Abstract
Previous studies have indicated that retinoic acid (RA) may be therapeutic for endometrial cancer. However, the downstream target genes and pathways triggered by ligand-activated RA receptor α (RARα) in endometrial cancer cells are largely unknown. In this study, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, flow cytometry, and immunoblotting assays were used to assess the roles of RA and the RA agonist (AM580) in the growth of endometrial cancer cells. Illumina-based microarray expression profiling of endometrial Ishikawa cells incubated with and without AM580 for 1, 3, and 6 h was performed. We found that both RA and AM580 markedly inhibited endometrial cancer cell proliferation, while knockdown of RARα could block AM580 inhibition. Knockdown of RARα significantly increased proliferating cell nuclear antigen and BCL2 protein levels. Incubation of Ishikawa cells with or without AM580 followed by microarray expression profiling showed that 12 768 genes out of 47 296 gene probes were differentially expressed with significant P values. We found that 90 genes were the most regulated genes with the most significant P value (P<0.0001) using F-test. We selected four highly regulated genes with diverse functions, namely G0S2, TNFAIP2, SMAD3, and NRIP1. Real-time PCR verified that AM580 highly regulated these genes, whereas chromatin immunoprecipitation-PCR assay demonstrated that ligand-activated RARα interacted with the promoter of these genes in intact endometrial cancer cells. AM580 also significantly altered 18 pathways including those related to cell growth, differentiation, and apoptosis. In conclusion, AM580 treatment of Ishikawa cells causes the differential expression of a number of RARα target genes and activation of signaling pathways. These pathways could, therefore, mediate the carcinogenesis of human endometrial cancer.
Introduction
Endometrial cancer is the most common gynecologic malignancy. It accounts for 6% of all cancers in women and is the eighth most common cause of cancer-related death in the United States (Obel et al. 2006). According to the American Cancer Society, 40 880 new cases of endometrial cancer were diagnosed and 7310 women died of this disease in 2005 (Obel et al. 2006). Estrogen and progesterone play important roles in the regulation of endometrial function and the pathogenesis of endometrial cancer. Estrogen causes thickening of the endometrium through epithelial proliferation that is abruptly blocked and switched to a state of stromal-epithelial differentiation upon the addition of progesterone. The opposing action of progesterone on estrogen forms the rationale for progestin-based therapeutics for endometrial cancers. However, a continuing clinical puzzle is the patient with recurrent or metastatic endometrial cancer who is poorly responsive to progestin treatment (Obel et al. 2006). Thus far, there is no evidence demonstrating that progesterone directly induces differentiation and apoptosis of endometrial epithelial cells using conventional culture methods (Pierro et al. 2001). Our laboratory, as well as other laboratories, found that progesterone downregulation of estrogen in the endometrial epithelial cell is mediated by the stimulation of 17β-hydroxysteroid dehydrogenase type 2 (HSD17B2), which is a key enzyme that oxidizes estradiol (E2) to estrone, testosterone to androstene-dione, and 20-α-dihydroprogesterone to progesterone (Bulun et al. 2002). Further studies have indicated that the effect of progesterone on HSD17B2 expression occurs via a paracrine mechanism, whereby stromal endometrial cells secrete paracrine retinoids, which in turn stimulate HSD17B2 transcription within endometrial epithelial cells (Yang et al. 2001, Cheng et al. 2006).
Retinoids are involved in the proliferation, differentiation, and apoptosis of various cell types (Lohnes et al. 1995, Meyer et al. 1996, Morriss-Kay & Ward 1999, Bastien & Rochette-Egly 2004). Active retinoids occur in three forms: alcohol (retinol), aldehyde (retinal or retinaldehyde), and acid (retinoic acid, RA). In the human body, retinol is the predominant form, but it must be converted to RA to show biological activity. RA plays important roles in development, growth, and differentiation by regulating the expression of its target genes. RA appears to directly regulate over 500 proteins (Lohnes et al. 1995, Meyer et al. 1996, Morriss-Kay & Ward 1999, Bastien & Rochette-Egly 2004). The RA signal is transduced by members of two families of nuclear hormone receptors, the RA receptors (RARs) and the retinoid X receptors (RXRs; Meyer et al. 1996, Morriss-Kay & Ward 1999, Bastien & Rochette-Egly 2004). All-trans RA (ATRA) acts as a ligand for RAR, while the isomer 9-cis RA can bind either RAR or RXR. For each receptor, there are three subtypes (α, β, and γ) and several isoforms, which differ in their tissue distribution. These receptors function as ligand-inducible transcription regulators by heterodimerizing and binding to specific DNA sequences called RAREs to modulate gene transcription.
Several lines of evidence strongly support the importance of retinoids for the maintenance of the differentiated phenotypes of endometrial epithelial tissues. Vitamin A deficiency leads to widespread hyperkeratinization, while high concentrations of retinoids promote secretory characteristics (Lohnes et al. 1994, Mendelsohn et al. 1994, Bucco et al. 1997). Retinol deficiency in rat leads to irregular estrous cycles, morphological changes in the uterine epithelium, failure to establish or complete pregnancy, and fetal malformations. Treatment with RA can restore normal uterine epithelium and maintain fertility (Bucco et al. 1997, Zheng et al. 2000). Studies from Loughney et al. have demonstrated that RA may be involved in the control of human endometrial differentiation by promoting secretory characteristics during the luteal phase of the menstrual cycle. Disruption of the genes encoding RAR and RXR in mice showed abnormalities consistent with fetal vitamin A deficiency, including agenesis of oviduct and uterus (Lohnes et al. 1994, Mendelsohn et al. 1994).
The role of retinoids as agents inducing differentiation has been under investigation for around three decades, and their use in the diet remains a promising therapy for the prevention of several types of cancer (Levi et al. 1993, Negri et al. 1996). Studies showed that β-carotene (the pro-vitamin form of RA) in the diet conferred a significant protection against the development of endometrial carcinoma (Levi et al. 1993, Negri et al. 1996). 13-cis RA combined with α-interferon has been used to treat metastatic endometrial cancer (Kudelka et al. 1993). Although it has been known for many years that retinoids are required for normal differentiation of reproductive epithelia, the role of retinoids in endometrial differentiation remains poorly understood. Studies from Carter et al. (1996) and Carter & Madden (2000) have demonstrated that RA could induce differentiation of human endometrial adenocarcinoma cell lines (CAC-1 and RL-95 cells). Further studies indicated that phosphatidylinositol 3-kinase and epidermal growth factor receptor cell signaling pathways were involved in RA-induced cell differentiation of human endometrial adenocarcinoma cells (Carter & Shaw 2000, Carter 2003). Saidi et al. (2006) have recently reported that PPARα agonist (Fenofibrate) inhibits proliferation and induces apoptosis in endometrial cancer cells in vitro, and these effects are potentiated by RA. However, the molecular mechanisms by which RA induces cell apoptosis and growth inhibition in endometrial cancer cells are not well defined.
All isoforms of RAR and RXR are expressed in the human endometrium. However, the predominant retinoid receptors found in the endometrium are RARα and RXRα (Kumarendran et al. 1996, Cheng et al. 2006). We previously showed that retinoids decreased estrogen production by inducing HSD17B2 expression in the endometrial Ishikawa cells (Cheng et al. 2008). Further studies indicated that RA activated RARα and stimulated HSD17B2 expression, and this action could be inhibited by RA-specific antagonist (ANG17230; Cheng et al. 2008). To delineate the specific roles of RARα in retinoid signaling, the identification of specific RARα target genes and pathways is crucial. Using DNA microarray analysis, we have identified critical downstream targets by ligand-activated RARα in the endometrial Ishikawa cells.
Materials and methods
Culture of endometrial malignant epithelial cells
Endometrial epithelial Ishikawa cell lines (a kind gift from Dr Masato Nishida (Kasumigaura National Hospital, Tsuchiura, Ibaraki, Japan)) were derived from human malignant endometrial epithelial cells. Previous studies indicated that Ishikawa cells treated with RA were responsive via RARα and RXRα (Bergeron et al. 1999, Dardes et al. 2002). The characterization of Ishikawa cells is described in detail elsewhere (Bergeron et al. 1999, Dardes et al. 2002). Ishikawa cells were grown in a mixture of DMEM and F12 (1:1) medium (Invitrogen Life Technologies, Inc.) with 2.5 mM l-glutamine, 1.2 g/l sodium bicarbonate, 1.5 mM HEPES, and 10% fetal bovine serum (FBS). ATRA, 9-cis RA, progesterone, and E2 were purchased from Sigma-Aldrich. AM580 was a gift from Dr Debabrate Chakravarti, and the dose and durations that we used in this study are from the following references: Chen et al. 1997, Schulman et al. 1997, and Brand et al. 2002.
Microarray hybridization and analysis
Expression profiling was performed using Sentrix human-6 Expression BeadChip arrays from Illumina (Illumina, Inc., San Diego, CA, USA). RNA was assessed for integrity using the Agilent Bioanalyzer 2100, and RNA integrity scores above 9.5 were present in all samples. Amplification was performed with 500 ng total RNA using the Illumina TotalPrep RNA Amplification kit (Ambion, Inc., Austin, TX, USA) with a 12 h in vitro transcription reaction period. The quantity and quality of biotin-UTP-incorporated cRNA were also assessed on the Agilent Bioanalyzer 2100. Amplified cRNA (1500 ng per array) was hybridized to Sentrix Human-6 Expression BeadChip arrays according to the manufacturer guidelines and detected with Fluorolink Streptavidin-Cy3 (Amersham Biosciences). Arrays were scanned using the Illumina BeadStation Scanner. The raw intensity values obtained for the scanned array images were compiled using the proprietary BeadStudio software and imported into GeneSpring GX v7.3.1 (Agilent Technologies, Santa Clara, CA, USA). A human Illumina probe set was defined in the GeneSpring Workgroup using the Illumina target IDs as the unique identifiers and annotated according to array content files supplied by Illumina (Illumina, Inc.). The data were repeated three times, and triplicate samples were used for each group.
Data normalization was performed by first setting all measurements <0.01–0.01, then applying per chip normalization to the 50th percentile and per gene normalization to the median. From an interpretation that included the three experiments, a non-parametric Welch ANOVA (where variances were not assumed equal) was performed on all 47 296 unique probes to find a subset of genes whose expression varied significantly in the presence and absence of AM580 treatment. A Benjamini and Hochberg false discovery rate multiple testing correction was applied to reduce the number of false positives (Hochberg & Benjamini 1990, Klipper-Aurbach et al. 1995, Letwin et al. 2006). For each gene, the expression level in the different treatment groups was compared with the expression level in the control group. Changes in the expression levels are reported as ‘ (treated/control) − 1’ (in the case of an increase) or ‘ − (control/treated) +1’ (in the case of a decrease). The significance of differential expression of each gene, relative to the respective control, was evaluated using an empirical Bayes shrinkage moderated t-statistic analysis (Smyth 2004). P values were adjusted with false discovery rate of multiple testing. Only genes with P count larger than 6 and the overall gene profile variance larger than the average of the triplicate variance were included in further analysis. Hierarchical clustering of the regulated genes was done using Pearson’s correlation (Meunier et al. 2007). Gene ontology (GO) analysis was based on differentiated gene list (Hosack et al. 2003), and gene set enrichment analysis was based on Kyoto Encyclopedia of genes and genomes (KEGG) pathways (Backes et al. 2007).
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay
Epithelial Ishikawa cells (3×102/well) were seeded in 96-well plates and grown to 70–80% confluence in a mixture of DMEM and F12 (1:1) medium containing 10% FBS. After overnight starvation, cells were continually cultured in FBS-free media for 48 h in the presence or absence of chemical regents. Cell growth was monitored using the Cell Proliferation 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) kit (Sigma-Aldrich) according to the manufacturer’s directions. Absorbance values at 560 and 660 nm were recorded by an ELISA reader, and the difference between these values was recorded as the optical density.
Trypan blue staining analysis
Ishikawa cells were transfected with control siRNA or siRNA specific against the RARα gene. Cells were treated with vehicle control or AM580 for 18 h. Cells were stained with trypan blue and counted.
Propidium iodide flow cytometry assay
After overnight starvation, Ishikawa cells were continually cultured in FBS-free media for 48 h in the presence or absence of chemical regents. Cultured cells were harvested using trypsin/EDTA and washed twice with PBS. After fixation with 70% ethanol for 2 h, cells were stained with propidium iodide (PI) solution (100 mg/ml RNase and 50 mg/ml in 1× PBS) for 30 min. Stained cells were subjected to flow cytometry.
siRNA transfection
RNA oligonucleotides directed against RARα (sense sequence: GGUAUUAAUUCUCGCUGGUtt) and RXRα (sense sequence: GGAGAUGCAUCUAUUUUAAtt) were purchased from Ambion (Ambion, Inc.). The efficiency of cell transfection was optimized using carboxyfluorescein-labeled negative control siRNA (Ambion, Inc.). More than 80% of the cells were transfected using 150 pmol of carboxyfluorescein-labeled siRNA per 104 cells in a six-well plate at 30–40% confluence. This amount of siRNA was used in all transfection experiments. Ishikawa cells were grown to 30–40% confluence in antibiotic-free medium in a six-well plate. siRNA (150 pmol) in 0.3 ml OptiMEM-1 (Invitrogen Life Technologies, Inc.) and 5 µl Lipofectamine 2000 (Invitrogen Life Technologies, Inc.) in 0.3 ml OptiMEM were incubated separately for 10 min at room temperature before incubating them together for 10 min at room temperature. The liposome/siRNA complex was added to the cells that were previously washed twice with 1 ml OptiMEM. The cells were incubated at 37 °C for 6 h and then were treated either with vehicle or with ATRA. A negative control siRNA was purchased from Ambion (Ambion, Inc.), which has no matches in the human genome.
Reverse transcription and real-time quantitative PCR
Total RNA was isolated from Ishikawa cells. First-strand cDNA synthesis was performed using SuperScript III Reverse Transcriptase (Invitrogen Life Technologies, Inc.) and oligo(dT) according to the manufacturer’s protocol. Real-time quantitative PCR (qPCR) was performed using a 7000 Sequence Detection System (Applied Biosystems) employing a SYBR Green Master Mix (Applied Biosystems). Primer sequences used for the detection of genes are listed in Table 1. Electro-phoresis of the real-time PCR products by 2% agarose gel electrophoresis yielded single bands of the predicted sizes (data not shown). In each instance, the amount of reverse transcription (RT)-PCR product for the gene of interest was normalized to the amount of GAPDH in the same sample. The experiments were repeated three times using triplicates in each group.
Table 1.
Primer sequences for quantitative reverse transcription (RT)-PCR and chromatin immunoprecipitation (ChlP)-PCR
| Genes |
Forward |
Reverse |
|---|---|---|
| Primers for RT-PCR (5′-3′) | ||
| DHRS3 | GATCTATCTGGTGGTGAAAGCA | CTCCTTCAGGCATTTCTCAGTC |
| TNFAIP2 | GAGCTCTGGAGCTAGAGGCA | GTAGCTCCTCAGGAACGCAG |
| SMAD3 | TTGTCCAGTCTCCCAACTGTAAC | GTCAACTGGTAGACAGCCTCAAA |
| CYP26A1 | TGATCACTTACCTGGGGCTC | AGTCTTCAGAGCAACCCGAA |
| MYEOV | CTTTAGAGTGGGCGTTGAGC | GGTCAGGCATGCTCTTCTTC |
| ZNF488 | AGCTGTCTGGACTCCTCAACA | GCACCCAGAAAAGTGCTACAG |
| NRIP1 | GTGGAACAAAGGTCATGAGTGA | CTCGAGAATACTGCTGCAAATG |
| EPHB2 | TACCTGGCCTTCCAG ACTAT | ATGGGTACATCCACCTCTTCC |
| C14orf4 | CATCGAATTCGTGATCGAGAC | CTGCTGTTGCTGTTGCTGTT |
| RAPGEF3 | CATGAGGGAGATGATTTTGGA | GCCTCCACATCCTTGATGATA |
| LAMA3 | GATGGCTCAGGCATATGTGTT | GGCAAAGCTGTTAGTGTGAGG |
| ELF3 | GCCATTGACTTCTCACGATGT | TCAGAAGAGCTG GAAGTGAG G |
| MGC3123 | GCTGATTCTAGCCTTGGTCCT | TCATTACCCTCTGATG GATG G |
| FLJ10640 | AGGGACATGGAGTTGCTCAG | GAGATGCTGATGACGAGCAG |
| SERPINB62 | CCAAATCTTGGTGCTTCCATA | GGACACTTCCACCTCCTCTTC |
| THRAP6 | CCAGTCGAGCAACTTATTCCA | AGCAATTTCTCGCCTCTCTTC |
| MAP4K2 | GCCCTGACCAAGAATCCTAAG | GGAGTGAATGGTGTCTGGAAA |
| FLJ10647 | AGGGACATGGAGTTGCTCAG | GAGATGCTGATGACGAGCAG |
| G0S2 | GGAAGATGGTGAAGCTGTACG | CTTGCTTCTGGAGAGCCTGT |
| CAPS | AGGTTTTTCCGCCAACTAGAC | AAGGAACTCCTCCAGATCCAG |
| RARα | CACATCTTCATCACCAGCAAAC | TTAATACCTGGGGTTGGTATGG |
| RARβ | CACCACTCGTGCAATAAGACAT | TCAGTCCAAAAACTAAGCAGCA |
| RARγ | TGATGCCTTCCACCCTTTACTC | GTCAATTCCACGGTGTAAACAA |
| GAPDH | CCATGGAGAAGGCTGGGG | CAAAGTTGTCATGGATGACC |
| Primers for ChIP-PCR (5′-3′) | ||
| TNFAIP2 | GTGGAATGCTCAACCCAAAT | CACTACCCTGTCCCCATCCT |
| SMAD3 | GGTGACAGCACTTGGAAAGG | GCAGCAGAAGTTTGGGTTTC |
| NRIP1 | TGGAAATCCTGTGGGTTAATG | GCCCAGTAATGTAAGGAAGCA |
| G0S2 | AATGTCAGGCTGTCCTGGAC | GGAAGAAGAGGAGGGGAAAA |
Preparation of cell protein extracts
Cell protein extracts were prepared from the endometrial Ishikawa cells using M-PER mammalian protein extraction kit (Pierce Biotechnology, Rockford, IL, USA) following the manufacturer’s instructions. The protein concentrations of the extracts were determined by the Bradford assay (Bio-Rad Laboratories, Inc.) using BSA as a standard. The protein extracts were aliquoted and stored at − 80 °C.
Immunoblotting
Equal amounts (50 µg) of protein extracted from Ishikawa cells were subjected to 7.5% SDS-PAGE. The blot membrane was blocked for 30 min in 5% instant non-fat dry milk, which was dissolved in Tris (hydroxymethyl) aminomethane-buffer saline containing 0.05% Tween-20 (TBST). The antisera used in the immunoblotting experiments included rabbit polyclonal antibodies raised against RARα (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), mouse monoclonal IgG raised against proliferating cell nuclear antigen (PCNA; Chemicon, Billerica, MA, USA), and rabbit polyclonal antibodies raised against BCL2 protein (Cell Signaling Technology, Danvers, MA, USA). The first antibody was diluted 1:5000 in milk-TBST and incubated overnight at room temperature. The blot was washed four times (5 min each wash) with TBST and incubated with secondary antiserum (HRP-coupled goat anti-rabbit IgG antiserum or HRP-coupled goat anti-mouse IgG antiserum; Chemicon) for 1 h. The blot was washed four times with TBST and developed with a chemiluminescence kit (Pierce Chemical Co.). To confirm equal loading of the samples, membranes were stripped and reprobed with a monoclonal mouse antibody specific for β-actin (Chemicon). Quantification of the blots was carried out with Quantity One software (Bio-Rad). Values given are means ± s.e.m. of three independent experiments.
Chromatin immunoprecipitation-PCR assays
Chromatin immunoprecipitation (ChIP) assays were performed using a kit following the manufacturer’s instructions (Upstate Biotechnology, Inc., Lake Placid, NY, USA). Ishikawa cells were incubated with AM580 (10−7 M) for 2 h, and then cross-linked with 1% formaldehyde. The cell lysate was sonicated to generate DNA fragments with an average length of 200–1000 bp. Genomic DNA enriched by antibody against RARα (Santa Cruz Biotechnology, Inc.) was purified by phenol extraction. The promoter regions of the target genes were detected by PCR amplification using primers listed in Table 1.
Statistical analysis
Statistical differences between samples were determined by ANOVA followed by post-hoc multiple comparison testing using the Neuman–Keuls procedure. Values are expressed as mean + s.e.m., and P<0.05 is considered statistically significant.
Results
RA inhibits endometrial epithelial cell proliferation
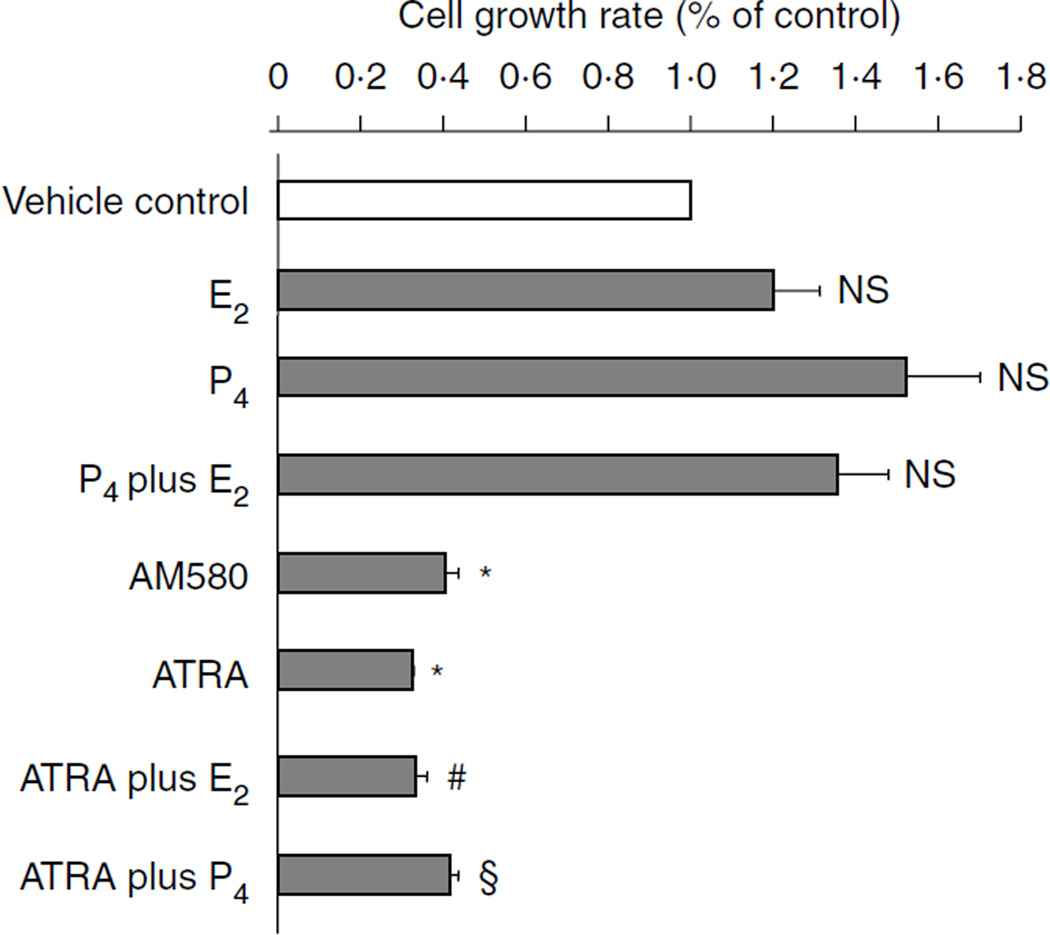
To determine whether RA inhibits endometrial epithelial cell proliferation, we treated epithelial Ishikawa cells with RA alone or in combination with either estrogen or progesterone for 48 h. Ishikawa cell proliferation was then assessed by MTT assay (Sigma–Aldrich). As shown in Fig. 1, estrogen had no significant effect, whereas progesterone slightly induced cell proliferation, the effects of steroids on cell growth had been further investigated by a wide range of concentration of E2 and progesterone in Ishikawa cells (Supplementary Figure 1, see section on supplementary data given at the end of this article). However, the addition of ATRA or RA agonist (AM580) significantly inhibited basal and hormone-stimulated Ishikawa cell proliferation by over 60%, suggesting that RA may directly modulate the hormone responsiveness of endometrial epithelial cells in vivo.
Figure 1.
MTT assay: RA but not progesterone inhibits endometrial epithelial Ishikawa cell proliferation. Ishikawa cells were plated in 96-well plates and grown to 70% confluence. After overnight starvation, the cells were exposed to vehicle (control) or ATRA(10−7M), or AM580 (10−7 M), or E2 and P4(10−7 M), or ATRA plus E2 and P4 for 48 h. MTT assay for cell proliferation was then performed. Data are reported as the mean + s.e.m. of three independent experiments. *P<0.01, compared with vehicle control; #P<0.01, compared with treatment with E2 alone; § P<0.01, compared with treatment with P4 alone; NS, not significant, compared with vehicle control; AM580, RA agonist; ATRA, all-trans RA; E2, estradiol and P4, progesterone.
RA induces endometrial epithelial cell cycle arrest at G1 phase
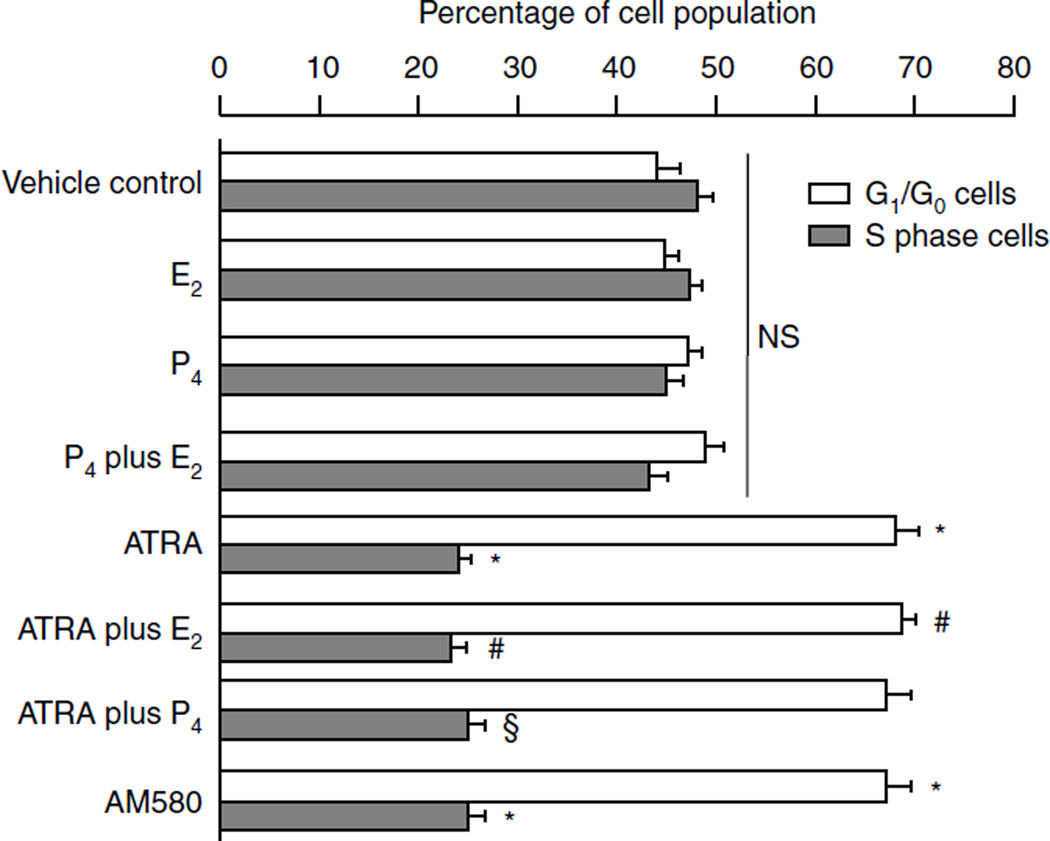
To understand further how RA and AM580 affect cell growth, we treated epithelial Ishikawa cells with RA and/or E2 and progesterone steroid hormones for 48 h. Ishikawa cell growth was assessed by PI flow cytometry assay. As shown in Fig. 2, both ATRA and AM580 significantly increased the G1/G0 cell population by 20% and decreased the S phase cells by 20%, suggesting that RA induces cell arrest at the G1 phase. Treatment with E2 or progesterone had no significant effect on Ishikawa endometrial cell cycle in vitro.
Figure 2.
Flow cytometry assay: RA induced epithelial Ishikawa cell growth arrest in the G phase. Ishikawa cells were plated in 100 mm dishes and grown to 70% confluence. After overnight starvation, Ishikawa cells were exposed to vehicle (control), or ATRA (10−7 M), or E2 and P4 (10−7 M), or ATRA plus E2 and P4, or AM580 (10−7 M) for 48 h. Cells were collected, and PI flow cytometry was performed. Distribution of cell cycle phase in asynchronously proliferating cells was examined at 48 h after treatment using PI staining. PI-stained DNA were analyzed with flow cytometer, and cell cycle analysis was performed using the ModFit program. The graphical results combine data from three independent experiments. *P<0.01, compared with vehicle control; #P<0.01, compared with treatment with E2 alone; §P<0.01, compared with treatment with P4 alone; NS, not significant, compared with vehicle control; ATRA, all-trans RA; E2, estradiol; P4, progesterone; and AM580, RA agonist.
RA regulates Ishikawa cell growth and apoptosis
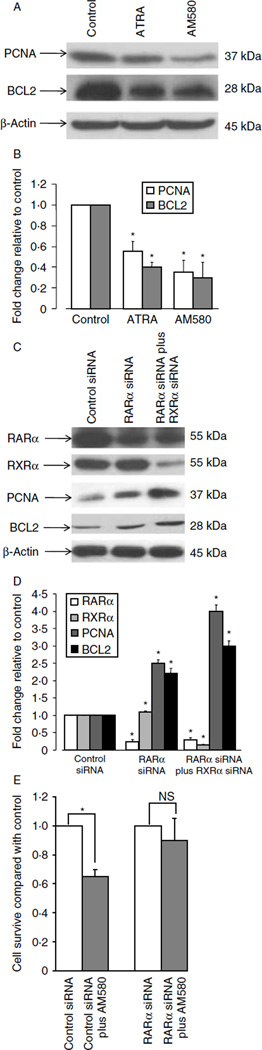
To determine whether RA and AM580 regulate the cell growth and apoptosis, we treated Ishikawa cells with ATRA and AM580 for 48 h. The protein levels of PCNA and anti-apoptotic BCL2 were determined by immunoblotting assay. As shown in Fig. 3A and B, both PCNA and BCL2 proteins levels were significantly decreased by ATRA and AM580, strongly suggesting that RA and AM580 may directly regulate endometrial Ishikawa cell proliferation and apoptosis.
Figure 3.
Immunoblotting assay: (A and B) Both ATRA and AM580 regulate cell proliferation, survival, and apoptosis. Ishikawa cells were plated in 100 mm dishes and grown to 70% confluence. After overnight starvation, Ishikawa cells were exposed to vehicle (control), AM580(10−7 M), or ATRA (10−7 M) for 48 h. Whole-cell protein was then isolated and used for immunoblotting (A). The protein levels of PCNA and BCL2 were quantitated and shown in B. (C and D) Ablation of RARα or RARα plus RXRα with siRNA significantly increased proliferating cell nuclear antigen (PCNA) and anti-apoptotic BCL2 protein levels in Ishikawa cells. Ishikawa cells were transfected with control siRNA or siRNA targeted against RARα for 24 h. Whole-cell protein was then isolated and used for immunoblotting (C). The protein levels have been quantitated and shown in D. β-Actin was used as an internal control. (E) Ishikawa cells were transfected with control siRNA or siRNA specific against RARα, and then treated with either vehicle control or AM580 for 48 h. Survival cells were counted using trypan blue staining method. The results are shown as the mean + s.e.m. of three independent experiments. NS, not significant and *P<0.01, compared with vehicle control.
siRNA ablation of RARα protein significantly increases Ishikawa cell proliferation and cell survival
To examine whether inhibition of epithelial cell growth by RA is mediated by RARα, the siRNA technique was used to knock down endogenous receptor expression in the Ishikawa endometrial epithelial cell line. Knockdown of RARα alone or both RARα and RXRα significantly increased PCNA (Fig. 3) and anti-apoptotic BCL2 protein levels, suggesting that RA signaling via RAR/RXR activation is critical for normal endometrial growth and differentiation. Knockdown of RARα and RXRα protein was confirmed by immunoblot (Fig. 3), and the decrease in RARα mRNA was also confirmed by real-time RT-PCR. Furthermore, knockdown of RARα did not significantly affect the RARβ and RARγ mRNA expression (Supplementary Figure 2, see section on supplementary data given at the end of this article). Figure 3E shows that knockdown of RARα could block AM580 inhibition of cell growth.
Microarray analysis of AM580-regulated genes in endometrial epithelial cells
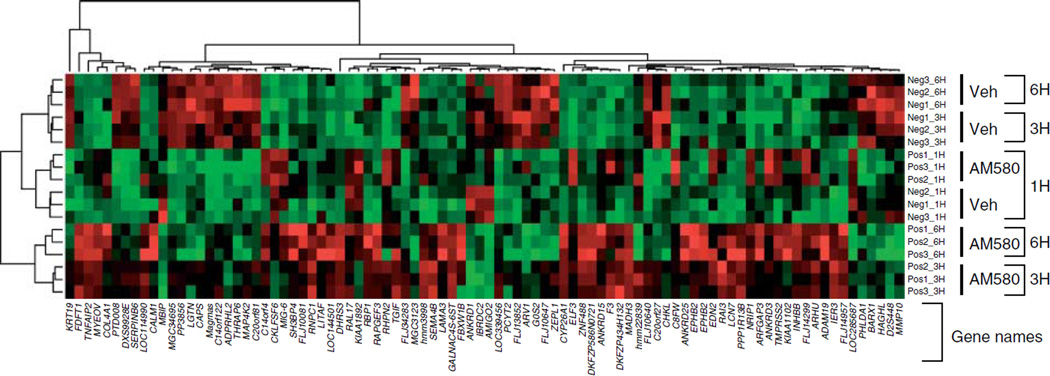
To identify RA target genes in endometrial epithelial cells, we performed triplicate microarray experiments comparing gene expression in Ishikawa cells treated continuously with 10−7 M of AM580 compound (a synthetic RARα-specific ligand; Chen et al. 1997, Schulman et al. 1997, Brand et al. 2002) for 1, 3, and 6 h. Gene expression profiles were studied using the Illumina Sentrix Human-6 Expression BeadChip Array (Biogem, La Jolla, CA, USA). Of 47 296 gene elements analyzed on the GeneChip, we identified 12 768 genes with P count larger than 6 and the overall gene profile variance larger than the average of the triplicate variance. Since there are multiple comparisons involved, we selected the genes based on F-test implemented in the limma Bioconductor package. Basically, we fit a linear model and constructed a contrast matrix with three comparisons, and then used F-test (P value) to select genes. These selected genes were differentiated in at least one comparison. Of 12 768 genes, 90 were the most regulated genes with the smallest P value using F-test (Fig. 4).
Figure 4.
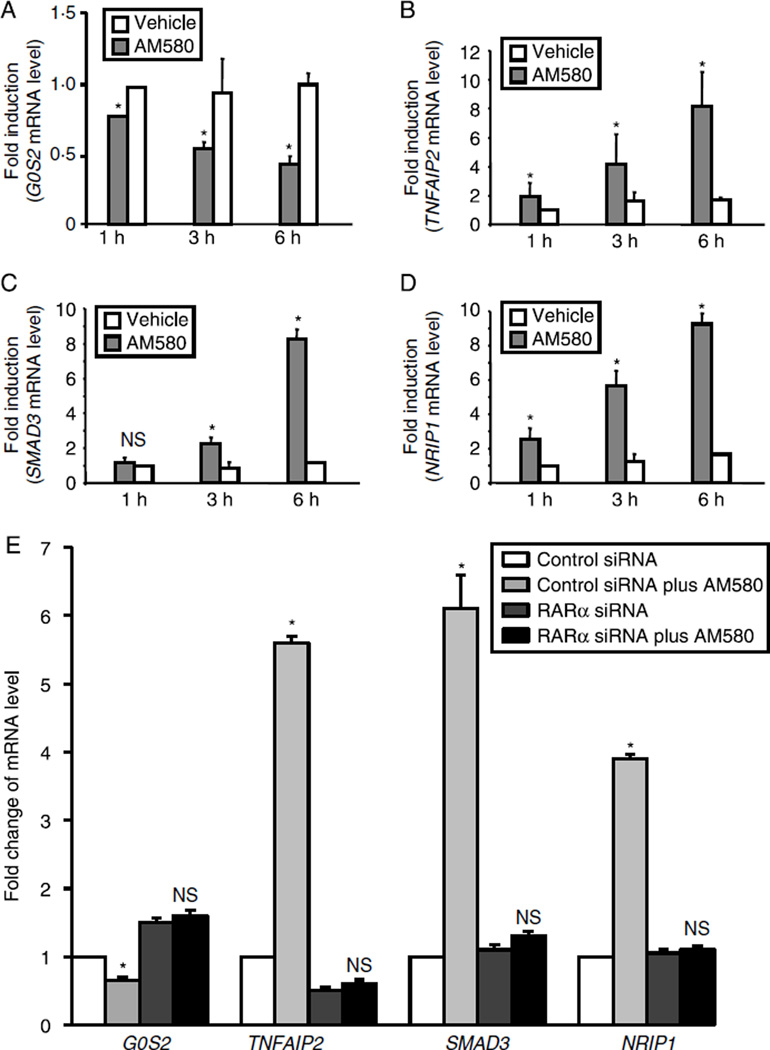
Microarray analysis: identification of differentiated genes in AM580-treated Ishikawa cells was done with a microarray analysis. Heat map of lllumina Sentrix Human-6 Expression BeadChip arrays hybridized with the RNA of Ishikawa cells that were incubated with ethanol control or AM580 (10−7 M) at three time points and each time point containing triplicate samples. Based on three comparisons at three time points, 90 genes from the test set were differentially expressed between control and AM580 treatment groups (F-test, P<0.0001). In the heat map, each column is a sample, and each row is a standardized gene expression profile with red, green, and black colors representing relative high, low, and close to average expression levels respectively. Both row and columns were hierarchically clustered using complete linkage method. Veh, vehicle; Neg, ethanol control; Pos, AM580 treatment; 1H, 1 h treatment; 3H, 3 h treatment; and 6H, 6 h treatment.
To assess the statistical significance of the difference between these groups, we represented the data as a volcano plot. The volcano plot was used to identify significance and magnitude of change in expression of a set of genes between the groups (Supplementary Figure 3, see section on supplementary data given at the end of this article). The plots display the negative log of P values from a t-test on the x-axis and the log2 of the change between two conditions on the y-axis. The coordinates represent the magnitude of the difference in expression of that gene and its significance. The data clearly demonstrate that there are many genes that were significantly different between the two groups, and that this was not due to chance. Most of the AM580 targeted genes showed a more gradual change in expression at 1, 3, and 6 h after AM580 treatment (Supplementary Figure 1). To validate the microarray data, qRT-PCR was employed to assess the expression of the selected genes (Table 2 and Fig. 5). To test the effect of endogenous RA on mRNA expression of the selected genes, we treated Ishikawa cells with ATRA for 1, 3, and 6 h. mRNA levels of the selected genes were measured by qRT-PCR. As shown in Table 3, ATRA has same effects as AM580 in Ishikawa cells. The full table of volcano plots, by significance level, is provided in the Supplementary Tables 1 and 2, see section on supplementary data given at the end of this article.
Table 2.
Comparison of the 20 most highly up- or downregulated genes by AM580 between microarray data and quantitative reverse transcription (qRT)-PCR in endometrial cancer cells
| Gene description |
GenBank Accession Numbers |
1 h |
3 h |
6 h |
|||
|---|---|---|---|---|---|---|---|
| Microarray |
qRT-PCR |
Microarray |
qRT-PCR |
Microarray |
qRT-PCR |
||
| DHRS3 | AF061741 | 1.3401 | 5.1 (*) | 2.4582 | 8.2 (*) | 3.2714 | 13.6 (*) |
| TNFAIP2 | BC128449 | 1.3115 | 1.9 (*) | 2.1012 | 4.1 (*) | 2.6756 | 8.1 (*) |
| SMAD3 | BC050743 | NS | 1.5 (NS) | 1.8594 | 3.9 (*) | 1.9913 | 8.3 (*) |
| CYP26A1 | AF005418 | NS | 10 (*) | 1.7661 | 28 (*) | 1 .9886 | 72 (*) |
| MYEOV | AJ223366 | NS | 1.3 (NS) | 1.5083 | 1.6 (NS) | 1.6419 | 1.8 (*) |
| ZNF488 | AK056666 | NS | 3.0 (*) | 1.2397 | 6.0 (*) | 1.3156 | 3.9 (*) |
| NRIP1 | X84373 | 1.2135 | 2.2 | 1.2211 | 2.9 (*) | 1.3134 | 4.1 (*) |
| EPHB2 | AF025304 | 1.1672 | 0.6 (NS) | 1.3274 | 1.0 (NS) | 1 .2344 | 0.85 (NS) |
| C14orf4 | AJ277365 | NS | 1.55 (NS) | NS | 0.95 (NS) | 1 .3542 | 1.0 (NS) |
| RAPGEF3 | AK092448 | NS | 1.2 (NS) | NS | 3.4 (*) | 1 .4287 | 2.0 (*) |
| LAMA3 | L34155 | NS | 2.3 (*) | NS | 2.1 (*) | 1.1085 | 1.0 (NS) |
| ELF3 | AF016295 | 1.2689 | 5.8 (*) | 1.2353 | 9.9 (*) | NS | 1.5 (NS) |
| MGC3123 | NM_177441 | NS | 1.4 (NS) | NS | 1.5 (NS) | −1.1441 | 0.75 (*) |
| FLJ10640 | AK001502 | NS | 1.1 (NS) | NS | 0.8 (NS) | −1.1458 | 0.6 (*) |
| SERPINB6 | Z22658 | NS | 1.4 (NS) | NS | 1.2 (NS) | −1.1515 | 1.5 (NS) |
| THRAP6 | AY083305 | NS | 1.1 (NS) | NS | 0.6 (*) | −1.1966 | 1.2 (NS) |
| MAP4K2 | BC047865 | NS | 1.05 (NS) | NS | 1.5 (NS) | −1.2109 | 0.95 (NS) |
| FLJ10647 | AK001509 | NS | 1.4 (NS) | −1.1237 | 0.95 (NS) | −1.2139 | 0.9 (NS) |
| G0S2 | NM_015714 | NS | 0.75 (*) | −1.1458 | 0.65 (*) | −1.2791 | 0.51 (*) |
| CAPS | X97966 | NS | 1.1 (NS) | −1.1515 | 1.2 (NS) | −1.3405 | 0.63 (*) |
NS, not significant.
P<0.01.
Figure 5.
(A–D) Analysis of AM580-regulated genes by quantitative RT-PCR: after overnight starvation, Ishikawa cells were exposed to AM580 (10−7 M) or vehicle for the indicated time. Total cellular RNA was isolated and subjected to quantitative RT-PCR. The amount of AM580 target genes was normalized to the amount of GAPDH in the same sample. (E) Ishikawa cells were transfected with either control siRNA or siRNA specific against RARα. Cells were then treated with vehicle or AM580 for 18 h. Total cellular RNA was isolated and subjected to quantitative RT-PCR. The amount of AM580 target genes was normalized to the amount of GAPDH in the same sample. The results are shown as the mean + s.e.m. of three independent experiments. NS, not significant, and *P<0.01.
Table 3.
Fold changes of mRNA level of 20 selected genes by all-trans retinoic acid in endometrial cancer cells
| Gene description |
Gen Bank accession numbers |
1 h |
2 h |
3 h |
|||
|---|---|---|---|---|---|---|---|
| Microarray |
P value |
Microarray |
P value |
Microarray |
P value |
||
| DHRS3 | AF061741 | 4.2 | <0.01 | 11.3 | <0.01 | 13.6 | <0.01 |
| TNFAIP2 | BC128449 | 2.8 | <0.01 | 8.1 | <0.01 | 7.9 | <0.01 |
| SMAD3 | BC050743 | 2.3 | <0.01 | 2.8 | <0.01 | 5.2 | <0.01 |
| CYP26A1 | AF005418 | 12 | <0.01 | 25 | <0.01 | 65 | <0.01 |
| MYEOV | AJ223366 | 1.2 | 0.56 | 1.3 | 0.29 | 1.5 | 0.25 |
| ZNF488 | AK056666 | 2.0 | <0.01 | 3.1 | <0.01 | 2.5 | <0.01 |
| NRIP1 | X84373 | 2.6 | <0.01 | 3.5 | <0.01 | 3.9 | <0.01 |
| EPHB2 | AF025304 | 0.9 | 0.23 | 1.0 | 0.46 | 1.1 | 0.49 |
| C14orf4 | AJ277365 | 1.3 | 0.39 | 1.5 | 0.45 | 1.3 | 0.44 |
| RAPGEF3 | AK092448 | 1.5 | 0.29 | 2.5 | <0.01 | 1.2 | 0.09 |
| LAMA3 | L34155 | 3.9 | <0.01 | 2.3 | <0.01 | 1.3 | 0.23 |
| ELF3 | AF016295 | 2.8 | <0.01 | 5.3 | <0.01 | 1.6 | 0.13 |
| MGC3123 | NM_177441 | 1.1 | 0.33 | 1.3 | 0.22 | 0.85 | 0.39 |
| FLJ10640 | AK001502 | 1.2 | 0.35 | 1.5 | 0.18 | 0.72 | 0.25 |
| SERPINB6 | Z22658 | 0.9 | 0.29 | 1.3 | 0.39 | 0.7 | 0.21 |
| THRAP6 | AY083305 | 1.2 | 0.39 | 0.72 | <0.05 | 0.69 | 0.11 |
| MAP4K2 | BC047865 | 1.5 | 0.41 | 1.9 | 0.22 | 0.55 | 0.99 |
| FLJ10647 | AK001509 | 1.3 | 0.32 | 1.5 | 0.17 | 0.68 | <0.05 |
| G0S2 | NM_015714 | 0.71 | <0.05 | 0.72 | <0.01 | 0.65 | <0.01 |
| CAPS | X97966 | 1.3 | 0.43 | 0.68 | 0.06 | 0.63 | <0.01 |
GO stores controlled vocabulary organized under the three independent ontologies, i) biological process; ii) molecular function; and iii) cellular component (Dennis et al. 2003). Using significantly up- and downregulated genes, hypergeometric test identified the GO categories altered, and we have shown a representative number of these processes in Table 3. Of 842 genes, 237 were identified with differential expression based on hypergeometric test (P<0.05). The full table of GO classifications, by significance level, is provided in the Supplementary Table 3, see section on supplementary data given at the end of this article.
In order to understand the higher order functional significance of the differentially expressed genes, KEGG pathway analysis was performed (Backes et al. 2007). We identified 18 pathways that were significantly affected by mRNA expression variances in the comparison between AM580 treatment and vehicle control (Table 4). These important pathways included the mitogen-activated protein kinase (MAPK) signaling pathway, focal adhesion, Wnt signaling pathway, extracellular matrix (ECM)-receptor interaction and cell communication, and cell apoptosis. The full table of RA target genes associated with KEGG pathway is provided in the Supplementary Table 4, see section on supplementary data given at the end of this article.
Table 4.
Gene ontology (GO) categories according to the involvement in biological processes
| GO term |
Total genes in GO |
Increased genes |
Decreased genes |
P value |
|---|---|---|---|---|
| Protein biosynthesis | 248 | 31 | 41 | 0.0000068 |
| Notch signaling pathway | 24 | 7 | 4 | 0.0009943 |
| Chromatin modification | 75 | 13 | 12 | 0.0011292 |
| Regulation of progression via cell cycle | 208 | 29 | 25 | 0.0022299 |
| DNA replication | 91 | 12 | 14 | 0.0042991 |
| DNA unwinding during replication | 11 | 6 | 0 | 0.0052119 |
| DNA repair | 125 | 12 | 22 | 0.0055608 |
| Positive regulation of transcription | 42 | 11 | 5 | 0.0078227 |
| DNA dependent apoptotic program | 9 | 4 | 1 | 0.0099475 |
| DNA topological change | 9 | 5 | 0 | 0.0099475 |
Details in Supplementary Table 3, see section on supplementary data given at the end of this article.
RA regulated the mRNA expression of the G0/G1 switch gene 2 (G0S2), SMAD3, nuclear receptor interacting protein 1 (NRIP1), and tumor necrosis factor-α-induced protein 2 (TNFAIP2) genes in the endometrial epithelial Ishikawa cells. To validate the microarray data, real-time RT-PCR was employed to assess the expression of the G0S2, SMAD3, NRIP1, and TNFAIP2 genes in the endometrial Ishikawa cells in the presence or absence of AM580. As shown in Fig. 5A, AM580 significantly inhibited G0S2 expression in the endometrial cancer cells. Figure 5B–D indicate that AM580 markedly increased SMAD3, NRIP1, and TNFAIP2 mRNA levels in the endometrial Ishikawa cells. Figure 5E indicates that induction of SMAD3, NRIP1, and TNFAIP2 mRNA expression by AM580 could be abolished by knockdown of RARα gene in Ishikawa cells. Interestingly, ablation of RARα gene could not block AM580 induction of G0S2 mRNA level in Ishikawa cells, suggesting that other nuclear transcriptional factor (s) may be involved in the regulation of G0S2 gene expression in the endometrial cells.
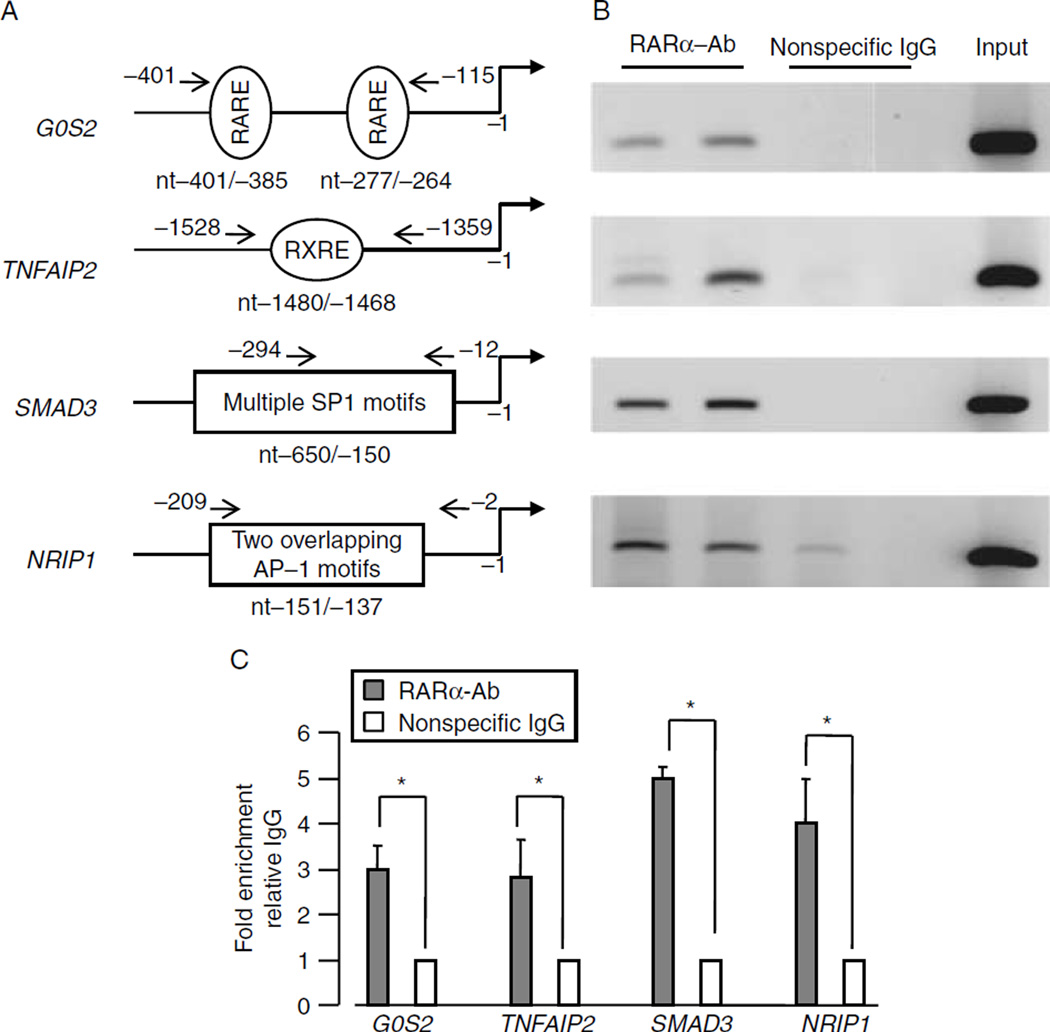
RA-activated RARα binds to the G0S2, SMAD3, NRIP1, and TNFAIP2 promoters in the endometrial epithelial Ishikawa cells. Based on the preliminary data from the microarray analysis, qRT-PCR analysis, and the documented functions of each RA target, we selected transcription factors (G0S2, SMAD3, NRIP1, and TNFAIP2) for more in-depth investigation. ChIP-PCR assays were performed to determine whether RARα protein interacts with these RA target genes. Ishikawa cells were exposed to AM580 (10−7 M) for 2 h, and then cross-linked with 1% formaldehyde. Sheared chromatin isolated from Ishikawa cells was immuno-precipitated with antibodies against RARα or normal IgG. As shown in Fig. 6, RARα could interact with the G0S2, SMAD3, NRIP1, and TNFAIP2 genes.
Figure 6.
ChIP-PCR assay: ChIP-PCR assays demonstrate that RARα interacts with the promoters of the RA target genes in epithelial Ishikawa cells. After overnight starvation, Ishikawa cells were exposed to AM580 compound (10−7 M) for 2 h. After cross-linking with 1% formaldehyde, the sheared cellular chromatins were used for ChIP-PCR assay. The physical structures of the RA-RARα target genes are shown in (A), the ChIP-PCR results are shown in (B), and quantitative fold enrichment relative to IgG is shown in (C). The results are shown as the mean + s.e.m. of three independent experiments.
Discussion
The biologically potent estrogen causes thickening of the endometrium through epithelial proliferation in vivo. However, proliferation effect of estrogen on cultured endometrial epithelial cell lines is not clear. Studies showed that longer (2–3 weeks) exposure to estrogen could induce Ishikawa endometrial cell growth in vitro (Holinka et al. 1986), but shorter time incubation of estrogen with these endometrial cells could not significantly stimulate cell proliferation (Pierro et al. 2001). These findings indicated that the regulation of cell growth by steroids in vitro has a substantially molecular difference comparing their in vivo status. The studies from tissue recombinant experiments in mice have indicated that epithelial steroid receptors are not required for the regulation of growth, differentiation, and apoptosis of normal epithelial cells (Kurita et al. 2001a,b). The growth, differentiation, and apoptosis of endometrial epithelial cells are regulated by stromal steroid receptors via a paracrine mechanism, whereby stromal endometrial cells secrete paracrine substances, which in turn, regulate targeted gene expressions within endometrial epithelial cells (Kurita et al. 2001a,b).
RA has the ability to induce differentiation and/or growth arrest in a variety of cancer cells (Morriss-Kay & Ward 1999, Bastien & Rochette-Egly 2004). In this study, we demonstrate that RA and RA agonist significantly inhibit basal and hormone-stimulated Ishikawa cell proliferation by over 60% using MTT assay. Cell cycle analysis indicated that both ATRA and AM580 significantly increased the G1/G0 cell population by 20% and decreased the S phase cells by 20%, suggesting that RA induces cell cycle arrest at the G1 phase. Treatment of Ishikawa cells with ATRA or AM580 could significantly decrease PCNA and anti-apoptotic BCL2 protein expression. Knockdown of RARα alone or both RARα and RXRα significantly increases PCNA and BCL2 protein levels in Ishikawa cells. These data strongly suggest that RA signaling via RAR/RXR activation may play a critical role in mediating the carcinogenesis of human endometrial cancer, and RARα may be an important new target for treatment of this disease.
RA has been used to treat metastatic endometrial cancer (Kudelka et al. 1993). However, the genes and pathways that mediate the biological effects of RA have not been fully elucidated. In this study, we used human DNA microarray to determine the time-dependent changes in gene expression in Ishikawa cells treated with control versus AM580, a RARα-specific ligand. Our results show a small number of changes in gene expression at the early time point (1 h treatment with AM580). RA target genes showed a more gradual change in expression at 1, 3, and 6 h after treatment with AM580. There was a major increase in the number of genes whose expression changed at 6 h of AM580 treatment. We also noted that at this time point, the number of genes that were downregulated (187 genes) outnumbered those that were upregulated (156 genes; Supplementary Tables 1 and 2). To our knowledge, this is the first reported study of time-dependent changes in the gene expression of a human endometrial cancer cell line treated with RA.
Among the identified GO groups, ontologies related to protein biosynthesis, chromatin modification, regulation of progression via cell cycle, and DNA replication and repair were the top four groups in the list of differentially expressed genes (Table 3 and Supplementary Table 3). Within these functional groups, genes that were up- or down-regulated were generally consistent with the experimental findings of inhibition of G1-S phase progression. The target genes classified in the cell cycle category include G0S2, genes for cell division cycle (CDC), cyclin-dependent kinase 10 (CDK-10), and MAPK3 (Supplementary Table 3). Many of these targets have been found to be aberrantly expressed in malignant cancer cells (Weinert & Hartwell 1993, Hartwell et al. 1994). Interestingly, studies from Saidi et al. (2006) indicated that fenofi-brate (an agonist of PPARα) downregulates cyclin D1 (CCND1) and other cell cycle-related gene expression in the endometrial Ishikawa cells.
KEGG pathway analysis of specific signaling pathways that mediate the effect of RA inhibition of cell proliferation and stimulation of differentiation provide a valuable profile for systemic biology capture of the properties of cellular activities. MAPK pathway is one of the most important pathways regulated by RA (Table 4 and Supplementary Table 4). Using KEGG pathway analysis, we identified 25 MAPK components that were significantly induced, while 10 MAPK components were decreased by AM580 treatment. These targets include DUSP3, DUSP5, MAPK1, MAPK3, MAP3K1, JunD, and EVI1 (Supplementary Table 4).
Cell apoptosis is the process of programmed cell death that involves a series of biochemical events leading to a characteristic cell morphological change and death. In the gene set enrichment analysis based on KEGG pathways, nine genes were significantly increased and four genes were decreased after AM580 treatment for 6 h (Table 4 and Supplementary Table 4). These genes may be potential drug targets for human endometrial cancer.
Of the 343 differentially expressed transcripts (Supplementary Tables 1 and 2), the genes for DHRS3, TNFAIP2, SMAD3, CYP26A1, and NRIP1 were the most upregulated, while the genes for CAPS, G0S2, and MAP4K2 were the most down-regulated (Table 2). These changes in gene expression by RA may directly regulate endometrial cell growth and differentiation. It has been known that both DHRS3 and CYP26A1 enzymes regulate cellular level of RA that is a significant self-protective mechanism in intact cells. However, increased retinoid metabolism by CYP26A1 causes the RA resistance that can be seen clinically with the RA treatment of human cancers (Patel et al. 2004, Njar et al. 2006). Studies have shown that CYP26A1 is a potential target for RA metabolism blocking agents.
SMAD3 is a member of the SMAD family of transcription factors that mediate intracellular signaling by the transforming growth factor-β (TGF-β) superfamily (Bruckheimer & Kyprianou 2000). Several lines of evidence strongly support the role of TGF-β in mediating growth, differentiation, and apoptosis of endometrial epithelial cells. Müllerian inhibiting substance, a glycoprotein closely related to the TGF-β family of molecules, is involved in the apoptotic regression of the Müllerian ducts in the reproductive organ development (Roberts et al. 1999). TGF-β stimulates normal cervical remodeling and induces apoptosis of cultured cervical cells (Jacobberger et al. 1995, Rorke & Jacobberger 1995, Woodworth et al. 1996, Rorke et al. 2000). Rotello et al. (1991) found that TGF-β1 regulates cell proliferation and apoptosis in cultured uterine epithelial cells. Wada et al. (1996) demonstrated that transcripts for TGF-β1, −2, and −3 and their type-II receptors are upregulated in mouse uterus after E2 withdrawal, and that administration of progesterone inhibits this upregulation. Our data showed that AM580 dramatically induces SMAD3 expression in the endometrial cancer cells (Fig. 5 and Table 2), strongly suggesting that RA-induced apoptosis of endometrial cells may occur, at least in part, via modulation of the TGF-β signaling pathway (Table 5).
Table 5.
Retinoic acid target cell signaling pathways based on Kyoto Encyclopedia of genes and genomes pathways
| Pathway ID |
Pathway name |
P value |
|---|---|---|
| Hsa00190 | Oxidative phosphorylation | 0.0000357 |
| Hsa03010 | Ribosome | 0.0004350 |
| Hsa04810 | Regulation of actin cytoskeleton | 0.0006072 |
| Hsa04010 | MAPK signaling pathway | 0.0015832 |
| Hsa04510 | Focal adhesion | 0.0020038 |
| Hsa00600 | Glycosphingolipid metabolism | 0.0088223 |
| Hsa04310 | Wnt signaling pathway | 0.0106631 |
| Hsa04530 | Tight junction | 0.0107101 |
| Hsa00062 | Fatty acid elongation in mitochondria |
0.0156252 |
| Hsa01430 | Cell communication | 0.0214341 |
| Hsa00561 | Glycerolipid metabolism | 0.0226540 |
| Hsa04514 | Cell adhesion molecules (CAMs) | 0.0227252 |
| Hsa00564 | Glycerophospholipid metabolism | 0.0291549 |
| Hsa04512 | ECM-receptor interaction | 0.0299668 |
| Hsa05040 | Huntington’s disease | 0.0347901 |
| Hsa04210 | Apoptosis | 0.0404347 |
| Hsa05050 | Dentatorubropallidoluysian atrophy (DRPLA) |
0.0419918 |
| Hsa00240 | Pyrimidine metabolism | 0.0482376 |
Details in Supplementary Table 4, see section on supplementary data given at the end of this article.
G0S2 is a member of the G0/G1 switch gene family, which was first reported as markedly induced in mononuclear hematopoietic cells following treatment with the lectin concanavalin A and the inhibitor of protein synthesis, cycloheximide (CHX; Cristillo et al. 1997, Bachner et al. 1998). Because its mRNA expression coincided with cell cycle progression from G0 to G1 by lectin or CHX treatment of lymphocytes and was induced during differentiation of adipocytes by PPAR ligands, the biological role of G0S2 may be involved in the cell cycle regulation and cell differentiation. In this study, we show that retinoids induce epithelial cell cycle arrest at the G0/G1 phase, and a RARα-specific ligand (AM580) inhibits G0S2 expression in the endometrial cancer cells (Fig. 5 and Table 2), indicating that retinoid inhibition of epithelial cell proliferation may occur via inhibition of G0S2 expression in the endometrial epithelial cells.
NRIP1 is a nuclear protein that specifically interacts with the hormone-dependent activation domain AF2 of nuclear receptors (Lee et al. 1998). Also known as RIP140, this protein may interact with the transcription factor AP1 to antagonize ER transcription in vivo (Lee et al. 1998, Teyssier et al. 2003). Given the fact that RA stimulates NRIP1 expression in the endometrial cancer cells (Fig. 5 and Table 2), it is possible that the anti-estrogenic effect of RA may occur via regulation of NRIP1 expression in the endometrium.
TNFAIP2 is a cytokine-induced early gene that was originally identified as a TNF-α-inducible transcript in human endothelial cells (Rusiniak et al. 2000). TNF-α-mediated induction of apoptosis has been well characterized in myeloid cells (Witcher et al. 2003). Considering that RA dramatically increases TNFAIP2 mRNA levels in Ishikawa cells (Fig. 5 and Table 2), TNFAIP2 may be involved in RA-induced apoptosis human endometrium.
Numerous studies strongly support the idea that ligand-activated RARα regulates the critical events in cellular differentiation and apoptosis (Bruckheimer & Kyprianou 2000, Witcher et al. 2003, Tarrade et al. 2005, Pavone et al. 2010). The molecular mechanisms underlying RARα regulation of its targets are of considerable interest. Classically, RARα functions as ligand-inducible transcription regulators by hetero-dimerizing and binding to specific DNA sequences called RAREs to modulate gene transcription. Numerous studies have also shown that RARα could interact with other transcriptional factors (e.g. SP1) to regulate target gene transcription. Using ChIP-PCR analysis, we demonstrated that ligand-activated RARα could interact with the promoter of RA target genes that bear a classic RARE (i.e. G0S2 and TNFAIP2) or a promoter that contains only SP1 (i.e. SP3) or AP1 (i.e. NRIP1) motifs (Fig. 6). The biological functions of these targets will be key issues for future studies.
In summary, RA treatment of endometrial cancer cells causes the differential expression of a number of target genes and KEGG pathways. Many of these pathways, most importantly MAPK cascade and TGF-β signaling, are involved in cell growth, proliferation, differentiation, and apoptosis and could, therefore, mediate carcinogenesis of human endometrial cancer.
Supplementary Material
Acknowledgements
We thank Drs Simon Lin and Pan Du in the Center for Biomedical Bioinformatics of Northwestern University Feinberg School of Medicine for assistance in bioinformatics analysis.
Funding
Supported by the NIH grant HD40093 and grants from the Friends of Prentice and AVON Foundation.
Footnotes
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1530/JME-10-0064.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution statement
Y H C performed the bioinformatics statistical analysis, drafted the manuscript, and supervised the project. H U designed and performed the molecular studies. M E P and P Y performed the molecular studies and helped to draft the manuscript. S E B participated in the data interpretation and supervised the project. All authors read and approved the final manuscript.
References
- Bachner D, Ahrens M, Schroder D, Hoffmann A, Lauber J, Betat N, Steinert P, Flohe L, Gross G. Bmp-2 downstream targets in mesenchymal development identified by subtractive cloning from recombinant mesenchymal progenitors (C3H10T1/2) Developmental Dynamics. 1998;213:398–411. doi: 10.1002/(SICI)1097-0177(199812)213:4<398::AID-AJA5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail – advanced gene set enrichment analysis. Nucleic Acids Research. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Bergeron RM, Thompson TB, Leonard LS, Pluta L, Gaido KW. Estrogenicity of bisphenol A in a human endometrial carcinoma cell line. Molecular and Cellular Endocrinology. 1999;150:179–187. doi: 10.1016/s0303-7207(98)00202-0. [DOI] [PubMed] [Google Scholar]
- Brand C, Segard P, Plouvier P, Formstecher P, Danze PM, Lefebvre P. Selective alteration of gene expression in response to natural and synthetic retinoids. BMC Pharmacology. 2002;2:13. doi: 10.1186/1471-2210-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckheimer EM, Kyprianou N. Apoptosis in prostate carcinogenesis. A growth regulator and a therapeutic target. Cell and Tissue Research. 2000;301:153–162. doi: 10.1007/s004410000196. [DOI] [PubMed] [Google Scholar]
- Bucco RA, Zheng WL, Davis JT, Sierra-Rivera E, Osteen KG, Chaudhary AK, Ong DE. Cellular retinoic acid-binding protein(II) presence in rat uterine epithelial cells correlates with their synthesis of retinoic acid. Biochemistry. 1997;36:4009–4014. doi: 10.1021/bi962094o. [DOI] [PubMed] [Google Scholar]
- Bulun SE, Yang S, Fang Z, Gurates B, Tamura M, Sebastian S. Estrogen production and metabolism in endometriosis. Annals of the New York Academy of Sciences. 2002;955:75–85. doi: 10.1111/j.1749-6632.2002.tb02767.x. [DOI] [PubMed] [Google Scholar]
- Carter CA. Retinoic acid signaling through PI3-kinase induces differentiation of human endometrial adenocarcinoma cells. Experimental and Molecular Pathology. 2003;75:34–44. doi: 10.1016/s0014-4800(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Carter CA, Madden VJ. A newly characterized human endometrial adenocarcinoma cell line (CAC-1) differentiates in response to retinoic acid treatment. Experimental and Molecular Pathology. 2000;69:175–191. doi: 10.1006/exmp.2000.2334. [DOI] [PubMed] [Google Scholar]
- Carter CA, Shaw BL. Retinoic acid affects the EGF-R signaling pathway during differentiation induction of human endometrial adenocarcinoma cells. Experimental and Molecular Pathology. 2000;68:170–186. doi: 10.1006/exmp.2000.2301. [DOI] [PubMed] [Google Scholar]
- Carter CA, Pogribny M, Davidson A, Jackson CD, McGarrity LJ, Morris SM. Effects of retinoic acid on cell differentiation and reversion toward normal in human endometrial adenocarcinoma (RL95-2) cells. Anticancer Research. 1996;16:17–24. [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Cheng YH, Imir A, Suzuki T, Fenkci V, Yilmaz B, Sasano H, Bulun SE. SP1 and SP3 mediate progesterone-dependent induction of the 17β-hydroxysteroid dehydrogenase type 2 gene in human endometrium. Biology of Reproduction. 2006;75:605–614. doi: 10.1095/biolreprod.106.051912. [DOI] [PubMed] [Google Scholar]
- Cheng YH, Yin P, Xue Q, Yilmaz B, Dawson MI, Bulun SE. Retinoic acid (RA) regulates l7β-hydroxysteroid dehydrogenase type 2 expression in endometrium: interaction of RA receptors with specificity protein (SP) 1/SP3 for estradiol metabolism. Journal of Clinical Endocrinology and Metabolism. 2008;93:1915–1923. doi: 10.1210/jc.2007-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristillo AD, Heximer SP, Russell L, Forsdyke DR. Cyclosporin A inhibits early mRNA expression of G0/G1 switch gene 2 (G0S2) in cultured human blood mononuclear cells. DNA and Cell Biology. 1997;16:1449–1458. doi: 10.1089/dna.1997.16.1449. [DOI] [PubMed] [Google Scholar]
- Dardes RC, Schafer JM, Pearce ST, Osipo C, Chen B, Jordan VC. Regulation of estrogen target genes and growth by selective estrogen-receptor modulators in endometrial cancer cells. Gynecologic Oncology. 2002;85:498–506. doi: 10.1006/gyno.2002.6659. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4:P3. [PubMed] [Google Scholar]
- Hartwell L, Weinert T, Kadyk L, Garvik B. Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harbor Symposia on Quantitative Biology. 1994;59:259–263. doi: 10.1101/sqb.1994.059.01.030. [DOI] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in Medicine. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Holinka CF, Hata H, Gravanis A, Kuramoto H, Gurpide E. Effects of estradiol on proliferation of endometrial adenocarcinoma cells (Ishikawa line) Journal of Steroid Biochemistry. 1986;25:781–786. doi: 10.1016/0022-4731(86)90308-0. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobberger JW, Sizemore N, Gorodeski G, Rorke EA. Transforming growth factor β regulation of epidermal growth factor receptor in ectocervical epithelial cells. Experimental Cell Research. 1995;220:390–396. doi: 10.1006/excr.1995.1330. [DOI] [PubMed] [Google Scholar]
- Klipper-Aurbach Y, Wasserman M, Braunspiegel-Weintrob N, Borstein D, Peleg S, Assa S, Karp M, Benjamini Y, Hochberg Y, Laron Z. Mathematical formulae for the prediction of the residual beta cell function during the first two years of disease in children and adolescents with insulin-dependent diabetes mellitus. Medical Hypotheses. 1995;45:486–490. doi: 10.1016/0306-9877(95)90228-7. [DOI] [PubMed] [Google Scholar]
- Kudelka AP, Freedman RS, Edwards CL, Lippman SM, Tornos CS, Krakoff IH, Kavanagh JJ. Metastatic adenocarcinoma of the endometrium treated with 13-cis-retinoic acid plus interferon-α. Anticancer Drugs. 1993;4:335–337. doi: 10.1097/00001813-199306000-00007. [DOI] [PubMed] [Google Scholar]
- Kumarendran MK, Loughney AD, Prentice A, Thomas EJ, Redfern CP. Nuclear retinoid receptor expression in normal human endometrium throughout the menstrual cycle. Molecular Human Reproduction. 1996;2:123–129. doi: 10.1093/molehr/2.2.123. [DOI] [PubMed] [Google Scholar]
- Kurita T, Wang YZ, Donjacour AA, Zhao C, Lydon JP, O’Malley BW, Isaacs JT, Dahiya R, Cunha GR. Paracrine regulation of apoptosis by steroid hormones in the male and female reproductive system. Cell Death and Differentiation. 2001a;8:192–200. doi: 10.1038/sj.cdd.4400797. [DOI] [PubMed] [Google Scholar]
- Kurita T, Cooke PS, Cunha GR. Epithelial-stromal tissue interaction in paramesonephric (Mullerian) epithelial differentiation. Developmental Biology. 2001b;240:194–211. doi: 10.1006/dbio.2001.0458. [DOI] [PubMed] [Google Scholar]
- Lee CH, Chinpaisal C, Wei LN. Cloning and characterization of mouse RIP140, a corepressor for nuclear orphan receptor TR2. Molecular and Cellular Biology. 1998;18:6745–6755. doi: 10.1128/mcb.18.11.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letwin NE, Kafkafi N, Benjamini Y, Mayo C, Frank BC, Luu T, Lee NH, Elmer GI. Combined application of behavior genetics and microarray analysis to identify regional expression themes and gene-behavior associations. Journal of Neuroscience. 2006;26:5277–5287. doi: 10.1523/JNEUROSCI.4602-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Franceschi S, Negri E, La Vecchia C. Dietary factors and the risk of endometrial cancer. Cancer. 1993;71:3575–3581. doi: 10.1002/1097-0142(19930601)71:11<3575::aid-cncr2820711119>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Decimo D, LeMeur M, Dierich A, Gorry P, Chambon P. Developmental roles of the retinoic acid receptors. Journal of Steroid Biochemistry and Molecular Biology. 1995;53:475–486. doi: 10.1016/0960-0760(95)00094-g. [DOI] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Meunier B, Dumas E, Piec I, Bechet D, Hebraud M, Hocquette JF. Assessment of hierarchical clustering methodologies for proteomic data mining. Journal of Proteome Research. 2007;6:358–366. doi: 10.1021/pr060343h. [DOI] [PubMed] [Google Scholar]
- Meyer M, Sonntag-Buck V, Keaveney M, Stunnenberg HG. Retinoid-dependent transcription: the RAR/RXR-TBP-EIA/EIA-LA connection. Biochemical Society Symposium. 1996;62:97–109. [PubMed] [Google Scholar]
- Morriss-Kay GM, Ward SJ. Retinoids and mammalian development. International Review of Cytology. 1999;188:73–131. doi: 10.1016/s0074-7696(08)61566-1. [DOI] [PubMed] [Google Scholar]
- Negri E, La Vecchia C, Franceschi S, Levi F, Parazzini F. Intake of selected micronutrients and the risk of endometrial carcinoma. Cancer. 1996;77:917–923. doi: 10.1002/(sici)1097-0142(19960301)77:5<917::aid-cncr17>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Njar VC, Gediya L, Purushottamachar P, Chopra P, Vasaitis TS, Khandelwal A, Mehta J, Huynh C, Belosay A, Patel J. Retinoic acid metabolism blocking agents (RAMBAs) for treatment of cancer and dermatological diseases. Bioorganic & Medicinal Chemistry. 2006;14:4323–4340. doi: 10.1016/j.bmc.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Obel JC, Friberg G, Fleming GF. Chemotherapy in endometrial cancer. Clinical Advances in Hematology and Oncology. 2006;4:459–468. [PubMed] [Google Scholar]
- Patel JB, Huynh CK, Handratta VD, Gediya LK, Brodie AM, Goloubeva OG, Clement OO, Nanne IP, Soprano DR, Njar VC. Novel retinoic acid metabolism blocking agents endowed with multiple biological activities are efficient growth inhibitors of human breast and prostate cancer cells in vitro and a human breast tumor xenograft in nude mice. Journal of Medicinal Chemistry. 2004;47:6716–6729. doi: 10.1021/jm0401457. [DOI] [PubMed] [Google Scholar]
- Pavone ME, Reierstad S, Sun H, Milad M, Bulun SE, Cheng YH. Altered retinoid uptake and action contributes to cell survival in endometriosis. Journal of Clinical Endocrinology and Metabolism. 2010;95:E300–E309. doi: 10.1210/jc.2010-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierro E, Minici F, Alesiani O, Miceli F, Proto C, Screpanti I, Mancuso S, Lanzone A. Stromal-epithelial interactions modulate estrogen responsiveness in normal human endometrium. Biology of Reproduction. 2001;64:831–838. doi: 10.1095/biolreprod64.3.831. [DOI] [PubMed] [Google Scholar]
- Roberts LM, Hirokawa Y, Nachtigal MW, Ingraham HA. Paracrine-mediated apoptosis in reproductive tract development. Developmental Biology. 1999;208:110–122. doi: 10.1006/dbio.1998.9190. [DOI] [PubMed] [Google Scholar]
- Rorke EA, Jacobberger JW. Transforming growth factor-β1 (TGFβ1) enhances apoptosis in human papillomavirus type 16-immortalized human ectocervical epithelial cells. Experimental Cell Research. 1995;216:65–72. doi: 10.1006/excr.1995.1008. [DOI] [PubMed] [Google Scholar]
- Rorke EA, Zhang D, Choo CK, Eckert RL, Jacobberger JW. TGF-β-mediated cell cycle arrest of HPV16-immortalized human ectocervical cells correlates with decreased E6/E7 mRNA and increased p53 and p21 (WAF-1) expression. Experimental Cell Research. 2000;259:149–157. doi: 10.1006/excr.2000.4953. [DOI] [PubMed] [Google Scholar]
- Rotello RJ, Lieberman RC, Purchio AF, Gerschenson LE. Coordinated regulation of apoptosis and cell proliferation by transforming growth factor β1 in cultured uterine epithelial cells. PNAS. 1991;88:3412–3415. doi: 10.1073/pnas.88.8.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiniak ME, Yu M, Ross DT, Tolhurst EC, Slack JL. Identification of B94 (TNFAIP2) as a potential retinoic acid target gene in acute promyelocytic leukemia. Cancer Research. 2000;60:1824–1829. [PubMed] [Google Scholar]
- Saidi SA, Holland CM, Charnock-Jones DS, Smith SK. In vitro and in vivo effects of the PPAR-α agonists fenofibrate and retinoic acid in endometrial cancer. Molecular Cancer. 2006;513 doi: 10.1186/1476-4598-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG, Li C, Schwabe JW, Evans RM. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes and Development. 1997;11:299–308. doi: 10.1101/gad.11.3.299. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Tarrade A, Bastien J, Bruck N, Bauer A, Gianni M, Rochette-Egly C. Retinoic acid and arsenic trioxide cooperate for apoptosis through phosphorylated RXRα. Oncogene. 2005;24:2277–2288. doi: 10.1038/sj.onc.1208402. [DOI] [PubMed] [Google Scholar]
- Teyssier C, Belguise K, Galtier F, Cavailles V, Chalbos D. Receptor-interacting protein 140 binds c-Jun and inhibits estradiol-induced activator protein-1 activity by reversing glucocorticoid receptor-interacting protein 1 effect. Molecular Endocrinology. 2003;17:287–299. doi: 10.1210/me.2002-0324. [DOI] [PubMed] [Google Scholar]
- Wada K, Nomura S, Morii E, Kitamura Y, Nishizawa Y, Miyake A, Terada N. Changes in levels of mRNAs of transforming growth factor (TGF)-beta1, -beta2, -beta3, TGF-beta type II receptor and sulfated glycoprotein-2 during apoptosis of mouse uterine epithelium. Journal of Steroid Biochemistry and Molecular Biology. 1996;59:367–375. doi: 10.1016/s0960-0760(96)00139-2. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witcher M, Ross DT, Rousseau C, Deluca L, Miller WH., Jr Synergy between all-trans retinoic acid and tumor necrosis factor pathways in acute leukemia cells. Blood. 2003;102:237–245. doi: 10.1182/blood-2002-09-2725. [DOI] [PubMed] [Google Scholar]
- Woodworth CD, Chung J, McMullin E, Plowman GD, Simpson S, Iglesias M. Transforming growth factor β1 supports autonomous growth of human papillomavirus-immortalized cervical keratinocytes under conditions promoting squamous differentiation. Cell Growth and Differentiation. 1996;7:811–820. [PubMed] [Google Scholar]
- Yang S, Fang Z, Gurates B, Tamura M, Miller J, Ferrer K, Bulun SE. Stromal PRs mediate induction of l7β-hydroxysteroid dehydrogenase type 2 expression in human endometrial epithelium: a paracrine mechanism for inactivation of E2 . Molecular Endocrinology. 2001;15:2093–2105. doi: 10.1210/mend.15.12.0742. [DOI] [PubMed] [Google Scholar]
- Zheng WL, Sierra-Rivera E, Luan J, Osteen KG, Ong DE. Retinoic acid synthesis and expression of cellular retinol-binding protein and cellular retinoic acid-binding protein type II are concurrent with decidualization of rat uterine stromal cells. Endocrinology. 2000;141:802–808. doi: 10.1210/endo.141.2.7323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.