g. We consider a sequence of nested edge sets across multiscales hs such that h0 = 0 ≤ h1 ≤ ··· ≤ hS and
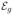 (h0) = {g} ⊂ ··· ⊂
(h0) = {g} ⊂ ··· ⊂
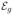 (hS). To learn the homogeneous patches, a general framework of Multiscale Adaptive Regression Model (MARM) developed in [13] is to maximize a sequence of weighted functions as follows:
(hS). To learn the homogeneous patches, a general framework of Multiscale Adaptive Regression Model (MARM) developed in [13] is to maximize a sequence of weighted functions as follows:
| (7) |
where ω(g, g′; h) characterizes the similarity between the data in vertexes g′ and g with ω(g, g; h) = 1. If ω(g, g′; h) ≈ 0, then the observations in vertex g′ do not provide information on β(g). Therefore, ω(g, g′; h) can prevent incorporation of vertexes whose data do not contain information on β(g) and preserve the edges of homogeneous regions. Let D1(g, g′) and D2(β̂(g; hs−1), β̂(g′; hs−1)) be, respectively, the spatial distance between vertexes g and g′ and a similarity measure between β̂(g; hs−1) and β̂(g′; hs−1). The ω(g, g′; hs) can be defined as
| (8) |
where
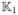 (·) and
(·) and
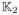 (·) are two nonnegative kernel functions and γn is a bandwidth parameter that may depend on n. See the detailed algorithm of MARM in [13]. After the iteration hs, we can obtain β̂(g; hS) and its covariance matrix, denoted by Cov(β̂(g; hs)), across all g ∈
(·) are two nonnegative kernel functions and γn is a bandwidth parameter that may depend on n. See the detailed algorithm of MARM in [13]. After the iteration hs, we can obtain β̂(g; hS) and its covariance matrix, denoted by Cov(β̂(g; hs)), across all g ∈
 and ω(g, g′; hs) for all g′ ∈
and ω(g, g′; hs) for all g′ ∈
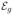 (hs) and g ∈
(hs) and g ∈
 . Finally, we calculate statistics wI,g based on β̂(g; hs) and Cov(β̂(g; hs)), such as the Wald test, and then we use a clustering algorithm, such as the K-mean algorithm, to group {β̂(g; hs): g ∈
. Finally, we calculate statistics wI,g based on β̂(g; hs) and Cov(β̂(g; hs)), such as the Wald test, and then we use a clustering algorithm, such as the K-mean algorithm, to group {β̂(g; hs): g ∈
 } into several homogeneous clusters, in which β̂(g; hs) varies very smoothly in each cluster. Moreover, each homogenous cluster can be a union of several homogeneous patches.
} into several homogeneous clusters, in which β̂(g; hs) varies very smoothly in each cluster. Moreover, each homogenous cluster can be a union of several homogeneous patches.
Spatial Weights
As discussed in Section 2.3, wE,gg′ often characterizes the degree of certain ‘similarity’ between vertexes g and g′. The locally spatial weighting matrix consists of non-negative weights assigned to the spatial neighboring vertexes of each vertex. It is assumed that
| (9) |
in which ω(g, g′; hs) is defined in (8). Therefore, wE,gg′= 0 for all g′ ∉
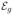 (hs) and
(hs) and
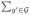 wE,gg′ = 1. The weights
wE,gg′ = 1. The weights
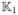 (D1(g, g′)/hs) give less weight to vertex g′ ∈
(D1(g, g′)/hs) give less weight to vertex g′ ∈
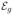 (hs), whose location is far from the vertex g. The weights
(hs), whose location is far from the vertex g. The weights
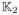 (u) down-weight the vertex g′ with large D2(β̂(g; hs), β̂(g′; hs)), which indicates a large difference between β̂(g′; hs) and β̂(g; hs). Moreover, by following [4, 13, 15, 16], we set
(u) down-weight the vertex g′ with large D2(β̂(g; hs), β̂(g′; hs)), which indicates a large difference between β̂(g′; hs) and β̂(g; hs). Moreover, by following [4, 13, 15, 16], we set
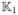 (x) = (1−x)+ and
(x) = (1−x)+ and
 (x) = exp(−x). Although m is often much larger than n, the computational burden associated with the local spatial weights is very minor when hs is relatively small.
(x) = exp(−x). Although m is often much larger than n, the computational burden associated with the local spatial weights is very minor when hs is relatively small.
3 Simulation Study
In this section, we conducted one set of simulation study corresponding to binary responses, in order to examine the finite-sample performance of SWPCR in the high-dimensional classification analysis. We demonstrate that SWPCR outperforms many state-of-the-art methods for at least in the simulated dataset.
We simulated 20 × 20 × 10 (x × y × z) 3D-images from a linear model given by
| (10) |
where yi is the class label coded as either 0 or 1 and εi(g) are random variables with zero mean. The true mean images of class yi = 0 and class yi = 1 are shown in Figure 2. Voxels in the red cuboid region have the maximum difference 1 between classes 0 and 1. The dimension of red cuboid is 3 × 3 × 4 and contains 36 voxels. In this case, m = 4, 000 and we set n = 100 with 60 images from Class 0 and the rest from Class 1. We consider three types of noise εi(g) in (10). First, were independently generated from a N(0, 22) generator across all voxels g. Second, were generated from in order to introduce the short range spatial correlation, where mg is the number of voxels in the set {||g′ − g|| ≤ 1}. Third, to introduce long range spatial correlation, were generated according to , where ξi,k for k = 1, 2, 3 were independenly generated from a N(0, 1) generator. Moreover, the noise variances in all voxels of the red cuboid region equal 4, 4/6, and 4{sin(πg1/10)2 +cos(πg2/10)2 +sin(πg3/5)2} +4 for Type I, II, and III noises, respectively. Therefore, among the three types of noise, Type III noise has the smallest signal-to-noise ratio and Type II noise has the largest one.
Fig. 2.
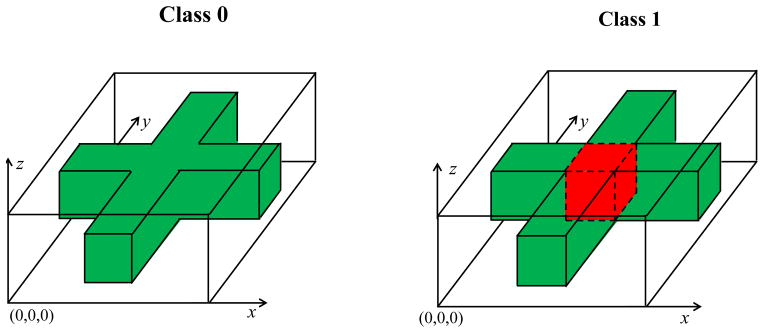
True mean images for the simulation study: Class 0 in the left panel and Class 1 in the right panel. The white, green, and red colors, respectively, correspond to 0, 1, and 2.
We ran the three stages of SWPCR as follows. In Stage 1, let {hℓ = 1.2ℓ, ℓ = 0, 1, …, S = 5}, and for each g ∈
 , wI,g = −m log(p(g))/[−
, wI,g = −m log(p(g))/[−
 log(p(g)), where p(g) is the p-value of Wald test B1(g) = 0 in (7) (β(g) = (B0(g), B1(g))T) for each voxel g. The spatial weight WE is given by (9). Here we haven’t used the simple Pearson correlation (1) for computing weights because it neglects the spatial correlation of the data set. In Stage 2, for each hℓ, we define QE,ℓ = WE and generate QI,ℓ through (2) and (3), where sI,ℓ thresholds out the wI,g with p(g) < 0.01. Then we extract different K principal components of GPCA to reconstruct the low dimensional representations of simulated images and then do classification analysis. The results are very stable for different number of principal components and here we let K = 5. In Stage 3, we tried different classification methods, including linear regression, k-Nearest Neighbor (k-NN) [11] and support vector machine (SVM) [14], on these low dimensional spaces. Based on the misclassification error for the leave-one-out cross validation, the linear regression is slight better than others. The linear regression uses class label yi as dependent variable and principal components as explanatory variables. If the prediction value is less than 0, the image is classified as 0. Otherwise, the image is classified as 1.
log(p(g)), where p(g) is the p-value of Wald test B1(g) = 0 in (7) (β(g) = (B0(g), B1(g))T) for each voxel g. The spatial weight WE is given by (9). Here we haven’t used the simple Pearson correlation (1) for computing weights because it neglects the spatial correlation of the data set. In Stage 2, for each hℓ, we define QE,ℓ = WE and generate QI,ℓ through (2) and (3), where sI,ℓ thresholds out the wI,g with p(g) < 0.01. Then we extract different K principal components of GPCA to reconstruct the low dimensional representations of simulated images and then do classification analysis. The results are very stable for different number of principal components and here we let K = 5. In Stage 3, we tried different classification methods, including linear regression, k-Nearest Neighbor (k-NN) [11] and support vector machine (SVM) [14], on these low dimensional spaces. Based on the misclassification error for the leave-one-out cross validation, the linear regression is slight better than others. The linear regression uses class label yi as dependent variable and principal components as explanatory variables. If the prediction value is less than 0, the image is classified as 0. Otherwise, the image is classified as 1.
We compared SWPCR with other state-of-the-art classification methods. The leave-one-out cross validation is used here to calculate the misclassification rates of the different methods. Other classification methods considered here include sparse linear discriminant analysis (sLDA) [6], sparse partial least squares (SPLS) analysis [5], sparse logistic regression (SLR) [20], SVM, and regularized optimal affine discriminant (ROAD) [8]. These methods are well known for their excellent performance in various simulated and real data sets. Inspecting Table 1 reveal that except SWPCR, all classification methods perform pretty poor, when the signal-to-noise ratio is low in those simulated datasets with Type I and II noises. Except SPLS, PCA, and SWPCR, all other methods are seem to be sensitive to the presence of the long-range correlation structure in Type III noise.
Table 1.
Classification results for the first set of simulations: comparison between SWPCR and other Classification Methods. sLDA denotes sparse linear discriminant analysis; SPLS denotes sparse partial least squares; SLR denotes sparse logistic regression; SVM denotes support vector machine; ROAD denotes regularized optimal affine discriminant; and PCA denotes principal component analysis.
| Noise | sLDA | SPLS | SLR | SVM | ROAD | PCA | SWPCR |
|---|---|---|---|---|---|---|---|
| Type I | 0.28 | 0.43 | 0.45 | 0.38 | 0.36 | 0.36 | 0.10 |
| Type II | 0.27 | 0.08 | 0.18 | 0.26 | 0.08 | 0.45 | 0.03 |
| Type III | 0.52 | 0.30 | 0.61 | 0.60 | 0.50 | 0.35 | 0.09 |
4 Real Data Analysis
4.1 ADNI PET Data
The real data set is the baseline fluorodeoxyglucose positron emission tomography (FDG-PET) data downloaded from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) web site (www.loni.ucla.edu/ADNI). The ADNI1 PET data set consists of 196 subjects (102 Normal Controls (NC) and 94 AD subjects). There are three subjects, missing the gender and age information. Among the rest of the subjects, there are 117 males whose mean age is 76.20 years with standard deviation 6.06 years and 76 females whose mean age is 75.29 years with standard deviation 6.29 years.
The dimension of the processed PET images is 79 × 95 × 69. Left panel in Figure 3 shows some selected slices of the processed PET images from 2 randomly selected AD subjects and 2 randomly selected NC subjects.
Fig. 3.
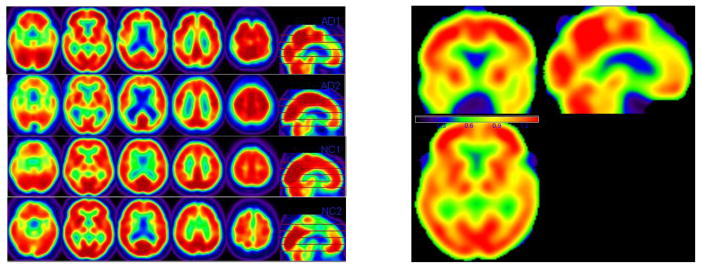
ADNI1 pet data and the important score weight matrix QI,ℓ in SWPCR. In the left panel, one row sequence of 2-D images belongs to one subject. The first two rows respectively belongs to AD subjects and the rest belongs to NC subjects. In the right panel, the three plots (left -right-bottom) are three view slices of the weight matrix QI,ℓ at the coordinate (40, 57, 26). The red region corresponds to large weight score and contains the most classification information.
4.2 Binary Classification
Our first goal is to apply SWPCR in classifying subjects from ADNI1 to AD or CN group based on their FDG-PET images. Such goal is associated with the second primary objective of ADNI aiming at developing new diagnostic methods for AD intervention, prevention, and treatment. Similar as in Section 3, SWPCR contains the three detailed stages that will not be repeated again. The right panel in Figure 3 is the three view slices of the weight matrix QI,ℓ at the coordinate (40, 57, 26) in the stage 2 of SWPCR. The red region in three slices corresponds to the large important score weight and contains the most classification information.
We compared SWPCR with six other classification methods including sLDA, SPLS, SLR, SVM, ROAD, and PCA. We used their leave-one-out cross validation rates. Table 2 shows the classification results of all the seven methods. sLDA performs much worse than all other six methods. ROAD performs slightly better than PCA. SPLS and SVM are comparable with each other, but they outperform SLR and ROAD. SWPCR outperforms all six classification methods. It suggests that the classification performance can be significantly improved by incorporating spatial smoothness and simple dimension reductions methods, such as PCA.
Table 2.
Misclassification Rates of Different Methods for ADNI 1 Pet Data
| sLDA | SPLS | SLR | SVM | ROAD | PCA | SWPCR |
|---|---|---|---|---|---|---|
| 0.255 | 0.163 | 0.179 | 0.168 | 0.189 | 0.194 | 0.117 |
4.3 Age Prediction
Our second goal is to apply SWPCR in predicting subjects’ age based on their FDG-PET images. The response variable y is the age of the subjects and the explanatory variables are the latent scores, extracted from image data. It is very interesting to use memory test scores as the response variable y. However, the data set here contains no such information. The three subjects without the age information are deleted and then we have 193 images left. yi in model (10) becomes age of the subjects. Here we will not repeat the detailed stages of SWPCR again, which is similar as in Section 3. The slight difference is stage 3. Here we run regression rather than classification methods between age and the SWPCR latent scores.
First, we compared SWPCR with three other dimensional reduction methods including PCA, weighted PCA (WPCA) [17], and supervised PCA (SPCA) [2]. We used the leave-one-out cross validation to compute the prediction errors of all the four methods. Let ŷi be the fitted response value based on the regression model, and the prediction error is defined as |ŷi − yi|/|yi|. Subsequently, we calculated the error difference between SWPCR and all three other methods across different numbers (K = 5, 7, 10) of principal components. Panels (a)–(c) in Figure 4 show the boxplots of the error difference between SWPCR and PCA, WPCA, and SPCA, respectively. The error differences are almost always less than 0 (under the dashed line) and these results show the better performance of SWPCR in dimension reduction.
Fig. 4.
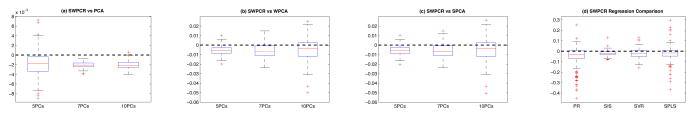
Performance of SWPCR Regression for ADNI 1 Pet Data. Panels (a)–(c) shows the boxplots of error difference between SWPCR and PCA (WPCA and SPCA). Panel (d) compares SWPCR regression with several other regression methods, including PR, SIS, SVR and SPLS.
Second, we compared SWPCR with several other high-dimensional regression methods including penalized regression (PR) [19], sure independence screening (SIS) regression [7], support vector regression (SVR) [3], and SPLS [5]. Panel (d) in Figure 4 shows the boxplots of the prediction error difference between SWPCR and all the other regression methods. The analysis results further confirm the better performance of SWPCR in regression.
5 Discussion
SWPCR enables a selective treatment of individual features, accommodates the complex dependence among features of graph data, and has the ability of utilizing the underlying spatial pattern possessed by image data. SWPCR integrates feature selection, smoothing, and feature extraction in a single framework. In the simulation studies and real data analysis, SWPCR shows substantial improvement over many state-of-the-art methods for high-dimensional problems.
Acknowledgments
This work was partially supported by the Startup Fund of University of South Florida, NIH grants MH086633, RR025747, and MH092335 and NSF grants SES-1357666 and DMS-1407655..
Contributor Information
Dan Shen, Email: danshen@usf.edu.
Hongtu Zhu, Email: htzhu@email.unc.edu.
References
- 1.Aharon M, Elad M, Bruckstein A. K-svd: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans on Signal Processing. 2006;54:4311–4322. [Google Scholar]
- 2.Bair E, Hastie T, Paul D, Tibshirani R. Prediction by supervised principal components. Journal of the American Statistical Association. 2006;101(473):119–137. [Google Scholar]
- 3.Basak D, Pal S, Patranabis DC. Support vector regression. Neural Information Processing-Letters and Reviews. 2007;11(10):203–224. [Google Scholar]
- 4.Buades A, Coll B, Morel JM. A non-local algorithm for image denoising. Computer Vision and Pattern Recognition, 2005. CVPR 2005. IEEE Computer Society Conference on; IEEE; 2005. pp. 60–65. [Google Scholar]
- 5.Chun H, Keles S. Sparse partial least squares regression for simultaneous dimension reduction and variable selection. J Roy Statist Soc Ser B. 2010;72:3–25. doi: 10.1111/j.1467-9868.2009.00723.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemmensen L, Hastie T, Witten D, Ersbøll B. Sparse discriminant analysis. Technometrics. 2011;53(4):406–413. [Google Scholar]
- 7.Fan J, Lv J. Sure independence screening for ultrahigh dimensional feature space. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2008;70(5):849–911. doi: 10.1111/j.1467-9868.2008.00674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan J, Feng Y, Tong X. A road to classification in high dimensional space: the regularized optimal affine discriminant. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2012;74(4):745–771. doi: 10.1111/j.1467-9868.2012.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friston KJ. Modalities, modes, and models in functional neuroimaging. Science. 2009;326:399–403. doi: 10.1126/science.1174521. [DOI] [PubMed] [Google Scholar]
- 10.Grenander U, Miller MI. Pattern Theory From Representation to Inference. Oxford University Press; 2007. [Google Scholar]
- 11.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2. Springer; Hoboken, New Jersey: 2009. [Google Scholar]
- 12.Lee M, Shen H, Huang JZ, Marron JS. Biclustering via sparse singular value decomposition. Biometrics. 2010;66:1087–1095. doi: 10.1111/j.1541-0420.2010.01392.x. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Zhu H, Shen D, Lin W, Gilmore JH, Ibrahim JG. Multiscale adaptive regression models for neuroimaging data. Journal of the Royal Statistical Society: Series B. 2011;73:559–578. doi: 10.1111/j.1467-9868.2010.00767.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y. Support vector machines and the bayes rule in classification. Data Mining and Knowledge Discovery. 2002;6:259–275. [Google Scholar]
- 15.Manjón JV, Carbonell-Caballero J, Lull JJ, García-Martí G, Martí-Bonmatí L, Robles M. MRI denoising using non-local means. Medical image analysis. 2008;12(4):514–523. doi: 10.1016/j.media.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 16.Polzehl J, Spokoiny VG. Propagation-separation approach for local likelihood estimation. Probab Theory Relat Fields. 2006;135:335–362. [Google Scholar]
- 17.Skočaj D, Leonardis A, Bischof H. Weighted and robust learning of subspace representations. Pattern recognition. 2007;40(5):1556–1569. [Google Scholar]
- 18.Taylor KM, Meyer FG. A random walk on image patches. SIAM J Imaging Sciences. 2012;5:688–725. [Google Scholar]
- 19.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society Series B (Methodological) 1996;58:267–288. [Google Scholar]
- 20.Yamashita O. Quick manual for sparse logistic regression toolbox ver1.2.1: software. 2011 at http://www.cns.atr.jp/~oyamashi/SLR_WEB/
- 21.Yan S, Xu D, Zhang B, Zhang HJ, Yang Q, Lin S. Graph embedding and extensions: a general framework for dimensionality reduction. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2007;29:40–51. doi: 10.1109/TPAMI.2007.12. [DOI] [PubMed] [Google Scholar]


