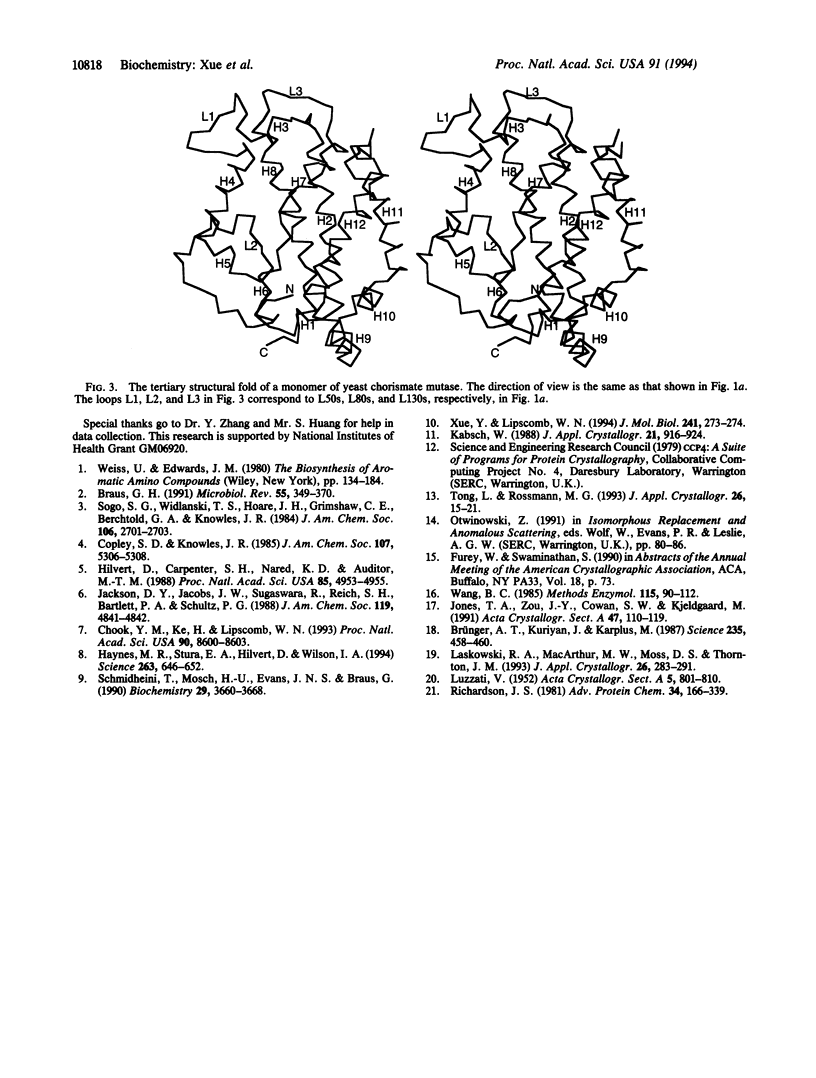
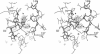Abstract
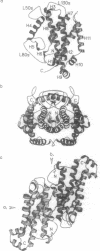
The crystal structure of an allosteric chorismate mutase, the Thr-226-->Ile mutant, from yeast Saccharomyces cerevisiae has been determined to 2.2-A resolution by using the multiple isomorphous replacement method. Solvent-flattening and electron-density modification were applied for phase improvement. The current crystallographic R factor is 0.196. The final model includes 504 of the 512 residues and 97 water molecules. In addition, two tryptophan molecules were identified in the interface between monomers. The overall structure is completely different from the reported structure of chorismate mutase from Bacillus subtilis. This structure showed 71% helices with essentially no beta-sheet structures.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braus G. H. Aromatic amino acid biosynthesis in the yeast Saccharomyces cerevisiae: a model system for the regulation of a eukaryotic biosynthetic pathway. Microbiol Rev. 1991 Sep;55(3):349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chook Y. M., Ke H., Lipscomb W. N. Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes M. R., Stura E. A., Hilvert D., Wilson I. A. Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart. Science. 1994 Feb 4;263(5147):646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- Hilvert D., Carpenter S. H., Nared K. D., Auditor M. T. Catalysis of concerted reactions by antibodies: the Claisen rearrangement. Proc Natl Acad Sci U S A. 1988 Jul;85(14):4953–4955. doi: 10.1073/pnas.85.14.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Schmidheini T., Mösch H. U., Evans J. N., Braus G. Yeast allosteric chorismate mutase is locked in the activated state by a single amino acid substitution. Biochemistry. 1990 Apr 17;29(15):3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- Wang B. C. Resolution of phase ambiguity in macromolecular crystallography. Methods Enzymol. 1985;115:90–112. doi: 10.1016/0076-6879(85)15009-3. [DOI] [PubMed] [Google Scholar]
- Xue Y., Lipscomb W. N. The crystallization and preliminary X-ray analysis of allosteric chorismate mutase. J Mol Biol. 1994 Aug 12;241(2):273–274. doi: 10.1006/jmbi.1994.1497. [DOI] [PubMed] [Google Scholar]