Summary
Induced pluripotency is a promising avenue for disease modeling and therapy, but the molecular principles underlying this process, particularly in human cells, remain poorly understood due to donor-to-donor variability and intercellular heterogeneity. Here we constructed and characterized a clonal, inducible human reprogramming system that provides a reliable source of cells at any stage of the process. This system enabled integrative transcriptional and epigenomic analysis across the human reprogramming timeline at high resolution. We observed distinct waves of gene network activation, including the ordered reactivation of broad developmental regulators followed by early embryonic patterning genes and culminating in the emergence of a signature reminiscent of pre-implantation stages. Moreover, complementary functional analyses allowed us to identify and validate novel regulators of the reprogramming process. Altogether, this study sheds light on the molecular underpinnings of induced pluripotency in human cells and provides a robust cell platform for further studies.
Introduction
Engineered reprogramming systems have provided useful tools for the study of induced pluripotency. In “secondary” reprogramming systems, somatic cells are first transduced with lentiviral constructs carrying drug-inducible transcription factors. Clonal induced pluripotent stem cells (IPSCs) are then derived and next differentiated back to a somatic state that can be reprogrammed a second time, often with greater efficiency (Hockemeyer et al., 2008; Stadtfeld et al., 2010; Wernig et al., 2008). Because the resulting somatic cells are clonal, this strategy eliminates biases and heterogeneity caused by variable lentiviral delivery and transgene stoichiometry present in “primary” reprogramming experiments (Stadtfeld et al., 2010).
Secondary reprogramming systems engineered from transgenic mouse embryonic fibroblasts (MEFs) have enabled large-scale genomic and epigenomic profiling studies of cells as they reacquire pluripotency (Hussein et al., 2014; Mikkelsen et al., 2008; Polo et al., 2012). These analyses have revealed that somatic identity is rapidly lost upon induction of the reprogramming factors and pluripotency is promoted by an early mesenchymal to epithelial transition (MET) (Li et al., 2010), a process accompanied by removal of several epigenetic roadblocks (Apostolou and Hochedlinger, 2013). It has, however, been difficult to directly compare the reprogramming MEFs to the same process in human cells, due to differences in culture conditions, differential expression of key markers and other factors. Under standard conditions, murine IPSCs also appear to reprogram with faster kinetics and higher efficiency than human IPSCs and reach a more naïve, pre-implantation-like cellular state (Hanna et al., 2010; Nichols and Smith, 2009). Moreover, analyses of intermediate states in previous systems have been complicated by heterogeneity in the initial cell populations and progressive loss of reprogramming capacity over serial passaging (Utikal et al., 2009). Thus, a well-controlled model system that generates intermediately- and fully-reprogrammed cells with consistent kinetics and efficiency even after extensive expansion in vitro would be a valuable asset for efforts to characterize reprogramming in human cells.
We hypothesized that senescence would be a major contributor to the variability and passage-dependent loss of reprogramming capacity that has been observed in previous attempts to generate human reprogramming systems (Park et al., 2008). We therefore sought to extend the lifespan of human secondary fibroblasts by overexpression of the telomerase gene (hTERT). Here, we employ this approach to generate a robust model system that enables continual propagation of clonal cells with a defined reprogramming capacity. We leverage this model to systematically characterize the transcriptional and epigenomic changes during reprogramming. Through integrative analyses, we find that OKMS induction leads to transient reactivation of genes in a pattern that is suggestive of a reversal of normal development. Unexpectedly, these changes culminate in the emergence of a subpopulation of cells with transcriptional and epigenomic signatures with pre-implantation-like characteristics. Finally, we demonstrate the utility of our secondary system for discovery and characterization of a variety of modulators of reprogramming in human cells.
Results
hTERT confers robustness to secondary reprogramming systems
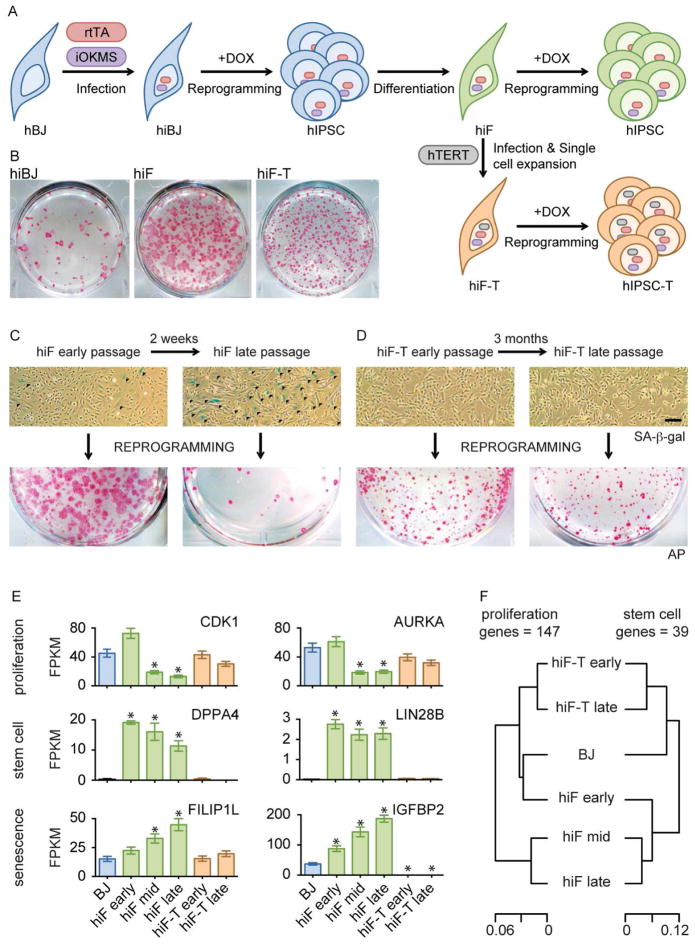
We generated human IPSCs (hIPSCs) from primary BJ foreskin fibroblasts using a doxycycline (DOX)-inducible, polycistronic human OCT4/KLF4/c-MYC/SOX2 (OKMS) cassette. We then differentiated these hIPSCs in a serum-based media (Park et al., 2008) to obtain human inducible fibroblasts-like cells (hiF) that could be subsequently reprogrammed by DOX treatment (Figure 1A). Consistent with previous attempts, both primary BJ cells and secondary hiF generated IPSC colonies that were highly heterogeneous in size and appeared asynchronously over three weeks following OKMS induction (Figure 1B). Moreover, secondary hiFs rapidly lost their reprogramming potential with successive passages in culture, which correlated with the appearance of senescent cells (Figure 1C). Foreskin fibroblasts from different donors also displayed variability in proliferation and senescence (Figure S1A–E), which influenced reprogramming efficiencies in a passage-dependent manner (Figure S1F). We observed similar variability across different batches of secondary cells generated from the same pluripotent stem cell (PSC) clone (dH1f or hiF; Figure S1A–F). These observations highlight the variable reprogramming efficiency of previous primary and secondary reprogramming systems.
Figure 1. An optimized secondary reprogramming system for human reprogramming.
A) Schematic representation of secondary reprogramming strategy and hiF-T engineering using inducible reprogramming factors (iOKMS) under the control of the reverse tetracycline transactivator (rtTA). Fibroblasts and IPSCs are shown as belonging to a primary hiBJ (blue), derived secondary hiF (green) and hiF-hTERT (brown) reprogramming system.
B) Alkaline Phosphatase (AP) staining of the indicated reprogrammed cells.
C–D) Representative bright fields of hiF and hiF-T cultures at different passages, after senescence-associated-beta-galactosidase (SA-b-GAL - upper panels) assay and corresponding alkaline phosphatase (AP) staining after 24 days of reprogramming (lower panels). Scale Bar, 100μm. Senescent cells are stained in blue and indicated with dark arrows.
E) Histograms showing absolute expression levels of selected proliferation, stemness and senescence related genes, as measured by RNA-seq. FPKM, fragments per kilobase per million fragments mapped. Error bars represent a 95% confidence interval around the average values. *, significant difference with respect to control BJ at FDR < 0.1%.
F) Hierarchical clustering of BJ, hiF and hiF-T cells according to expression levels of proliferation (left) or stemness-related (right) genes.
To increase the expansion potential of hiF cells, we delivered a lentiviral vector driving constitutive human telomerase (hTERT) expression (Stewart et al., 2003) and derived clonal cell lines (hiF-T; Figure 1A lower scheme). Inclusion of hTERT in reprogramming cocktails is known to be compatible with IPSC generation (Park et al., 2008). hiF-Ts displayed a lower reprogramming efficiency than early passage hiFs. But unlike both hiF and primary cells, they showed no evidence of senescence and maintained a stable reprogramming efficiency even after three months of continuous culture (Figure 1D, Figure S1F). At the same time, hiF-Ts showed growth and apoptosis rates that were equivalent to healthy primary cells and were negative in assays of cellular transformation (Figure S1A–E). Thus, hTERT expression also appears to reduce a key source of experimental variability in reprogramming systems.
To better understand the differences between primary (BJ), secondary (hIF) and secondary immortalized (hiF-T) reprogramming systems, we performed expression profiling by RNA-seq. We found that hiF cells down-regulated proliferative genes after only limited passaging, while hiF-Ts maintained expression of these genes in long-term cultures (Figure 1E). Moreover, hiF cultures expressed high levels of genes associated with “stemness”, even after the emergence of senescent cells, indicating either the persistence of primed or privileged subpopulations or incomplete differentiation. Either case might explain the high reprogramming efficiency of early passage hiFs. In contrast, the stemness genes were silenced in BJ and hiF-T cells. The consistency of hiF-T reprogramming may therefore reflect hTERT’s ability to block senescence, but may also ensure that the secondary cells can be cultured long enough to acquire a fully differentiated state (Figure 1F).
Importantly, hiF-T cells showed high levels of genomic and transcriptional stability (Figure S1G–H). Moreover, gene expression profiles of hIPSCs derived from reprogrammed hiF-T cells (hIPSC-T) are equivalent to reference PSCs, showing expression of expected protein markers and complete silencing of all transgenes, including hTERT (Figure S1I and S2). hIPSC-Ts also maintain the capacity to form all three embryonic germ layers in vitro through directed differentiation (Figure S1J–K). We therefore conclude that hIF-T secondary cells provide a faithful and reliable model system for large-scale studies of human reprogramming.
Genome-wide profiling of reprogramming cells
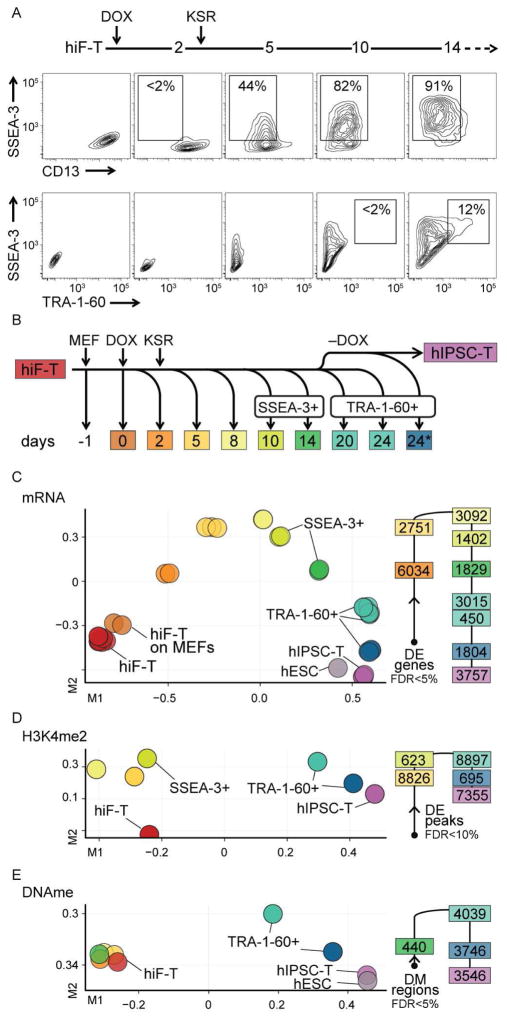
We next leveraged the increased proliferative capacity and decreased heterogeneity of hiF-Ts to collect large numbers of cells for comprehensive immunophenotypic, genomic and epigenomic analyses of reprogramming. hiF-Ts rapidly lost the somatic cell marker CD13 upon OKMS induction, and then acquired the embryonic marker SSEA-3. A subset of the most SSEA-3 positive cells subsequently acquired the pluripotency-associated marker TRA-1-60 (Figure 2A). We collected cells from key stages throughout this process. In the most advanced stages, we fractionated cells based on SSEA-3 or TRA-1-60 expression to isolate those that were transitioning towards pluripotency (Figure 2B). We then profiled mRNA and small RNA by RNA-seq, major histone modifications (H3K4me1, me2, me3; H3K27ac, me3; H3K36me3) by ChIP-seq and DNA methylation by Reduced Representation Bisulfite Sequencing (RRBS).
Figure 2. Integrative analysis of human cellular reprogramming.
A) Flow cytometry analysis of surface markers during reprogramming of hiF-T cells.
B) Schematic representation of time course collection of reprogramming intermediates, including fractionation by the indicated surface markers. For reprogramming, hiF-T cells were seeded on MEF feeder layer (MEF). Reprogramming was initiated at day 0 (DOX) and a switch to KSR-hESC media was performed at day 2 (KSR). The collection point labeled 24* represents cells reprogrammed for 20 days in DOX followed by 4 days without DOX.
C) MDS analysis of RNA-seq data (left) along with the number of differentially expressed (DE) genes associated with each transition (right). More comparisons are shown in Figure S2A.
D–E) MDS analyses of epigenomic data from regions differentially enriched (DE) in H3K4me2 or differentially DNA methylated (DM).
In all the representations, samples are color coded to the reference time points in panel B.
To characterize the major transitions in cellular states during reprogramming, we performed multidimensional scaling (MDS) analyses on the resulting data (Figure 2C–E).
MDS of the RNA-Seq data revealed a continuous trajectory of transcriptional changes beginning with uninduced hiF-Ts and ending with established hIPSC-Ts (Figure 2C). Cells with higher SSEA3 or TRA-1-60 expression were closer to the hIPSC-T state than cells with lower expression at the same day (Figure S2A), confirming the specificity of these surface antigens for reprogramming cells. Notably, the expression patterns of transgene-expressing TRA-1-60+ cells isolated at days 20–24 were clearly distinct from those observed after DOX withdrawal and establishment of hIPSC-Ts, as well as from reference hESCs.
MDS of H3K4me2 (Figure 2D), a histone methylation mark associated with open chromatin and active regulatory elements (Zhou et al., 2011), suggested two major transitions in its distribution: One occurring during the first five days, and a second coinciding with the acquisition of TRA-1-60. In contrast, genome-wide DNA methylation patterns (Figure 2E) appeared to remain largely constant throughout the first 14 days of the reprogramming course, but then changed rapidly upon TRA-1-60 acquisition and again upon removal of DOX and derivation of hIPSC-T lines, eventually reaching a pattern equivalent to that of reference hESCs.
The different genome-wide profiles therefore all suggested that TRA-1-60+ cells obtained at the end of the reprogramming course were qualitatively different from both derived hIPSC-T cell lines and reference hESCs when maintained under standard culture conditions. In the following sections, we characterize these differences and the preceding dynamics in more detail.
Ordered re-activation of developmental and pluripotency pathways
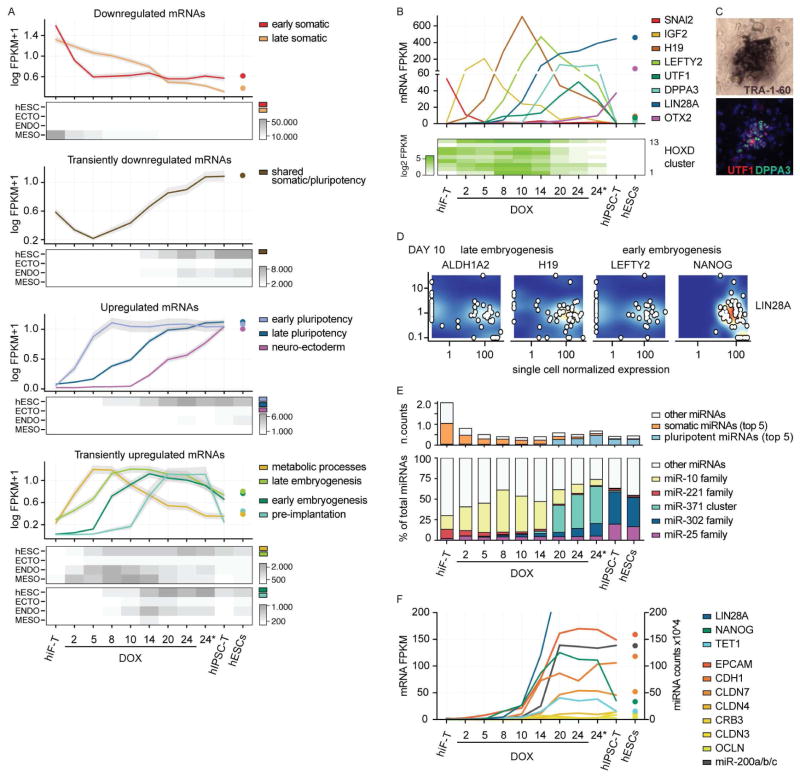
Clustering analysis of the RNA-seq data identified 10 major dynamic expression patterns (Figure 3A). We applied two complementary approaches to interpret these clusters: i) a comparison with gene expression signatures obtained from hESCs in their undifferentiated state or upon differentiation towards the three major germ layers; and ii) gene ontology enrichment analysis for both biological processes and developmental cell identity (Edgar et al., 2013) (Figure S2D; representative genes in Figure 3B and Figure S2B).
Figure 3. Transcriptional dynamics during human cellular reprogramming.
A) Line plots showing expression dynamics of differentially expressed genes during reprogramming, grouped by k-medoids clustering. Refer to Figure 2B for reprogramming time points. Grey shades represent a 95% bootstrap confidence interval around the median values. For each cluster the median expression value in hESC using 18 reference hESC lines is also reported. Heatmaps below each set of clusters (up-regulated, down-regulated and transients) show the total expression of genes in each cluster with respect to gene sets that define pluripotent (hESC) or embryonic germ layer-specific cells (ECTOderm, ENDOderm, MESOderm).
B) Absolute expression levels (FPKM) of selected dynamic genes reported as line plot or heatmap.
C) Representative colony of reprogrammed hIF-T cells identified by TRA-1-60 chromogenic staining in bright field and overlapping UTF1 and DPPA3 fluorescent staining. Complete field and staining controls are reported in Figure S3A.
D) Co-expression relationships between representative markers of the identified developmental transitions with respect to LIN28A as reference pluripotent marker, measured by single-cell RNA-seq. Additional single-cell data are shown in Figure S3D.
D) Absolute expression levels of categorized miRNAs (as normalized counts – see methods) (upper panel) and the relative expression levels of specific miRNA families with respect to total miRNA abundance (lower panel) at the indicated time points and in hESC line HUES64.
E) Line plot showing absolute expression values of mRNA and miRNA involved in MET during reprogramming and in reference hESCs. Pluripotent genes with different onset during reprogramming (LIN28A, NANOG, TET1) are also included to illustrate the relationship between epithelialization and acquisition of pluripotency.
Similar to murine reprogramming systems (Mikkelsen et al., 2008; Polo et al., 2012), OKMS rapidly downregulated mesenchymal signature genes, including genes encoding structural components like collagen and transcription factors like SNAI2 and FOSL1. We also observed rapid, but transient, down-regulation of genes that are known to be expressed in both stem cells and terminally differentiated cells, most notably the bHLH Inhibitor of DNA-binding (ID) proteins. Pluripotency-related genes were subsequently activated in two waves, distinguishing early pluripotency signature genes already detected by day 5, such as LIN28B, from late core regulators, like LIN28A (Chan et al., 2009) that were fully activated only in TRA-1-60+ cells at day 20. A final set of genes reached maximal levels only after derivation of hIPSC-T. This set included neuro-ectodermal and epiblast-related factors like SOX3 and OTX2, likely reflecting a priming of human PSCs under standard culture conditions (Tesar et al., 2007).
While rapid down-regulation of somatic genes and subsequent activation of the pluripotency network have been described (Chan et al., 2009; Park et al., 2008), comprehensive characterization of the transitions between these states has been limited by the heterogeneity of previous reprogramming systems. We found that the more synchronized changes of the hiF-T system allowed us to characterize several transient waves of gene activation (last panel in Figure 3A).
The first transient wave peaked at day 5 and was enriched for genes promoting proliferation and metabolic changes. Genes characteristic of this wave included IGF2, AFP, GSN and ALDH1A1, which are known to exert complementary proliferative and/or anti-apoptotic functions. The second wave, which peaked in the SSEA3+ fraction at day 10, included genes expressed during body patterning in late embryogenesis, such as HOX genes (see also Figure 3B), as well as markers of developing mesoderm (e.g. H19) and endoderm (e.g. HNF transcription factors). The third wave brought activation of genes associated with early embryogenesis and primitive endoderm, including NANOG, UTF1, LEFTY2, NODAL and GDF3. Similar to murine reprogramming, NANOG activation reached its peak prior to activation of the core pluripotency network (Samavarchi-Tehrani et al., 2010). Finally, and in parallel with activation of the core pluripotency network in TRA-1-60+ cells around day 20, we detected substantial expression of pre-implantation- or trophoblast-associated markers (e.g. DPPA2/3/5, DNMT3L, ALPPL2, FGF4, and TFCP2L1) and lowly expressed primitive streak genes (e.g. T, CER1, MIXL1). Notably, these markers were lost upon withdrawal of DOX and derivation of clonal hIPSC-T lines (e.g. DPPA3/STELLA (Hayashi et al., 2008) in Figure 3B–C and Figure S3A). This is consistent with the final step of hIPSC-T derivation from TRA-1-60+ colonies being accompanied by a shift from a gene expression program with pre-implantation-like characteristics to a program with more post-implantation-like characteristics (Figure S3B–C).
To examine whether the patterns we observed from bulk RNA profiling actually reflected changes that occurred within individual cells, or were in fact averages over multiple distinct subpopulations, we collected single cell RNA-seq profiles from 52 unfractionated hiF-T cells after 10 days of reprogramming (Figure 3D). We found low expression of genes from the previously defined somatic cluster and high expression of genes from the developmental clusters across the majority of the cells (Figure S3D). Notably, consistent with the bulk profiles, many of the single cells displayed simultaneous expression of genes associated with early and late embryogenesis, as well as LIN28A and other pluripotency markers (Figure 3D and S3D).
To test whether the expression dynamics in hiF-Ts were representative of other reprogramming systems, we also profiled non-immortalized hiFs at multiple time points (Figure S2C). These secondary cells were derived similarly to the frequently used dH1f (Park et al., 2008). We found that all patterns described above, including the late expression of pre-implantation-associated markers, also emerged from the hiFs, although with lower magnitude. Our data therefore suggest a general model of human OKMS-mediated reprogramming where cells first enter a highly proliferative state and lose their somatic identity, then display preferential re-activation of late, followed by early developmental genes, finally leading to the emergence of a distinct transient expression program with pre-implantation-like characteristics in the TRA-1-60+ subpopulation.
Analysis of miRNA captured in the small RNA-seq data (Figure 3E) reinforced this model. We similarly observed the rapid loss of somatic miRNAs (e.g. the miR-221 family), followed by up-regulation of miRNAs under developmental control (e.g. miR-10) and eventually of pluripotency-associated families (the miR-371 cluster and the miR-302 family). Strikingly, while many different miRNA families were expressed at comparable levels in hiF-Ts, each phase of the reprogramming process was defined by only one specific miRNA family that alone accounted for almost 50% of the sequenced molecules from that phase. Notably, miR-10b alone represented 49% of total miRNAs sequenced during the reactivation of late mesoderm pathways, which is consistent with its location within the HOXD cluster that is activated during this phase. The miR-371 cluster accounted for the majority of the mature miRNAs in TRA-1-60+ cells after day 20, while the miR-302 family, which is enriched in the post-implantation mouse embryo (Parchem et al., 2014), dominated in derived hIPSC-Ts. Consistent with neuro-ectodermal priming, derived hIPSC-Ts also expressed higher levels of miRNAs associated with neuronal progenitors (e.g. members of miR-25 family) (Nowakowski et al., 2013).
A possible driver of the differences in gene expression between TRA-1-60+ cells in DOX-containing media and derived hIPSC-Ts is elevated expression of KLF4 in the former. While the combined expression levels of the lentiviral and endogenous OCT4, SOX2 and c-MYC genes were largely similar, KLF4 expression was ~100-fold higher in the TRA-1-60+ cells (Figure S2B). This would be consistent with recent reports that higher expression of KLF proteins can push IPSC colonies to a more pre-implantation-like state (Gafni et al., 2013; Takashima et al., 2014).
Interestingly, while a mesenchymal to epithelial transition (MET) has been reported as a critical early event in reprogramming of mouse cells (Li et al., 2010; Samavarchi-Tehrani et al., 2010), we did not observe clear enrichment of epithelial associations in the early phases of hiF-T reprogramming in our gene ontology analysis. We therefore looked specifically for the activation of key epithelial marker genes (Figure 3F). In mouse cells, activation of these markers precedes activation of Nanog and the core pluripotency network (Samavarchi-Tehrani et al., 2010). In contrast, during hiF-T reprogramming, these markers appeared to be activated at the same time as NANOG, LIN28A, TET1 and other core regulators of pluripotency.
A pre-implantation-like chromatin state is transiently established during reprogramming
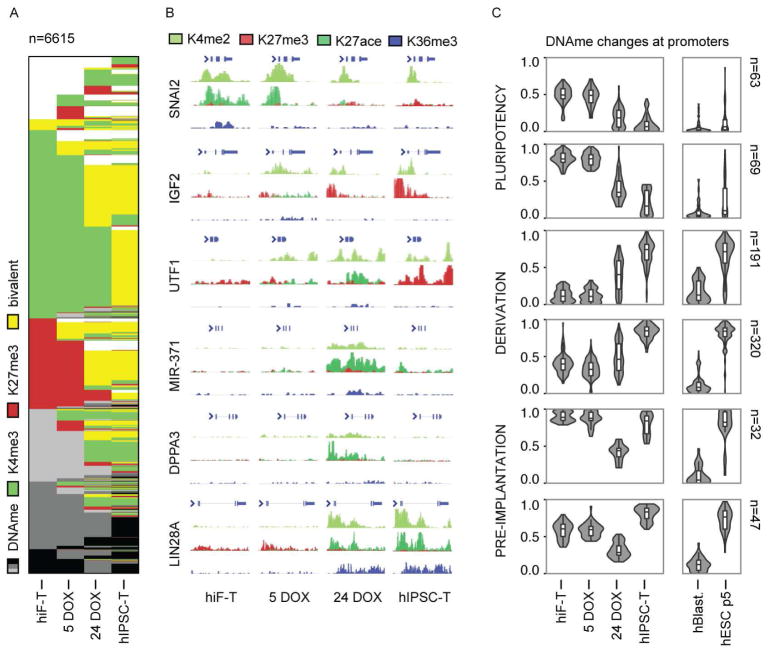
A characteristic feature of pluripotent cell lines is that the promoters of many developmental regulatory genes display a bivalent chromatin state, where histone marks associated both with activation (H3K4me2/me3) and repression (H3K27me3) co-occur (Zhou et al., 2011). To study the acquisition of bivalency during hiF-T reprogramming, we focused on 6,615 promoters that showed a significant change in either H3K4me3 or H3K27me3 signal between day 0 and the iPSC state (Figure 4A–B). As expected, bivalent promoters were rare at the somatic and early stages. The reprogrammed TRA-1-60+ subpopulation at 24 days displayed a significant number of bivalent promoters, but only approximately half of that found in clonal hIPSC-Ts derived from them. Interestingly, other groups have recently reported that an increase in bivalency is a key feature of the transition between naïve and primed states in ESCs (Marks et al., 2012; Theunissen et al., 2014), although the extent to which this reflects epigenetic changes occurring during human embryogenesis remains unknown.
Figure 4. Changes in bivalency and DNA methylation during reprogramming.
A) Chromatin state maps of 6,595 dynamic promoter regions, showing active (H3K4me3), repressed (H3K27me3) or bivalent regions (functionally poised by the co-enrichment of both H3K4me3 and H3K27me3). Promoters with none of these histone marks are marked by different degrees of DNA methylation (DNAme, 3 shades of gray for the ranges 25–50% – 50–75% and 75–100%).
B) Histone methylation at representative 5–50 kb loci from the major transcriptional clusters in Figure 3B. A similar map of the broad pre-implantation region around the miR-371 cluster is also reported in Figure S4C–D.
C) Violin plots showing promoter DNA methylation dynamics across the indicated reprogramming time points and sample types, grouped by k-means clustering. n, size of each cluster. The box plots show the first and the third quartiles, along with the medians.
The distinction between TRA-1-60+ cells and established hIPSC-T was, however, further clarified by analysis of promoter DNA methylation. Consistent with the genome-wide MDS analysis (Figure 2E), we found that the vast majority of changes in promoter DNA methylation took place during the late stages of reprogramming (Figure 2E and 4C). A comparison of our data with methylation profiles from human blastocysts and reference hESCs (Smith et al., 2014) allowed us to identify 722 differentially methylated promoters that each followed one of three major patterns: I) promoters with high methylation in fibroblasts that were hypomethylated in all reprogrammed or pluripotent samples (24 days TRA-1-60+ cells, derivative hIPSC-T, in vivo blastocysts and in-vitro derived hESCs); II) promoters with low to moderate methylation in fibroblasts that became hypermethylated upon hIPSC-T derivation were also hypermethylated in hESCs, but showed lower methylation in both 24 days TRA-1-60+ cells and blastocysts; and III) hypermethylated promoters that were transiently de-methylated in 24 days TRA-1-60+ cells showed low methylation in blastocysts but hypermethylation in both hIPSC-Ts and hESCs. This third pattern included promoters of key pre-implantation markers, such as DNTM3L, DPPA3 and MIR-371. Although we did not observe global DNA hypomethylation, the differences in promoter DNA methylation between TRA-1-60+ cells at 24 days of reprogramming and derived hIPSC-Ts are in agreement with those previously described between human pre-implantation blastocyst and derived hESCs (Smith et al., 2014).
Transient chromatin remodeling at lineage-specific regulatory elements
To corroborate our finding that OKMS transiently re-activates diverse developmental pathways prior to acquisition of pluripotency, we next examined changes in H3K4me2, a histone methylation mark associated with both active promoters and enhancers (Zhou et al., 2011).
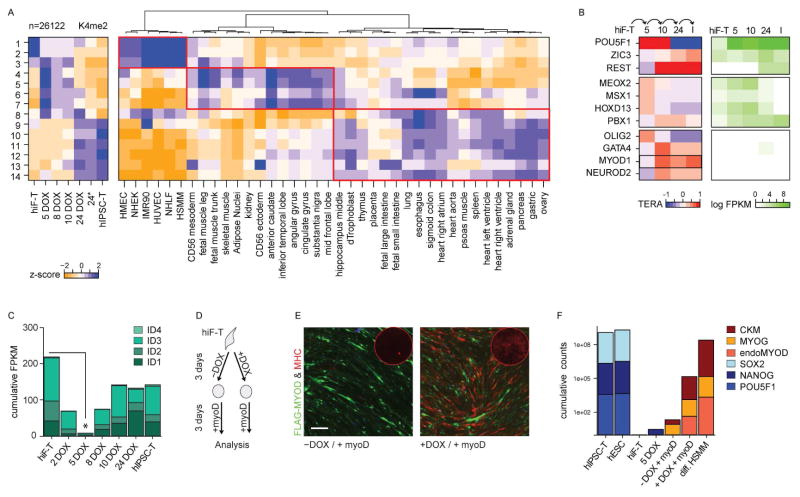
We detected altered H3K4me2 levels in 26,122 distinct genomic regions throughout hiF-T reprogramming. These regions could be grouped into 14 dynamic clusters (Figure 5A and S4A–B). 4,815 regions (clusters 1–2) displayed H3K4me2 in hiF-Ts, but lost this mark within 5 days of OKMS induction. 8,794 regions (clusters 9–14) were unmarked until day 10 or later, with clusters 10 and 12 showing maximal signals only after derivation of hIPSC-Ts. The remaining 12,513 regions (clusters 3–8) showed variable patterns of transient H3K4me2, many with maximal signal at day 5.
Figure 5. Transient chromatin remodeling and epigenetic priming during reprogramming.
A) Left: Heatmap showing the z-scores of the mean H3K4me2 enrichment in 26,122 dynamic genomic regions, grouped into 14 clusters. For full representation of the clusters and corresponding H3K27me3 dynamics refer to Figure S4A–B. Right: Heatmap showing the corresponding z-scores of the mean H3K27ac enrichment across tissues of different identity.
B) Heatmap showing the TERA score of selected transcription factors predicted to be activated during the indicated reprogramming transitions, based on H3K4me2 footprints. Corresponding absolute gene expression values (FPKM) during reprogramming are reported on the right. A full list of the top transcription factors groups and their predicted co-binding relationships are shown in Figure S5.
C) Bar plot showing the cumulative absolute expression values (FPKM) of the ID gene family during reprogramming.
D) Schematic representation of the OKMS-enhanced MYOD reprogramming of hiF-Ts.
E) Representative field of MYOD-mediated myogenic conversion without (−DOX) or with (+DOX) prior OKMS activation for 3 days. Cells positive for ectopic FLAG-MYOD are green while cells positive for the late muscle marker MHC are red. The corresponding MHC whole-well staining is shown in the corner of each condition. Scale Bar, 200μm
F) Bar plot showing the normalized cumulative expression counts of pluripotent (SOX2, NANOG, POU5F1) and muscle -specific genes (muscle creatine kinase – CKM, myogenin – MYOG, endogenous MYOD, endoMYOD). Controls are reference PSCs, hiF-T reprogramming time points and differentiated human skeletal muscle myoblasts (HSMM).
Comparing these dynamic H3K4me2 patterns to reference chromatin maps from various human cell types and tissues (Roadmap Epigenomics Consortium et al., 2015) (Figure 5A), we found that regions that were marked by H3K4me2 in hiF-T cells but rapidly lost this mark upon OKMS induction were largely specific to in-vitro derived somatic cell lines. Regions that gained H3K4me2 during the early stages of reprogramming often displayed active chromatin marks in tissues of both mesodermal and ectodermal origin, while regions that gained H3K4me2 in the late stages often also display active chromatin marks in tissues of endodermal origin. We therefore conclude that the transient activation of developmental pathways during reprogramming is accompanied by chromatin remodeling at regulatory elements associated with lineages of different developmental stages.
OKMS induction supports direct lineage conversion
The transient re-activation of developmental pathways and regulatory elements suggested to us that OKMS expression might induce epigenetic changes that could enable direct reprogramming to alternative lineages in addition to pluripotency. In support of this, using the Transcription factor Epigenetic Remodeling Activity (TERA) framework (Ziller et al., 2015), we found that the DNA sequences of genomic regions showing changing H3K4me2 were associated with potential binding sites for numerous transcription factor families involved not only in pluripotency, but also in developmental patterning and differentiation (Figure 5B and Figure S5).
The TERA analysis revealed that regions that gained H3K4me2 at early time points frequently coincided with OCT4 and SOX2 binding sites, which is consistent with a more dominant role for these transgenes in chromatin remodeling during the early stages of reprogramming relative to the late stages of IPSC establishment (Soufi et al., 2012). In contrast, regions that gained H3K4me2 at later time points were enriched for potential binding sites for other transcription factors that were up-regulated towards the end of the time course, such as ZIC3 and REST. In agreement with our previous analyses, the transient waves of developmental gene expression were accompanied by exposure of binding sites for transcription factors acting in fetal development like HOXD13 and PBX1. Interestingly, some factors which binding sites were also enriched in transiently H3K4me2-marked regions, including bHLH transcription factors MYOD, NEUROD2 and OLIG2, were not detected by RNA-seq at any time point. This enrichment might therefore reflect lineage priming by other factors. In addition, we noticed that OKMS induction led to a transient down-regulation of the ID transcriptional repressor family (Figure 5C), which are known to restrict lineage commitment by inhibition of bHLH activity (Perk et al., 2005).
To test our hypothesis of epigenetic priming towards alternative fates, we performed ectopic expression of MYOD, a master regulator of skeletal muscle cell maturation. MYOD was first discovered as a factor that reprogrammed somatic cells towards a myotube fate (Fong and Tapscott, 2013), although the efficiency of this phenomenon differs across cell types (including fibroblasts subtypes - (Salvatori et al., 1995)), due to variation in epigenetic states and expression of inhibitors such as ID1 (Perk et al., 2005). We found that introduction of a MYOD lentivirus led to a very low rate of myogenic conversion of otherwise unperturbed hiF-T cells, as judged by low expression of skeletal muscle markers at both the protein and mRNA levels (Fig. 5D–F). In contrast, introduction of MYOD after a 3-day pulse of OKMS expression drove efficient conversion to myosin heavy chain (MHC) positive cells and induced key muscle genes to levels that approached those of mature skeletal muscle cells within another 3 days. Notably, introduction of MYOD after OKMS induction uniquely activated endogenous MYOD, which may support stabilization of the converted state through its auto-regulation (Hanna et al., 2010). The rapid rate of conversion and the complete absence of pluripotency markers throughout the time course (Fig 5F) strongly suggest that it did not involve transition through an intermediate pluripotent state. We therefore conclude that OKMS rapidly drives cells into an epigenetic state that is conducive not only to derivation of pluripotent cells, but also to direct conversion into alternate lineages.
Identification of regulatory genes that inhibit reprogramming
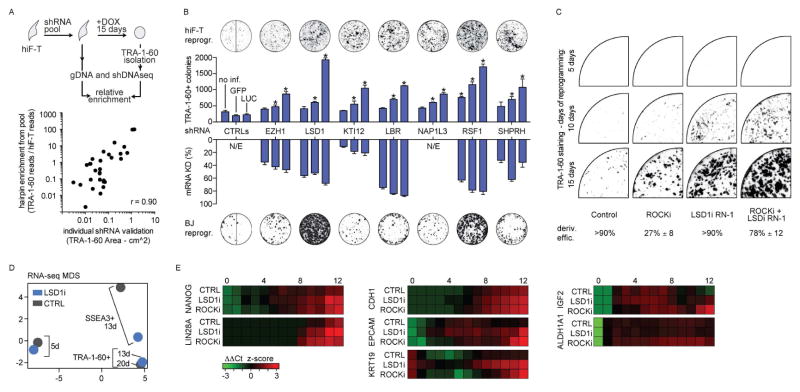
We next attempted to identify regulatory factors that might inhibit or delay reprogramming to pluripotency. We again leveraged the expansion capacity and uniformity of hiF-Ts to perform a quantitative RNAi screen (Luo et al., 2008) using a pooled lentiviral library encoding ~2,900 shRNAs targeting 370 distinct regulatory genes with known or putative roles in chromatin remodeling and other epigenetic processes. The library complexity was chosen to maximize the representation of neutral shRNAs in the final TRA-1-60+ population despite the bottleneck imposed by limited reprogramming efficiency (see methods).
We noted that hiF-T cultures infected with the lentiviral pool generated TRA-1-60+ colonies much more efficiently than controls. To identify the genes for which knock-down led to enhanced reprogramming, we compared shRNA abundances before (hiF-T) and after (TRA-1-60+) reprogramming using deep sequencing (Fig. 6A). This primary screen identified 23 candidate genes with at least one strongly enriched hairpin. In a secondary screen, we confirmed seven of these candidates (EZH1, KTI12, LBR, NAP1L3, RSF1, SHPRH and LSD1) based on consistent phenotypes from three distinct shRNAs (Figure 6B). The majority of the validated regulators have not been well characterized, but they span a variety of different chromatin modification and remodeling processes. Six of the seven have to our knowledge not been reported as barriers to human reprogramming (Onder et al., 2012). The positive effect of inhibiting the Polycomb complex member EZH1 is surprising given that its homolog EZH2 is required for reprogramming (Onder et al., 2012), but we note that the two appear to regulate different targets (Shen et al., 2008). Inhibition of the histone lysine demethylase LSD1 has been reported to enhance reprogramming (Li et al., 2009), but its mode of action remains unknown.
Figure 6. Characterization of negative regulators of reprogramming.
A) Schematic representation of the pooled shRNA screening strategy.
B) Scatter plot showing comparison of selected reprogramming efficiencies in shRNA-perturbed hIF-T cells at day 15 in a pooled screening format (Y axis - enrichment of shRNA sequence reads from TRA-1-60+ cells versus cells prior to induction of reprogramming) versus an arrayed format (X axis - area of TRA-1-60+ colonies). The reported values are the mean of biological duplicates.
C) Bar plots showing reprogramming efficiency (number of TRA-1-60+ colonies) upon shRNA-mediated perturbation of candidate regulators (upper histogram) and the corresponding change in mRNA expression levels in hIF-T cells relative to the effect of a control shRNA targeting luciferase (LUC) mRNA (lower histogram). Additional controls are shRNAs targeting GFP mRNA or uninfected cells. Three distinct hairpins were tested for each gene and representative TRA-1-60 stainings for each shRNA group are displayed above each set (control lane shows wells of both shGFP and shLUC treatments). Similar effects were observed in primary BJ reprogramming experiments, as shown by TRA-1-60 stainings below each set. Error bars indicate s.d. from the average. All reported values in histograms are significant with respect to controls at FDR < 5%. *, significant difference with respect to control at FDR < 1%.
C) Representative TRA-1-60 staining at indicated time points of hiF-T reprogrammed in the continuous presence of the indicated inhibitors.
D) Gene expression profiles, from RNA-seq, of hiF-T reprogramming with or without LSD1 inhibition (LSD1i and CTRL respectively), represented as points in two-dimensional MDS component space. A gene set enrichment analysis is described in Figure S6C.
E) Heatmap showing expression (normalized z-score) of indicated genes during reprogramming in different conditions (untreated control, LSD1 inhibition, ROCK inhibition). Key time points for reprogramming transitions are indicated (0, 4, 8, 12). Corresponding MDS plot of the RNA-seq time course utilized to identify these genes is reported in Figure S6D.
Comparison of the effects of LSD1 and ROCK1 inhibition on reprogramming
As LSD1 was the only validated hit in our RNAi screen with available chemical inhibitors, we sought study the effects of its perturbation in more detail. We found that inhibition of LSD1 with either a standard inhibitor (parnate) or a potent analog (Histone Lysine Demethylase Inhibitor RN-1 (Neelamegam et al., 2012) dramatically enhanced hiF-T reprogramming. A 10 nM dose of RN-1 over the first five days was sufficient to both accelerate and increase the efficiency of reprogramming (Figure S6A), generating TRA-1-60+ cells in less than 10 days (Figure 6C), as opposed to at least 15 days in untreated cultures. This effect was higher than the previously reported effect of ROCK1 inhibitor Y-27632, which also requires much higher concentrations (≥1 μM). In addition, chemical inhibition of LSD1 enhanced reprogramming even in the presence of saturating doses of the ROCK1 inhibitor (Figure S6B), which suggests synergistic modulation of distinct pathways.
Notably, TRA-1-60+ cells from LSD1i-treated cultures at day 13 were indistinguishable from those from untreated cultures from day 20, as judged by RNA-seq profiling (Figure 6D), and could be used to generate stable hIPSC-T clones. The efficiency of hIPSC-T derivation from LSD1i-treated cells was significantly higher than that of ROCK1i-treated cells, which indicates more consistent and complete reprogramming (Figure 6C lower panel). Moreover, the accelerated reprogramming caused by LSD1 inhibition did not appear to simply be a result of increased proliferation or decreased apoptosis (Figure S6C), as has been suggested for ROCK1 inhibition (Watanabe et al., 2007). On the contrary, the replication rate of LSD1i treated hiF-T is not affected in short term cultures.
To gain deeper insights into the effects of LSD1 and ROCK1 inhibition, we collected additional data from each of the first 12 days of a new reprogramming experiment with LSD1i- and ROCK1i-treated and untreated hiF-T cells using RNA-seq. We were particularly interested in whether the gene expression changes in the treated cells followed the same trajectory as untreated cells. MDS analysis suggested that this was indeed the case (Figure S6D). Both LSD1i and ROCK1i treatments led to the similar patterns of rapid down-regulation of somatic genes, followed by transient up-regulation of various developmental and embryonic genes. The expression of early and late markers of pluripotency, such as NANOG and LIN28A, were not accelerated with respect to the control cells in early time points, but they became significantly upregulated with respect to untreated cells by day 8 (Figure 6E).
Interestingly, the only significant change in gene expression unique to LSD1i treated cells in the early stages of the reprogramming was an accelerated up-regulation of a small set of genes enriched in epithelial markers, most notably e-cadherin (CDH1), EPCAM, KRT19 and CLDN10. CDH1 is known to be both a major driver of the MET process and essential for maintenance of the pluripotent state (Samavarchi-Tehrani et al., 2010). In fact, CDH1, EPCAM and KRT19 are among the most highly expressed epithelial genes in human PSCs (not shown). This suggests that LSD1 inhibition may enhance reprogramming through epithelialization, a phenomenon that we found to be delayed under standard conditions (see Figure 3F).
In contrast, the ROCK1i treatment was characterized by elevated and persistent expression of growth promoting genes like IGF2 and ALDH1A1 during the later stages (Figure 6E). This is consistent with increased proliferation and survival, but could also explain the lower efficiency of hIPSC-T derivation from ROCK1i treated cells (Figure 6C lower panel). IGF2 and ALDHA1 has both been found to be highly expressed in cancer stem cells (Baccelli and Trumpp, 2012; Pollak, 2008) and persistent IGF2 expression has recently been reported as a maker of transformation in vivo during cellular reprogramming (Ohnishi et al., 2014). This indicates that prolonged ROCKi treatment may favor the emergence of an aberrant reprogramming environment suggesting to avoid its prolonged use in reprogramming approaches.
Discussion
Ever since the first successful attempts to reprogram mouse and human cells were reported (Takahashi and Yamanaka, 2006), it has been clear that there are significant species-specific differences in these processes. The hiF-T secondary reprogramming system now provides a convenient and representative model system for dissection of reprogramming in human cells.
Recent work by Yamanaka and colleagues has shown that reprogramming human cells enter into an early mesendodermal state just prior to acquisition of pluripotency (Takahashi et al., 2014). Extending this observation, we found evidence of multiple transient waves of gene expression changes that begin with a rapid loss of somatic identity, followed by re-activation of early developmental pathways and embryonic patterning genes in the reverse order of normal development, eventually reaching a pre-implantation-like state that is only lost upon derivation of IPSC lines under standard conditions.
In fully committed somatic cells, the reprogramming factors appear to facilitate re-activation of related developmental lineages that reflect their ontology. Thus, fibroblast-like hiF-T cells largely de-differentiate to first express a broad mesodermal signature and then features of early development. We hypothesize that somatic cells of different origins might activate different transient gene sets. Eventually, only a small fraction of those cells will activate the core pluripotency network. A key remaining question is whether passing through either one of these transient cell states is strictly required for successful derivation of human IPSC lines.
Several recent studies have argued that human PSCs can be brought into a more naïve state of pluripotency with cocktails of chemical inhibitors and/or transcription factors (Gafni et al., 2013; Takashima et al., 2014; Theunissen et al., 2014). Our data show that OKMS expression alone is sufficient to induce key mRNA and miRNA genes that are uniquely expressed in the pre-implantation embryo, and to reduce DNA methylation of promoters that are known to be hypomethylated in the inner cell mass but hypermethylated in standard hESC cultures. We also show that prior to reaching the stabilized pluripotent state, OKMS expression induces a less restricted epigenetic state that is particularly amenable to direct lineage conversion. This is consistent with recent reports of derivation of mesodermal and endodermal cell types from the early stages of reprogramming cell populations (Efe et al., 2011; Zhu et al., 2014).
We expect that the consistency and virtually unlimited expansion potential of the hIF-T system will enable new approaches to dissection of human cellular reprogramming.
Methods
Cell culture and reprogramming
Human fibroblasts were cultured in an optimized DMEM/F12 culture media supplemented with 10% FBS. Pluripotent stem cells were cultured in 20% KSR-based DMEM/F12 culture media with irradiated MEF feeders or mTeSR1 or Essential 8 media without feeders. Reprogramming was performed on a confluent irradiated MEF layer using the KSR media formulation and doxycycline as indicated. BJ fibroblasts were first reprogrammed with a dox-inducible, polycistronic OKMS lentiviral vector (Addgene 51543). This gave rise to the first hIPSC line, which was then differentiated in vitro (Park et al., 2008) to obtain the hiF line. Infection of the hiF line with a CMV-hTERT lentivirus (Applied Biological Materials) and clonal isolation generated the final hiF-T. Directed differentiation of hIPSC-T was performed as previously reported (Gifford et al., 2013). Additional details of cell culture and media formulations, reprogramming and sampling are reported in the Extended Experimental Procedures.
Cellular assays
Senescence (beta-galactosidase), alkaline phosphatase, flow-cytometry and immunostaining analyses were performed with commercial kits and antibodies, as detailed in the Extended Experimental Procedures. Cell proliferation, senescence and apoptosis during fibroblasts expansion, and reprogramming efficiency, were assessed using manual cell or colony counting as indicated. For quantitative analysis of reprogramming efficiency in some comparative approaches (RNAi screening, LSD1 and ROCK1 inhibition), digital acquisition of chromogenic TRA-1-60 staining was performed and followed by colony identification and counting by ImageJ (http://imagej.nih.gov/ij/). Further details are reported in Extended Experimental Procedures.
Genomic and epigenomic profiling
Cells were prepared for profiling using MEF depletion, and in some instances SSEA-3 or TRA-1-60 enrichment/depletion using magnetic beads separation (Miltenyi Biotec) as indicated in the text. Bulk mRNA-Seq and small RNA-Seq were performed using TruSeq kits (Illumina). Single cell RNA-Seq was performed using the Smart-Seq2 protocol with minor modifications. RRBS and ChIP-Seq were performed as previously described (Mikkelsen et al., 2010; Boyle et al., 2012). Assessment of MYOD-mediated direct differentiation was performed using a NanoString nCounter with a custom codeset. High-throughput 3′ DGE was performed using a modified SCRB-Seq protocol with barcoded poly-dT RT primers and a hybrid Nextera/TruSeq sequencing strategy. Details of all the library construction and sequencing procedures as well as downstream bioinformatics analyses are reported in Extended Experimental Procedures. All sequencing-derived data are available through GEO accession GSE62777.
RNAi screening
RNAi screening was performed by infecting at least 6x107 hiF-T cells with a pool of The RNAi Consortium (TRC) lentiviral shRNA constructs targeting 370 distinct epigenetic regulators and then reprogramming the infected cells for 15 days. Integrated shRNA templates were recovered from TRA-1-60+ cell gDNA by PCR and counted using Illumina sequencing. Further details are reported in Extended Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to Martin Sauvageau and Alexander Tsankov for technical assistance and for helpful discussion. DC is a Human Frontier Science Program Fellow. CT is a Damon Runyon Fellow. MC is an EMBO and Leukemia and Lymphoma Society Fellow. MS was supported by a Swiss National Science Foundation postdoctoral fellowship and DC, MS, SHS and TSM were supported by the Harvard Stem Cell Institute. This work was supported by a grant from NIGMS (P01GM099117) to TSM, AM and JLR.
Footnotes
Author contributions
TSM and DC conceived the experimental plan and wrote the manuscript. DC derived and reprogrammed the hiF and hiF-T cell lines. SR, ZW, VA, CAG and JD assisted with cell culture and screening. DC, MS, MC, JD, XZ performed library preps and sequencing. CT, MJZ, DC, RK, ZDS and SHS processed and analyzed data. JLR, GQD, AM, ESL, TSM provided mentoring and assisted with data interpretation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502:462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli I, Trumpp A. The evolving concept of cancer and metastasis stem cells. J Cell Biol. 2012;198:281–293. doi: 10.1083/jcb.201202014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, Holmes L, Meldrim J, Kelley F, Gnirke A, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13:R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EM, Ratanasirintrawoot S, Park IH, Manos PD, Loh YH, Huo H, Miller JD, Hartung O, Rho J, Ince TA, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- Edgar R, Mazor Y, Rinon A, Blumenthal J, Golan Y, Buzhor E, Livnat I, Ben-Ari S, Lieder I, Shitrit A, et al. LifeMap Discovery™: the embryonic development, stem cells, and regenerative medicine research portal. PLoS ONE. 2013;8:e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nature Publishing Group. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- Fong AP, Tapscott SJ. Skeletal muscle programming and re-programming. Curr Opin Genet Dev. 2013;23:568–573. doi: 10.1016/j.gde.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–1163. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Lopes SMC de S, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Cook EG, Gao Q, Mitalipova M, Jaenisch R. A drug-inducible system for direct reprogramming of human somatic cells to pluripotency. Cell Stem Cell. 2008;3:346–353. doi: 10.1016/j.stem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein SMI, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, Mosbergen R, Li M, Lee DS, Cloonan N, et al. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206. doi: 10.1038/nature14046. [DOI] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Schöler HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. Highly parallel identification of essential genes in cancer cells. Proc Natl Acad Sci USa. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegam R, Ricq EL, Malvaez M, Patnaik D, Norton S, Carlin SM, Hill IT, Wood MA, Haggarty SJ, Hooker JM. Brain-penetrant LSD1 inhibitors can block memory consolidation. ACS Chem Neurosci. 2012;3:120–128. doi: 10.1021/cn200104y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nowakowski TJ, Fotaki V, Pollock A, Sun T, Pratt T, Price DJ. MicroRNA-92b regulates the development of intermediate cortical progenitors in embryonic mouse brain. Proc Natl Acad Sci USa. 2013;110:7056–7061. doi: 10.1073/pnas.1219385110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]
- Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, Larussa MF, Krishnakumar R, Blelloch A, Oldham MC, Blelloch R. Two miRNA Clusters Reveal Alternative Paths in Late-Stage Reprogramming. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008;8:915–928. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatori G, Lattanzi L, Coletta M, Aguanno S, Vivarelli E, Kelly R, Ferrari G, Harris AJ, Mavilio F, Molinaro M. Myogenic conversion of mammalian fibroblasts induced by differentiating muscle cells. J Cell Sci. 1995;108(Pt 8):2733–2739. doi: 10.1242/jcs.108.8.2733. [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Borkent M, Hochedlinger K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat Methods. 2010;7:53–55. doi: 10.1038/nmeth.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SA, Ben-Porath I, Carey VJ, O’Connor BF, Hahn WC, Weinberg RA. Erosion of the telomeric single-strand overhang at replicative senescence. Nat Genet. 2003;33:492–496. doi: 10.1038/ng1127. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Sasaki A, Yamamoto M, Nakamura M, Sutou K, Osafune K, Yamanaka S. Induction of pluripotency in human somatic cells via a transient state resembling primitive streak-like mesendoderm. Nat Commun. 2014;5:3678. doi: 10.1038/ncomms4678. [DOI] [PubMed] [Google Scholar]
- Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell. 2014;158:1254–1269. doi: 10.1016/j.cell.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:471–487. doi: 10.1016/j.stem.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa SI, Muguruma K, et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Wernig M, Lengner CJ, Hanna J, Lodato MA, Steine E, Foreman R, Staerk J, Markoulaki S, Jaenisch R. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nature Reviews Genetics. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhu S, Rezvani M, Harbell J, Mattis AN, Wolfe AR, Benet LZ, Willenbring H, Ding S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014 doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, Issner R, Gifford CA, Goren A, Xing J, et al. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature. 2015;518:355–359. doi: 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.