Abstract
Klebsiella variicola strain DX120E (=CGMCC 1.14935) is an endophytic nitrogen-fixing bacterium isolated from sugarcane crops grown in Guangxi, China and promotes sugarcane growth. Here we summarize the features of the strain DX120E and describe its complete genome sequence. The genome contains one circular chromosome and two plasmids, and contains 5,718,434 nucleotides with 57.1% GC content, 5,172 protein-coding genes, 25 rRNA genes, 87 tRNA genes, 7 ncRNA genes, 25 pseudo genes, and 2 CRISPR repeats.
Keywords: Endophyte, Klebsiella variicola , Klebsiella pneumoniae , Nitrogen fixation, Pathogenicity, Plant growth-promoting bacteria, Sugarcane
Introduction
The species Klebsiella variicola was classified in 2004 and consisted of clinical and plant-associated isolates [1].The species K. singaporensis was classified in 2004 based on a single soil isolate [2] and was recently identified as a later junior heterotypic synonym of K. variicola [3]. K. variicola is able to fix N2[1]. K. variicola strain At-22, one of the dominant bacteria in the fungus gardens of leaf-cutter ants, provides nitrogen source by N2 fixation [4] and carbon source by degrading leaf polymers to the ant-fungus symbiotic system [5]. Former K. pneumoniae strain 342 (Kp342), which is phylogenomically close to strain At-22 [6],[7] and has been identified as a strain of K. variicola [3], is able to colonize in plants and to provide small but critical amounts of fixed nitrogen to plant hosts [8].
K. variicola strain DX120E was isolated from roots of sugarcane grown in Guangxi, the major sugarcane production area in China [9]. It is able to colonize in sugarcane roots and shoots, to fix N2 in association with sugarcane plants, and to promote sugarcane growth [10], and thus shows a potential as a biofertilizer. Here we present a summary of the features of the K. variicola strain DX120E (=CGMCC 1.14935) and its complete genome sequence, and thus provide a genetic background to understand its endophytic lifestyle, plant growth-promoting potentials, and similarities and differences to other plant-associated and clinical K. variicola isolates.
Organism information
Classification and general features
K. variicola strain DX120E is a Gram-negative, non-spore-forming, non-motile rod (Figure 1). It grows aerobically but reduces N2 to NH3 at a low pO2. It is able to grow and fix N2 on media containing 10% (w/v) cane sugar or sucrose. It forms circular, convex, smooth colonies with entire margins on the solid high-sugar content media. It grows best around 30°C and pH 7 (Table 1).
Figure 1.
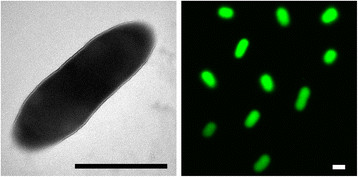
Morphology of Klebsiella variicola DX120E cells. Transmission electron micrograph (left) shows a DX120E cell stained by uranyl acetate; laser scanning confocal micrograph (right) shows DX120E cells tagged by green fluorescent protein. The scale bars represent 1 μm.
Table 1.
Classification and general features of Klebsiella variicola strain DX120E according to the MIGS recommendations[11]
| MIGS ID | Property | Term | Evidence code a |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [12] | |
| Phylum Proteobacteria | TAS [13] | ||
| Class Gammaproteobacteria | TAS [14],[15] | ||
| Order Enterobacteriales | TAS [16] | ||
| Family Enterobacteriaceae | TAS [17],[18] | ||
| Genus Klebsiella | TAS [18],[19] | ||
| Species Klebsiella variicola | TAS [1],[20] | ||
| Type strain:F2R9T (ATCC BAA-830 = DSM 15968) | TAS [1] | ||
| Gram stain | Negative | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Non-sporulating | IDA | |
| Temperature range | 4–40°C | IDA | |
| Optimum temperature | 28–32°C | IDA | |
| pH range; Optimum | 3.5–8.5; 7.0 | IDA | |
| Carbon source | Sucrose, citrate, fructose, galactose, glucose, lactose, malate, maltose, mannitol, mannose, rhamnose, & sorbitol | IDA | |
| MIGS-6 | Habitat | Soil, plants | IDA |
| MIGS-6.3 | Salinity | 0 – 5% NaCl (w/v) | IDA |
| MIGS-22 | Oxygen requirement | Aerobic | IDA |
| MIGS-15 | Biotic relationship | Free-living, endophytic | IDA |
| MIGS-14 | Pathogenicity | Not reported | |
| MIGS-4 | Geographic location | Daxin, Guangxi, China | TAS [9] |
| MIGS-5 | Sample collection | 2008 | TAS [9] |
| MIGS-4.1 MIGS-4.2 | Longitude | 107°20’E | NAS |
| Latitude | 22°80’N | NAS | |
| MIGS-4.3 | Depth | 0.1 – 0.2 m below the surface | IDA |
| MIGS-4.4 | Altitude | 320 m | NAS |
aEvidence codes – IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [21].
Phylogenetic analysis of the 16S rRNA gene sequences from strain DX120E and strain Kp342, the type strains of the species in the genera Klebsiella and Raoultella , and the type strain of the type species of the type genus of the family Enterobacteriaceae (Escherichia coli ATCC11775T) showed that K. variicola strains (type strain F2R9, Kp342, DX120E and LX3) were most closely related and formed a monophyletic group with K. pneumoniae and K. quasipneumoniae (Figure 2).
Figure 2.
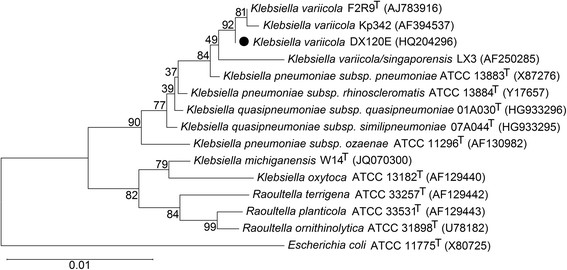
Phylogenetic tree of 16S rRNA gene sequences showing the position of Klebsiella variicola strain DX120E (●). Escherichia coli ATCC 11775T is used as an outgroup. The sequences were aligned with the CLUSTAL W program and were constructed with the neighbor-joining algorithm and the Kimura 2-parameter model integrated in the MEGA 5.2 program [22]. The phylogenetic tree was tested with 1,000 bootstrap replicates. Bootstrap values are shown at the nodes. The GenBank accession numbers of the sequences are indicated in parentheses. The scale bar represents a 1% nucleotide sequence divergence. Note that the genome of strain DX120E, F2R9T (DSM 15968T), or Kp342 contains eight copies of 16S rRNA genes; these gene sequences are generally not identical but phylogenetically grouped together (data not shown). The sequence of DX120E (HQ204296) used to construct the phylogenetic tree is identical to the sequence of locus tag KR75_09260 (CP009274:1935034–1936587).
Like typical members in the genera Klebsiella , K. variicola DX120E utilizes alanine, arabinose, D-arabitol, L-aspartate, D-cellobiose, citrate, D-fructose, L-fucose, D-galactose, gentiobiose, glucose, glycerol, myo-inositol, lactate, lactose, malate, maltose, D-mannitol, D-mannose, D-melibiose, L-proline, D-raffinose, L-rhamnose, L-serine, D-sorbitol, sucrose, and D-trehalose [23]. DX120E does not utilize adonitol (also known as ribitol), which is a distinctive characteristic from K. pneumoniae [1].
Genome sequencing information
Genome project history
K. variicola DX120E was selected for sequencing because it is a plant growth-promoting endophyte [10]. Its 16S rRNA gene sequence is deposited in GenBank under the accession number HQ204296. Its genome sequences are deposited in GenBank under the accession numbers CP009274, CP009275, and CP009276. A summary of the genome sequencing project information and its association with MIGS version 2.0 [11] is shown in Table 2.
Table 2.
Genome sequencing project information for Klebsiella variicola strain DX120E
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | PacBio 4 –10Kb library |
| Illumina 500 bp library | ||
| MIGS-29 | Sequencing platforms | PacBio RS II |
| Illumina HiSeq 2000 | ||
| MIGS-31.2 | Fold coverage | PacBio 96 × |
| Illumina 106 × | ||
| MIGS-30 | Assemblers | HGAP in smrtanalysis-2.1.1SOAPdenovo 2.04 |
| MIGS-32 | Gene calling method | GeneMarkS+ |
| Locus Tag | KR75 | |
| Genbank ID | CP009274 (Chromosome) | |
| CP009275 (plasmid pKV1) | ||
| CP009276 (plasmid pKV2) | ||
| Genbank Date of Release | January 1, 2015 | |
| GOLD ID | Gi0078577 | |
| BIOPROJECT | PRJNA259590 | |
| MIGS 13 | Source Material Identifier | CGMCC 1.14935 |
| Project relevance | Agriculture, plant-microbe interactions |
Growth conditions and DNA isolation
K. variicola DX120E was grown in liquid Luria-Bertani (LB) medium at 30°C to early stationary phase. The genome DNA was extracted from the cells by using a TIANamp bacterial DNA kit (Tiangen Biotech, Beijing, China). DNA quality and quantity were determined with a Nanodrop spectrometer (Thermo Scientific, Wilmington, USA).
Genome sequencing and assembly
The genome DNA of K. variicola DX120E was constructed into a 4 – 10 kb insert library and sequenced by the Pacific Biosciences’ (PacBio) Single Molecule, Real-Time (SMRT) sequencing technology [24] at the Duke University Genome Sequencing & Analysis Core Resource. Sequencing was run on single SMRT cell and resulted in 91,190 high-quality filtered reads with an average length of 6,196 bp. High-quality read bases were assembled by the Hierarchical Genome Assembly Process (HGAP) with smrtanalysis-2.1.1. The resulting draft genome consisted of 5,719,400 nucleotides and 5 contigs.
The genome DNA of K. variicola DX120E was also constructed into a 500-bp insert library and sequenced by an Illumina HiSeq 2000 sequencing system at BGI Tech, Shenzhen, China. The Illumina HiSeq 2000 sequencing resulted in 6,699,933 high-quality filtered reads with an average length of 90 bp. The sequencing data were assembled by the Short Oligonucleotide Analysis Package (SOAPdenovo 2.04) [25]. The resulting draft genome consisted of 5,695,362 nucleotides and 27 scaffolds.
The two draft genomes were aligned by Mauve [26]. The Illumina scaffold 1 bridged the PacBio contig 1 and contig 2; the Illumina scaffold 3 bridged the PacBio contig 1, contig 2, and contig 3; the Illumina scaffold 11 bridged the circular PacBio contig 4; the Illumina scaffold 16 bridged the circular PacBio contig 5. The genome sequencing was completed by PCR and Sanger sequencing to close the contig gaps of the PacBio-sequenced genome.
Genome annotation
Automated genome annotation was completed by the NCBI Prokaryotic Genome Annotation Pipeline. Product description annotations were obtained by searching against the KEGG, InterPro, and COG databases. Genes with signal peptides were predicted by SignalP [27]. Genes with transmembrane helices were predicted by TMHMM [28]. Genes for tRNA were found by tRNAScanSE [29]. Ribosomal RNAs were found by BLASTN vs. ribosomal RNA databases; 5S rRNA hits were further refined by Cmsearch [30]. Thirteen disrupted genes were replaced by the complete gene sequences obtained from the Illumina HiSeq 2000 sequencing.
Genome properties
The genome of K. variicola DX120E contains one circular chromosome and two plasmids (pKV1 and pKV2) (Table 3, Figure 3). The chromosome contains 5,501,013 nucleotides with 57.3% G + C content. The plasmid pKV1 contains 162,706 nucleotides with 50.7% G + C content. The plasmid pKV2 contains 54,715 nucleotides with 53.1% G + C content. The genome contains 5,316 predicted genes, 5,172 protein-coding genes, 119 RNA genes (25 rRNA genes, 87 tRNA genes, and 7 ncRNA genes), 25 pseudo genes, and 2 CRISPR repeats. The chromosome, pKV1, and pKV2 contain 4990, 131, and 51 protein-coding genes with coding density of 87.3%, 74.2%, and 83.9%, respectively. Among the 5,172 protein-coding genes, 4,511 genes (87.2%) have been assigned functions, while 661 genes (12.8%) have been annotated as hypothetical or unknown proteins (Table 4). The distribution of genes into COGs functional categories is presented in Table 5.
Table 3.
Summary of genome: one chromosome and two plasmids
| Label | Size (bp) | Topology | INSDC identifier | RefSeq ID |
|---|---|---|---|---|
| Chromosome | 5,501,013 | Circular | CP009274.1 | NZ_CP009274.1 |
| Plasmid pKV1 | 162,706 | Circular | CP009275.1 | NZ_CP009275.1 |
| Plasmid pKV2 | 54,715 | Circular | CP009276.1 | NZ_CP009276.1 |
Figure 3.
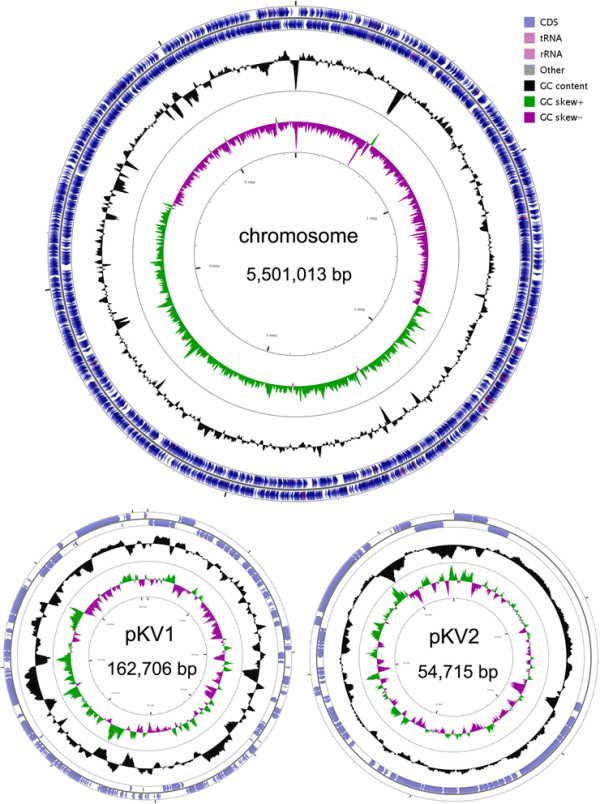
Circular map of the chromosome and plasmids of Klebsiella variicola strain DX120E. From outside to the center: genes on forward strand, genes on reverse strand, GC content, GC skew. Circular map was generated by CGView [31].
Table 4.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 5,718,434 | 100 |
| DNA coding (bp) | 4,930,539 | 86.22 |
| DNA G + C (bp) | 3,265,303 | 57.10 |
| DNA scaffolds | 3 | 100 |
| Total genes | 5,316 | 100 |
| Protein-coding genes | 5,172 | 97.29 |
| RNA genes | 112 | 2.12 |
| Pseudo genes | 25 | 0.47 |
| Genes with function prediction | 4,623 | 87.00 |
| Genes assigned to COGs | 4,398 | 82.73 |
| Genes with Pfam domains | 4,631 | 87.11 |
| Genes with signal peptides | 526 | 9.89 |
| Genes with transmembrane helices | 1,289 | 24.25 |
| CRISPR repeats | 2 | 0.04 |
Table 5.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 198 | 3.83 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.02 | RNA processing and modification |
| K | 489 | 9.45 | Transcription |
| L | 159 | 3.07 | Replication, recombination and repair |
| B | 1 | 0.02 | Chromatin structure and dynamics |
| D | 43 | 0.83 | Cell cycle control, cell division, chromosomepartitioning |
| V | 71 | 1.37 | Defense mechanisms |
| T | 235 | 4.54 | Signal transduction mechanisms |
| M | 260 | 5.03 | Cell wall/membrane biogenesis |
| N | 62 | 1.20 | Cell motility |
| U | 111 | 2.15 | Intracellular trafficking and secretion |
| O | 158 | 3.05 | Posttranslational modification, protein turnover, chaperones |
| C | 342 | 6.61 | Energy production and conversion |
| G | 583 | 11.27 | Carbohydrate transport and metabolism |
| E | 538 | 10.40 | Amino acid transport and metabolism |
| F | 102 | 1.97 | Nucleotide transport and metabolism |
| H | 215 | 4.16 | Coenzyme transport and metabolism |
| I | 130 | 2.51 | Lipid transport and metabolism |
| P | 344 | 6.65 | Inorganic ion transport and metabolism |
| Q | 112 | 2.17 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 541 | 10.46 | General function prediction only |
| S | 414 | 8.00 | Function unknown |
| - | 774 | 14.97 | Not in COGs |
The total is based on the total number of protein coding genes in the genome.
Insights from the genome sequence
The genome of K. variicola DX120E contains genes contributing to multiple plant-beneficial functions. In accordance with previously detected N2 fixation, indole-3-acetic acid production, siderophore production, and phosphate solubilization [9], the genome of K. variicola DX120E contains nif cluster, indole-3-pyruvate decarboxylase, siderophore enterobactin synthesis genes (entABCDEF) and enterobactin exporter gene (entS), and pyrroloquinoline quinone synthesis genes (pqqBCDEF) contributing to these functions. Moreover, the genome of K. variicola DX120E contains the budABC operon for the synthesis of acetoin and 2,3-butanediol [32], and thus may induce plant systemic resistance to pathogens [33].
DX120E contains plasmids similar to those in Klebsiella relatives. The plasmid pKV1 is most similar to the plasmid pKp5-1 of the K. pneumoniae strain 5–1 (Kp5-1) [34] with a 97% identity of 56% coverage (Additional file 1: Figure S1); the similar regions mainly encode transposase/recombinases and proteins functioning in plasmid replication, partitioning, and conjugal transfer. The plasmid pKV2 is most similar to the plasmid pKOXM1C of the K. oxytoca strain M1 with a 96% identity of 89% coverage (Additional file 2: Figure S2); the similar regions mainly encode proteins for plasmid partitioning and phage functions.
The genome of K. variicola DX120E has high average nucleotide identities (ANI) [35] about 99% to the available genomes of K. variicola strains DSM 15968T, At-22, Bz19, and Kp342. Bz19 was isolated from faeces of a hospitalized patient [6]. The plant-beneficial strain Kp342 is able to infect mouse organs, although it is less virulent than typical clinical K. pneumoniae isolates [36]. Kp5-1, which has the plasmid pKp5-1 close to pKV1, is a cotton pathogen causing boll-rot disease [34]. The genome of strain Kp5-1 has ANI values about 99% to the genomes of the known K. variicola strains and thus belongs to K. variicola . These drive concerns about potential pathogenicity of DX120E to animals and plants. Therefore, DX120E’s pathogenic potentials to animals and plants should be determined before using DX120E as a biofertilizer in the field.
Conclusions
The complete genome sequence of K. variicola DX120E provides the genetic background for understanding the bacterial mechanisms to adapt endophytic life and to promote plant growth. The high degree of whole-genome and plasmid similarities between DX120E and phytopathogenic and clinical Klebsiella isolates suggests the risk of using DX120E as a biofertilizer. The available genome sequences of the K. variicola strains allow an in-depth comparative analysis to understand the subtle pathogenicity mechanisms of the pathogens and to predict pathogenic risks for the plant-beneficial strain DX120E.
Competing interests
The authors declare that they have no competing interests.
Authors' contribution
LL did the microbiological studies and obtained the organism information; CW assembled the Illumina sequencing data; MC assembled the PacBio sequencing data; HW and YYL completed the genome analysis; YRL, LY, and QA designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Additional files
Supplementary Material
Comparison of plasmid pKV1 of Klebsiella variicola strain DX120E with plasmid pKp5-1 of K. pneumoniae strain 5–1.
Comparison of plasmid pKV2 of Klebsiella variicola strain DX120E with plasmid pKOXM1C of K. oxytoca strain M1.
Li Lin, Chunyan Wei and Mingyue Chen contributed equally to this work.
Contributor Information
Li Lin, Email: linligx@163.com.
Chunyan Wei, Email: weichunyan025@126.com.
Mingyue Chen, Email: chenmy2007525@163.com.
Hongcheng Wang, Email: whc512@foxmail.com.
Yuanyuan Li, Email: liyuanyuan2283@163.com.
Yangrui Li, Email: liyr@gxaas.net.
Litao Yang, Email: liyr@gxu.edu.cn.
Qianli An, Email: an@zju.edu.cn.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31171504 and 31240056), Guangxi Provincial Natural Science Foundation of China (2011GXNSFF018002 and 2013GXNSFBA019061), Science and Technology Development Foundation of Guangxi Academy of Agricultural Sciences (2013JZ11), Guangxi Special Funds for Bagui Scholars and Distinguished Experts (2013).
References
- Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. System Appl Microbiol. 2004;27:27–35. doi: 10.1078/0723-2020-00261. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang D, Chen F, Ma J, Dong Y, Zhang L. Klebsiella singaporensis sp. nov., a novel isomaltulose-producing bacterium. Int J Syst Evol Microbiol. 2004;54:2131–6. doi: 10.1099/ijs.0.02690-0. [DOI] [PubMed] [Google Scholar]
- Brisse S, Passet V, Grimont PA. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol. 2014;64:3146–52. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- Pinto-Tomás AA, Anderson MA, Suen G, Stevenson DM, Chu FS, Cleland WW. et al. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutter ants. Science. 2009;326:1120–3. doi: 10.1126/science.1173036. [DOI] [PubMed] [Google Scholar]
- Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomás AA. et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010;6:e1001129. doi: 10.1371/journal.pgen.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade BG, de Veiga RN, Abanto Marin MF, Fonseca EL, Vicente AC. The genome of a clinical Klebsiella variicola strain reveals virulence associated traits and a pl9-like plasmid. FEMS Microbiol Lett. 2014;360:13–6. doi: 10.1111/1574-6968.12583. [DOI] [PubMed] [Google Scholar]
- Hazen TH, Zhao L, Sahl JW, Robinson G, Harris AD, Rasko DA. et al. Characterization of Klebsiella sp. strain 10982, a colonizer of humans that contains novel antibiotic resistance alleles and exhibits genetic similarities to plant and clinical Klebsiella isolates. Antimicrob Agents Chemother. 2014;58:1879–88. doi: 10.1128/AAC.01605-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez AL, Dong Y, Triplett EW. Nitrogen fixation in wheat provided by Klebsiella pneumoniae 342. Mol Plant Microbe Interact. 2004;17:1078–85. doi: 10.1094/MPMI.2004.17.10.1078. [DOI] [PubMed] [Google Scholar]
- Lin L, Li Z, Hu C, Zhang X, Chang S, Yang L. et al. Plant growth-promoting nitrogen-fixing enterobacteria are in association with sugarcane plants growing in Guangxi, China. Microbes Environ. 2012;27:391–8. doi: 10.1264/jsme2.ME11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Lin L, Luo L, Xing Y, Hu C, Yang L. et al. Endophytic nitrogen-fixing Klebsiella variicola strain DX120E promotes sugarcane growth. Biol Fertil Soils. 2014;50:657–66. [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P. et al. Minimum Information about a Genome Sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–9. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn T. In: Bergey’s Manual of Systematic Bacteriology. 2. Garrity GM, Brenner DJ, Krieg NR, Staley JT, editor. Springer, New York; 2005. Phylum XIV. Proteobacteria phyl. nov; p. 1. [DOI] [Google Scholar]
- Garrity GM, Bell JA, Lilburn T. In: Bergey’s Manual of Systematic Bacteriology. 2. Garrity GM, Brenner DJ, Krieg NR, Staley JT, editor. Springer, New York; 2005. Class III. Gammaproteobacteria class. nov; p. 1. [DOI] [Google Scholar]
- Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol. 2005;55:2235–2238. PubMed 10.1099/ijs.0.64108-0. [DOI] [PubMed]
- Garrity GM, Holt JG. In: Bergey’s Manual of Systematic Bacteriology. 2. Garrity GM, Boone DR, Castenholz RW, editor. Springer, New York; 2001. Taxonomic Outline of the Archaea and Bacteria; pp. 155–66. [Google Scholar]
- Rahn O. New principles for the classification of bacteria. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene Abteilung II. 1937;96:273–86. [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol. 1980;30:225–420. [PubMed] [Google Scholar]
- Trevisan V. Carratteri di alcuni nuovi generi di Batteriacee. Atti della Accademia Fisica-Medica-Statistica in Milano, Series 4. 1885;3:92–107. [Google Scholar]
- Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 97. Int J Syst Evol Microbiol. 2004;54:631–632. PubMed 10.1099/ijs.0.63263-0. [DOI] [PubMed]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisse S, Grimont F, Grimont PAD. In: The Prokaryotes Volume 6: Proteobacteria: Gamma Subclass. Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editor. Springer, New York; 2006. The genus Klebsiella; pp. 159–96. [Google Scholar]
- Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G. et al. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323:133–8. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J. et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. t-RNAscan-SE: a program for improved detection of transfer RNA gene in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmsearch http://manpages.ubuntu.com/manpages/raring/man1/cmsearch.1.html.
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–9. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- Taghavi S, van der Lelie D, Hoffman A, Zhang Y-B, Walla MD, Vangronsveld J. et al. Genome sequence of the plant growth promoting endophytic bacterium Enterobacter sp. 638. PLoS Genet. 2010;6:e1000943. doi: 10.1371/journal.pgen.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Paré PW. et al. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:4927–32. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano EG, Forray MM, Bell AA. Complete genome sequence of a Klebsiella pneumoniae strain isolated from a known cotton insect boll vector. Genome Announc. 2014;2:e00850–914. doi: 10.1128/genomeA.00850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- Fouts DE, Tyler HL, DeBoy RT, Daugherty S, Ren Q, Badger JH. et al. Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 2008;4:e1000141. doi: 10.1371/journal.pgen.1000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of plasmid pKV1 of Klebsiella variicola strain DX120E with plasmid pKp5-1 of K. pneumoniae strain 5–1.
Comparison of plasmid pKV2 of Klebsiella variicola strain DX120E with plasmid pKOXM1C of K. oxytoca strain M1.


