Abstract
Leucobacter chironomi strain MM2LBT (Halpern et al., Int J Syst Evol Microbiol 59:665-70 2009) is a Gram-positive, rod shaped, non-motile, aerobic, chemoorganotroph bacterium. L. chironomi belongs to the family Microbacteriaceae, a family within the class Actinobacteria. Strain MM2LBT was isolated from a chironomid (Diptera; Chironomidae) egg mass that was sampled from a waste stabilization pond in northern Israel. In a phylogenetic tree based on 16S rRNA gene sequences, strain MM2LBT formed a distinct branch within the radiation encompassing the genus Leucobacter. Here we describe the features of this organism, together with the complete genome sequence and annotation. The DNA GC content is 69.90%. The chromosome length is 2,964,712 bp. It encodes 2,690 proteins and 61 RNA genes. L. chironomi genome is part of the Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes (KMG) project.
Keywords: Leucobacter chironomi , Microbacteriaceae , Chironomid, Chironomus , Egg mass, Hexavalent chromium
Introduction
Strain MM2LBT (=DSM 19883T = JCM 17022T = LMG 24399T), is the type strain of the species Leucobacter chironomi [1]. The genus Leucobacter was formed by Takeuchi et al. [2] and currently includes 15 species. Members of this genus were found in a verity of environments including air [2],[3], nematodes [4],[5], sediments with chromium contamination [6], cow dung [7], compost [8], fermented seafood [9], phyllosphere [10] and chironomids (Diptera) [1].
L. chironomi MM2LBT, was isolated from an insect egg mass (Chironomus sp.) that was sampled from a waste stabilization pond in northern Israel [1]. Chironomids (Insecta; Diptera; Chironomidae; Chironomus sp.), also known as the non biting midges, are aquatic insects. They undergo a complete metamorphosis of four life stages; egg, larva, pupa and adult that emerges into the air. The eggs are deposited by the adult female at the water’s edge in egg masses which contain hundreds of eggs [11]. Chironomid egg masses were found as natural reservoirs of Vibrio cholerae and Aeromonas species [11]-[17]. Strain MM2LBT was isolated in the course of a study that explored the endogenous bacterial communities in chironomid egg masses [1]. Using 454-pyrosequencing technique, Senderovich & Halpern [18], showed that the prevalence of Leucobacter in chironomid egg masses and larval endogenous bacterial communities is 0.1% and 0.2%, respectively.
Here we describe a summary classification and a set of the features of L. chironomi , together with the genome sequence description and annotation.
Organism Information
Classification and features
A taxonomic study using a polyphasic approach placed L. chironomi strain MM2LBT in the genus Leucobacter within the family Microbacteriaceae (order; Actinomyecetales, class; Actinobacteria , phylum; Actinobacteria ) (Figure 1). The family Microbacteriaceae comprises more than 40 genera and a large variety of species and phenotypes.
Figure 1.
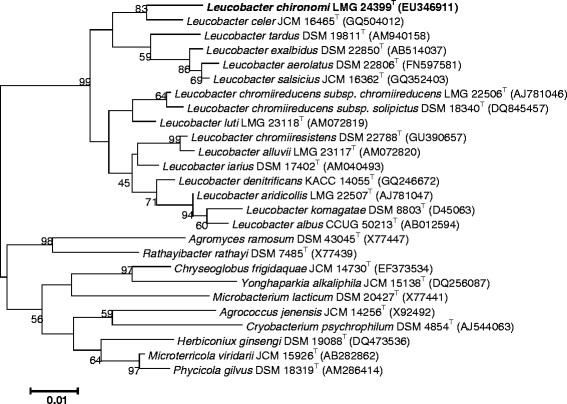
Phylogenetic tree highlighting the position of Leucobacter chironomi relative to the type strains of the other species within the genus Leucobacter. The sequence alignments were performed by using the CLUSTAL W program and the tree was generated using the maximum likelihood method in MEGA 5 software [41]. Bootstrap values (from 1,000 replicates) greater than 50% are shown at the branch points. The bar indicates a 1% sequence divergence.
L. chironomi strain MM2LBT is a Gram-positive, aerobic, chemo-organotrophic, non-motile single cell rod (Figure 2). After 48 h incubation on LB agar at 30°C, colonies are opaque, circular, with entire margins and yellow-coloured [1]. Growth is observed at 17–37°C (optimum 30°C), with 0–7% (w/v) NaCl (optimum 0–1.0% NaCl) and at pH 4.0–9.5 (optimum pH 6.0–8.0) (Table 1). Oxidase reaction is negative; catalase reaction is weakly positive. Strain MM2LBT produces acetoin and reduces nitrate to nitrogen; H2S and indole are not produced; urea and gelatin are not hydrolyzed; citrate is not utilized; β-galactosidase, arginine dihydrolase, lysine decarboxylase, ornithine decarboxylase and tryptophan deaminase activities are absent; Putrescine and glycerol are utilized [1].
Figure 2.
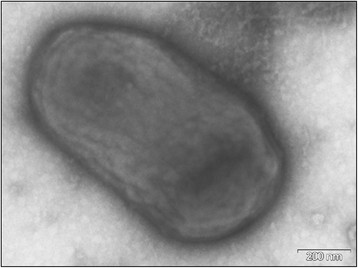
Electron micrograph of negatively stained cells of strain MM2LBT. Cells are nonflagellated rods. Bar, 200 nm.
Table 1.
Classification and general features ofLeucobacter chironomi strain MM2LBTaccording to the MIGS recommendations[42]], published by the Genome Standards Consortium[43]and the Names for Life database[44]
| MIGS ID | Property | Term | Evidence code a |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [44],[45] | |
| Phylum Actinobacteria | TAS [46] | ||
| Class Actinobacteria | TAS [47] | ||
| Order Actinomycetales | TAS [47]-[50] | ||
| Family Microbacteriaceae | TAS [47],[48],[51],[52] | ||
| Genus Leucobacter | TAS [2] | ||
| Species Leucobacter chironomi | TAS [1] | ||
| Type strain MM2LBT | TAS [1] | ||
| Gram stain | positive | TAS [1] | |
| Cell shape | rod | TAS [1] | |
| Motility | Non-motile | TAS [1] | |
| Sporulation | Non-sporulating | IDS | |
| Temperature range | 17-37°C | TAS [1] | |
| Optimum Temperature | 30°C | TAS [1] | |
| pH range | 4.0-9.5 | TAS [1] | |
| Optimum pH | 6.0-8.0 | TAS [1] | |
| Carbon source | Putrescine and Glycerolb | TAS [1] | |
| MIGS-6 | Habitat | Aquatic/Insect host | TAS [1] |
| MIGS-6.3 | Salinity | 0-7.0% NaCl (w/v) | TAS [1] |
| MIGS-22 | Oxygen requirement | Aerobic | TAS [1] |
| MIGS-15 | Biotic relationship | Commensal (Insect, chironomid) | TAS [1] |
| MIGS-14 | Pathogenicity | None | NAS |
| MIGS-4 | Geographic location | Northern Israel | TAS [1] |
| MIGS-5 | Sample collection | July 2006 | TAS [1] |
| MIGS-4.1 | Latitude | 32.669167 | IDS |
| MIGS-4.2 | Longitude | 35.128639 | IDS |
| MIGS-4.4 | Altitude | 40 m | TAS [1] |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). Evidence codes are from the Gene Ontology project [53].
bThe only carbon source that was positive for this strain, out of all carbon sources that were tested (strain MM2LBT does not use carbohydrates, not even glucose) [1].
Chemotaxonomic data
The dominant cellular fatty acids are anteiso-C15:0, anteiso-C17:0 and iso-C16:0. Cell-wall amino acids are alanine, glycine, threonine, DAB, γ-aminobutyric acid and glutamic acid. Strain MM2LBT has a B-type crosslinked peptidoglycan. The major menaquinone is MK-11; MK-10 and MK-12 occur in minor amounts [1]. Strain MM2LBT is able to grow in the presence of up to 18.0 mM Cr(VI) [1].
Genome sequencing information
Genome project history
L. chironomi MM2LBT, was selected for sequencing due to its phylogenetic position [19]-[21], and is part of Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes (KMG) study [22] which aims not only to increase the sequencing coverage of key reference microbial genomes [23] but also to generate a large genomic basis for the discovery of genes encoding novel enzymes [24]. The sequencing project is accessible in the Genomes OnLine Database [25] and the genome sequence is deposited in GenBank. Sequencing, finishing and annotation were accomplished by the DOE Joint Genome Institute (JGI) [26] using state of the art genome sequencing technology [27]. The project information is summarized in Table 2.
Table 2.
Genome sequencing project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31.1 | Sequencing quality | Level 2: High-Quality Draft |
| MIGS-28 | Libraries used | Illumina Std. shotgun library |
| MIGS 29 | Sequencing method | Illumina HiSeq 2000 |
| MIGS 31.2 | Fold coverage | 122.1X |
| MIGS 30 | Assemblers | Velvet (v. 1.1.04), ALLPATHS –LG (v. r42328) |
| MIGS 32 | Gene calling method | Prodigal 2.5 |
| Locus Tag | H629 | |
| Genbank ID | ATXU00000000 | |
| Genbank Date of Release | 12-DEC-2013 | |
| GOLD ID | Gp0013907 | |
| BIOPROJECT | PRJNA188922 | |
| MIGS-13 | Source Material Identifier | DSM 19883T |
| Project relevance | GEBA-KMG, Tree of: Life |
Growth conditions and genomic DNA preparation
L. chironomi MM2LBT, DSM 19883, was grown in Trypticase Soy Yeast Extract medium (DSMZ medium 92) at 28°C [28]. DNA was isolated from 0.5-1.0 g of cell paste using Masterpure DNA purification kit (Epicentre MGP04100) following the standard protocol as recommended by the manufacturer with additional 7.5 units of each of the following enzymes achromopeptidase, lysostaphin, mutanolysin and 2100 units of lysozyme, incubated for one hour at 37°C, followed by addition of 1 μl proteinase K and incubation for 20 min at 70°C for cell lysis. DNA is available through the DNA Bank Network [29].
Genome sequencing and assembly
The draft genome of L. chironomi DSM 19883T was generated at the DOE Joint genome Institute (JGI) using the Illumina technology [30]. An Illumina standard shotgun library was constructed and sequenced using the Illumina HiSeq 2000 platform which generated 13,901,154 reads totaling 2,085.2 Mb. All general aspects of library construction and sequencing performed at the JGI can be found at the Institute web site [25]. All raw Illumina sequence data was passed through DUK, a filtering program developed at JGI, which removes known Illumina sequencing and library preparation artifacts (Mingkun L, et al., unpublished, 2011). Following steps were then performed for assembly: (1) filtered Illumina reads were assembled using Velvet [31], (2) 1–3 kb simulated paired end reads were created from Velvet contigs using wgsim [32], (3) Illumina reads were assembled with simulated read pairs using Allpaths–LG [33]. Parameters for assembly steps were: (1) Velvet (velveth: 63 –shortPaired and velvetg: −very clean yes –exportFiltered yes –min contig lgth 500 –scaffolding no –cov cutoff 10) (2) wgsim (−e 0 –1 100 –2 100 –r 0 –R 0 –X 0) (3) Allpaths–LG (PrepareAllpathsInputs: PHRED 64 = 1 PLOIDY = 1 FRAG COVERAGE = 125 JUMP COVERAGE = 25 LONG JUMP COV = 50, RunAllpathsLG: THREADS = 8 RUN = std shredpairs TARGETS = standard VAPI WARN ONLY = True OVERWRITE = True). The final draft assembly contained 27 contigs in 27 scaffolds and is based on 361.8 Mb of Illumina data, which provides an average 122.1X coverage of the genome.
Genome annotation
Genes were detected using the Prodigal software [34] at the DOE-JGI Genome Annotation pipeline [35],[36]. The CDSs predicted were translated and searched against the Integrated Microbial Genomes (IMG) non-redundant database, UniProt, TIGRFam, Pfam, PRIAM, KEGG, COG, and InterPro databases. Additional gene prediction and functional annotation analysis was carried out in the Integrated Microbial Genomes (IMG-ER) platform [37].
Genome properties
The assembly of the draft genome sequence consists of 27 scaffolds amounting to 2,964,712 bp, and the G + C content is 69.9% (Table 3). Of the 2,751 genes predicted, 2,690 were protein-coding genes, and 61 RNAs. The majority of the protein-coding genes (79.5%) were assigned a putative function while the remaining ones were annotated as hypothetical proteins. The distribution of genes into COGs functional categories is presented in Table 4.
Table 3.
Genome statistics
| Attribute | Value | % of total |
|---|---|---|
| Genome size (bp) | 2,964,712 | 100.00 |
| DNA coding (bp) | 2,686,984 | 90.60 |
| DNA G + C (bp) | 2,072,411 | 69.90 |
| DNA scaffolds | 27 | 100.00 |
| Total genes | 2,751 | 100.00 |
| Protein coding genes | 2,690 | 97.78 |
| RNA genes | 61 | 2.22 |
| Pseudo genes | 0 | 0 |
| Genes in internal clusters | 2,248 | 81.7 |
| Genes with function prediction | 2,188 | 79.53 |
| Genes assigned to COGs | 1,842 | 66.96 |
| Genes with Pfam domains | 2,249 | 81.75 |
| Genes with signal peptides | 158 | 5.74 |
| Genes with transmembrane helices | 755 | 27.44 |
| CRISPR repeats | 0 | 0 |
Table 4.
Number of genes associated with the general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 161 | 8.39 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.05 | RNA processing and modification |
| K | 173 | 8.39 | Transcription |
| L | 108 | 5.24 | Replication, recombination and repair |
| B | 1 | 0.05 | Chromatin structure and dynamics |
| D | 19 | 0.92 | Cell cycle control, cell division, chromosome partitioning |
| V | 34 | 1.65 | Defense mechanisms |
| T | 77 | 3.74 | Signal transduction mechanisms |
| M | 90 | 4.37 | Cell wall/membrane biogenesis |
| N | 0 | 0 | Cell motility |
| U | 23 | 1.12 | Intracellular trafficking, secretion and vesicular transport |
| O | 60 | 2.91 | Posttranslational modification, protein turnover, chaperones |
| C | 109 | 5.29 | Energy production and conversion |
| G | 111 | 5.39 | Carbohydrate transport and metabolism |
| E | 285 | 13.83 | Amino acid transport and metabolism |
| F | 70 | 3.40 | Nucleotide transport and metabolism |
| H | 88 | 4.27 | Coenzyme transport and metabolism |
| I | 83 | 4.03 | Lipid transport and metabolism |
| P | 142 | 6.89 | Inorganic ion transport and metabolism |
| Q | 52 | 2.52 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 240 | 11.64 | General function prediction only |
| S | 144 | 6.99 | Function unknown |
| - | 909 | 33.04 | Not in COGs |
Insights from the genome sequence
Senderovich and Halpern [18],[38], demonstrated that endogenous bacteria in chironomids have a role in protecting their insect host from toxic metals. L. chironomi strain MM2LBT, which was isolated from a chironomid egg mass was found to tolerate up to 18 mM Cr(VI) [1]. Other Leucobacter species like L. alluvii ,L. aridicollis ,L. chromiireducens ,L. chromiiresistens ,L. komagatae ,L. luti and L. salisicius, have also been found to be resistant to hexavalent chromium [1],[2],[5],[39],[40]. A chromate membrane transport protein A (ChrA) was detected in the genome of the chromate-resistant bacterium, L. salsicius M1-8T[40]. However, this gene or other genes with chromium reduction predicted functions were not identified in L. chironomi MM2LBT genome. Nevertheless, three genes for ABC-type metal ion transport system (permease, ATPase and periplasmic components), were detected in the genome of strain MM2LBT. These genes may have a role in L. chironomi chromium tolerance.
More genes that may indicate the potential of strain MM2LBT to tolerate or detoxify metals, were also detected. Among them are genes for arsenical resistance: arsenical-resistance protein (arsB); arsenite efflux pump ACR3 and related permeases. Other genes suggest the potential of L. chironomi to survive in the presence of other toxic metals: copper chaperone; copper-(or silver)-translocating P-type ATPase; heavy metal-(Cd/Co/Hg/Pb/Zn)-translocating P-type ATPase and transcriptional regulator (ArsR family) which is involved in stress-response to heavy metal ions.
Three genes encoding drug resistance transporters are found in strain MM2LBT genome: drug resistance transporter Bcr/CflA subfamily; multidrug resistance efflux transporter and drug resistance transporter EmrB/QacA subfamily. Four copies of Beta-lactamase class C and other penicillin binding proteins were also found in three different domains of strain’s MM2LBT genome.
One gene encoding the two component transcriptional regulator LuxR family is present in the genome of strain MM2LBT and demonstrates quorum sensing skills.
Tolerance of up to 7.0% NaCl was described for strain MM2LBT[1]. Three genes for ABC-type proline/glycine betaine transport system (ATP binding subunit, permease and periplasmatic components), that seem to be located in the same operon, are present in strain MM2LBT genome. The accumulation of glycine betaine and other solutes offer osmoprotection, thus, this transport system is probably involved in osmoregulation.
Three genes in L. chironomi had best hits with genes from Eukaryotes, indicating a possible horizontal transfer of genes from Eukaryotes to L. chironomi . These genes were: Exodeoxyribonuclease VII small subunit and a protein from PAC2 family, both form Anopheles gambiae origin and a hypothetical protein from Drosophila williston origin. Anopheles and Drosophila as well as Chironomids belong to the Diptera order. L. chironomi was isolated from chironomids. Since chironomid species have not yet been sequenced, the horizontal gene transfer from the Diperan origin to L. chironomi may point toward the ancient relationships between this bacterium and its chironomid host.
The genome sequences of three more Leucobacter isolates have recently been published; L. chromiiresistens, isolated from a soil sample [40]; Leucobacter sp. UCD-THU isolated from a residential toilet [54]; and Leucobacter salsicius isolated from Korean salt-fermented seafood [39]. Chromate resistance was reported for some of these species (L. chironomi , L. chromiiresistens and L. salsicius ) [1],[39],[40]. The genome analysis of L. salsicius detected chromate transport protein A (ChrA) that confers heavy metal tolerance via chromate ion efflux from the cytoplasm [39]. In contrast, this gene is not present in the genome of L. chironomi and L. chromiiresistens . However, in both strains, other genes for metals tolerance or ion efflux, are present. Interestingly, we have detected a chromate transporter (Chr) gene in the genome of Leucobacter sp. UCD-THU, although no evidence for chromate resistance was reported in vivo for this strain [54]. Another interesting feature is the differences in the horizontal gene transfer found in all four Leucobacter species genomes. While no horizontal gene transfer from Eukaryotes was detected for Leucobacter sp. UCD-THU, we detected horizontal gene transfer from fungi belonging to the phyla Basidiomycota and Ascomycota in L. salsicius and L. chromiiresistens genomes, respectively. For L. chromiiresistens , which was isolated from seafood, genes transfer from the phylum Chordata was also found. Horizontal gene transfer from insects was detected for L. chironomi in the current study, confirming the fact that chironomid insects are L. chironomi hosts.
Conclusions
In the current study, we characterized the genome of L. chironomi strain MM2LBT that was isolated from a chironomid egg mass [1]. Recently, we have demonstrated that endogenous bacteria in chironomids have a role in protecting their insect host from toxic metals [18],[38]. Genes indicating the potential role of strain L. chironomi to tolerate or detoxify metals, where detected in its genome, demonstrating that indeed, L. chironomi which inhabits chironomids has a part in protecting its host from toxicants. Genes for ABC-type proline/glycine betaine transport system that were found in the genome may explain the salt tolerance properties of L. chironomi . Evidence of horizontal transfer of genes from Diperan origin to L. chironomi , implies toward an ancient relationships between L. chironomi and its chironomid host.
Abbreviations
KMG: One thousand microbial genomes
GEBA: Genomic encyclopedia of Bacteria and Archaea
MIGS: Minimum information about a genome sequence
DOE JGI: Department of Energy, Joint Genome Institute
TAS: Traceable
NAS: Non-traceable
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MH and RP isolated and characterized strain MM2LBT; SL, MH, HPK and NCK drafted the manuscript. AL, AC, TBKR, MH, AP, NNI, VMM and TW sequenced, assembled and annotated the genome. All authors read and approved the final manuscript.
Contributor Information
Sivan Laviad, Email: sivan_laviad@hotmail.com.
Alla Lapidus, Email: ALapidus@lbl.gov.
Alex Copeland, Email: ACCopeland@lbl.gov.
TBK Reddy, Email: tbreddy@lbl.gov.
Marcel Huntemann, Email: MHuntemann@lbl.gov.
Amrita Pati, Email: APati@lbl.gov.
Natalia N Ivanova, Email: NNIvanova@lbl.gov.
Victor M Markowitz, Email: VMMarkowitz@lbl.gov.
Rüdiger Pukall, Email: rpu@dsmz.de.
Hans-Peter Klenk, Email: hpk@dsmz.de.
Tanja Woyke, Email: twoyke@lbl.gov.
Nikos C Kyrpides, Email: nckyrpides@lbl.gov.
Malka Halpern, Email: mhalpern@research.haifa.ac.il.
Acknowledgements
The authors gratefully acknowledge the assistance of Iris Brandes for growing L. chironomi cultures and Evelyne-Marie Brambilla for DNA extraction and quality control (both at the DSMZ). This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory under contract No. DE-AC02-05CH11231, A.L. The work was supported in part by Russian Ministry of Science Mega-grant no.11.G34.31.0068 (PI. Dr Stephen J O'Brien) and by a grant from the US Civilian Research and Development Foundation (CRDF grant no. ILB1-7045-HA) (PI. Prof. Malka Halpern).
References
- Halpern M, Shaked T, Pukall R, Schumann P. Leucobacter chironomi sp. nov., a chromate resistant bacterium isolated from a chironomid egg mass. Int J Syst Evol Microbiol. 2009;59:665–670. doi: 10.1099/ijs.0.004663-0. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Weiss N, Schuman P, Yokota A. Leucobacter komagatae gen. nov., sp. nov., a new aerobic gram-positive, nonsporulating rod with 2,4-diaminobutyric acid in the cell wall. Int J Syst Bacteriol. 1996;46:967–971. doi: 10.1099/00207713-46-4-967. [DOI] [PubMed] [Google Scholar]
- Martin E, Lodders N, Jackel U, Schuman P, Kampfer P. Leucobacter aerolatus sp. nov., from the air of a duck barn. Int J Syst Evol Microbiol. 2010;60:2838–2842. doi: 10.1099/ijs.0.021303-0. [DOI] [PubMed] [Google Scholar]
- Somvanshi VS, Lang E, Schumann P, Pukall R, Kroppenstedt RM, Ganguly S. et al. Leucobacter iariussp. nov., in the family Microbacteriaceae. Int J Syst Evol Microbiol. 2007;57:682–686. doi: 10.1099/ijs.0.64683-0. [DOI] [PubMed] [Google Scholar]
- Muir RE, Tan MW. Leucobacter chromiireducens subsp. solipictus subsp. nov., a pigmented bacterium isolated from the nematode Caenorhabditis elegans, and emended description of L. chromiireducens. Int J Syst Evol Microbiol. 2007;57:2770–2776. doi: 10.1099/ijs.0.64822-0. [DOI] [PubMed] [Google Scholar]
- Morais PV, Paulo C, Francisco R, Branco R, Paula Chung A, Da Costa MS. Leucobacter luti sp. nov., and Leucobacter alluvii sp. nov., two new species of the genus Leucobacter isolated under chromium stress. Syst Appl Microbiol. 2006;29:414–421. doi: 10.1016/j.syapm.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Weon HY, Anandham R, Tamura T, Hamada M, Kim SJ, Kim YS. et al. Leucobacter denitrificans sp. nov., isolated from cow dung. J Microbiol. 2012;50:161–165. doi: 10.1007/s12275-012-1324-1. [DOI] [PubMed] [Google Scholar]
- Ue H. Leucobacter exalbidus sp. nov., an actinobacterium isolated from a mixed culture from compost. J Gen Appl Microbiol. 2011;57:27–33. doi: 10.2323/jgam.57.27. [DOI] [PubMed] [Google Scholar]
- Shin NR, Kim MS, Jung MJ, Roh SW, Nam YD, Park EJ. et al. Leucobacter celer sp. nov., isolated from Korean fermented seafood. Int J Syst Evol Microbiol. 2011;61:2353–2357. doi: 10.1099/ijs.0.026211-0. [DOI] [PubMed] [Google Scholar]
- Behrendt U, Ulrich A, Schuman P. Leucobacter tardus sp. nov., isolated from the phyllosphere of Solanum tuberosum L. Int J Syst Evol Microbiol. 2008;58:2574–2578. doi: 10.1099/ijs.0.2008/001065-0. [DOI] [PubMed] [Google Scholar]
- Halpern M, Landsberg O, Raats D, Rosenberg E. Culturable and VBNC Vibrio cholerae: Interactions with Chironomid egg masses and their bacterial population. Microb Ecol. 2007;53:285–293. doi: 10.1007/s00248-006-9094-0. [DOI] [PubMed] [Google Scholar]
- Broza M, Halpern M. Chironomid egg masses and Vibrio cholerae. Nature. 2001;412:40. doi: 10.1038/35083691. [DOI] [PubMed] [Google Scholar]
- Halpern M, Broza YB, Mittler S, Arakawa E, Broza M. Chironomid egg masses as a natural reservoir of Vibrio cholerae non-O1 and non-O139 in freshwater habitats. Microb Ecol. 2004;47:341–349. doi: 10.1007/s00248-003-2007-6. [DOI] [PubMed] [Google Scholar]
- Halpern M, Raats D, Lavion R, Mittler S. Dependent population dynamics between chironomids (non-biting midges) and Vibrio cholerae. FEMS Microbiol Ecol. 2006;55:98–104. doi: 10.1111/j.1574-6941.2005.00020.x. [DOI] [PubMed] [Google Scholar]
- Senderovich Y, Gershtein Y, Halewa E, Halpern M. Vibrio cholerae and Aeromonas; do they share a mutual host? ISME J. 2008;2:276–283. doi: 10.1038/ismej.2007.114. [DOI] [PubMed] [Google Scholar]
- Figueras MJ, Beaz-Hidalgo R, Senderovich Y, Laviad S, Halpern M. Re-identification of Aeromonas isolates from chironomid egg masses as the potential pathogenic bacteria Aeromonas aquariorum. Environ Microbiol Rep. 2011;3:239–244. doi: 10.1111/j.1758-2229.2010.00216.x. [DOI] [PubMed] [Google Scholar]
- Beaz-Hidalgo R, Shakèd T, Laviad S, Halpern M, Figueras M. Chironomid egg masses harbour the clinical species Aeromonas taiwanensis and Aeromonas sanarellii. FEMS Microbiol Lett. 2012;337:48–54. doi: 10.1111/1574-6968.12003. [DOI] [PubMed] [Google Scholar]
- Senderovich Y, Halpern M. The protective role of endogenous bacterial communities in chironomid egg masses and larvae. ISME J. 2013;7:2147–2158. doi: 10.1038/ismej.2013.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HP, Göker M. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol. 2010;33:175–182. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Göker M, Klenk HP. Phylogeny-driven target selection for large-scale genome-sequencing (and other) projects. Stand Genomic Sci. 2013;8:360–374. doi: 10.4056/sigs.3446951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN. et al. A phylogeny-driven Genomic Encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Woyke T, Eisen JA, Garrity G, Lilburn TG, Beck BJ. et al. Genomic Encyclopedia of Type Strains, Phase I: the one thousand microbial genomes (KMG-I) project. Stand Genomic Sci. 2013;9:628–634. doi: 10.4056/sigs.5068949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides NC, Hugenholtz P, Eisen JA, Woyke T, Göker M, Parker CT. et al. Genomic encyclopedia of Bacteria and Archaea: sequencing a myriad of type strains. PLoS Biol. 2014;8 doi: 10.1371/journal.pbio.1001920. Article ID e1001920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao H, Froula J, Du C, Kim TW, Hawley E, Bauer S. et al. Identification of novel biomass-degrading enzymes from microbial dark matter: populating genome sequence space with functional annotation. Biotechnol Bioeng. 2014;111:1550–1565. doi: 10.1002/bit.25250. [DOI] [PubMed] [Google Scholar]
- Reddy TB, Thomas AD, Stamatis D, Bertsch J, Isbandi M, Jansson J, et al. The Genomes OnLine Database (GOLD) v.5: a metadata management system based on a four level (meta) genome project classification. Nucleic Acids Res 2015;43:D1099-106. [DOI] [PMC free article] [PubMed]
- DOE Joint Genome Institute. http://www.jgi.doe.gov.
- Mavromatis K, Land ML, Brettin TS, Quest DJ, Copeland A, Clum A. et al. The fast changing landscape of sequencing technologies and their impact on microbial genome assemblies and annotation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0048837. Article ID e48837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- List of growth media used at the DSMZ. [ http://www.dsmz.de/catalogues/catalogue-microorganisms/culture-technology/list-of-media-for-microorganisms.html]
- Gemeinholzer B, Dröge G, Zetzsche H, Haszprunar G, Klenk HP, Güntsch AM. et al. The DNA Bank Network: the start from a German initiative. Biopreserv Biobank. 2011;9:51–55. doi: 10.1089/bio.2010.0029. [DOI] [PubMed] [Google Scholar]
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- Zerbino D, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wgsim. https://github.com/lh3/wgsim.
- Gnerre S, MacCallum I. High–quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108(4):1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavromatis K, Ivanova NN, Chen IM, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard operating procedure for the annotations of microbial genomes. Stand Genomic Sci. 2009;1:63–67. doi: 10.4056/sigs.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IM, Markowitz VM, Chu K, Anderson I, Mavromatis K, Kyrpides NC. et al. Improving microbial genome annotations in an integrated database context. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054859. Article ID e54859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- Senderovich Y, Halpern M. Bacterial community composition associated with chironomid egg masses. J Insect Sci. 2012;12:148. doi: 10.1673/031.012.14901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J-H, Cho Y-J, Chun J, Hyun D-W, Bae J-W. Genome sequence of the chromate-resistant bacterium Leucobacter salsicius type strain M1-8T. Stand Genomic Sci. 2014;9:495–504. doi: 10.4056/sigs.4708537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm G, Buchta K, Kurz T, Rensing SA, Gescher J. Draft genome sequence of Leucobacter chromiiresistens, an extremely chromium-tolerant strain. J Bacteriol. 2012;194:540–541. doi: 10.1128/JB.06413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;10:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Garrity GM, Gray T, Morrison N, Selengut J, Sterk P. et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Amaral-Zettler L, Cochrane G, Cole JR, Dawyndt P, Garrity GM. et al. The Genomic Standards Consortium. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001088. Article ID e1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM. Names for Life Browser Tool takes expertise out of the database and puts it right in the browser. Microbiol Today. 2010;37:9. [Google Scholar]
- Woese CR, Kandler O, Weelis ML. Towards a natural system of organisms. Proposal for the domains Archaea and Bacteria. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Holt JG. In: Bergey’s Manual of Systematic Bacteriology. 2. Garrity GM, Boone DR, Castenholz RW, editor. Springer, New York; 2001. The Road Map to the Manual; pp. 119–169. [DOI] [Google Scholar]
- Stackebrandt ERF, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol. 1997;47:479–491. doi: 10.1099/00207713-47-2-479. [DOI] [Google Scholar]
- Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol. 2009;59:589–608. doi: 10.1099/ijs.0.65780-0. [DOI] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- Buchanan RE. Studies in the nomenclature and classification of bacteria. II. The primary subdivisions of the Schizomycetes. J Bacteriol. 1917;2:155–164. doi: 10.1128/jb.2.2.155-164.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Validation of the publication of new names and new combinations previously effectively pub-lished outside the IJSB. List No. 53. Int J Syst Bacteriol. 1995;45:418–419. doi: 10.1099/00207713-45-2-418. [DOI] [PubMed] [Google Scholar]
- Park YH, Suzuki K, Yim DG, Lee KC, Kim E, Yoon J. et al. Suprageneric classification of peptidoglycan group B actinomycetes by nucleotide sequencing of 5S ribosomal RNA. Antonie Van Leeuwenhoek. 1993;64:307–313. doi: 10.1007/BF00873089. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. et al. Gene ontology: tool for the unification of biology. Gene Ontology Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland-Moritz HE, Bevans DR, Lang JM, Darling AE, Eisen JA, Coil DA. Draft Genome Sequence of Leucobacter sp. Strain UCD-THU (Phylum Actinobacteria) Genome Announc. 2013;1:e00325–e13. doi: 10.1128/genomeA.00325-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


