Summary
It is now well accepted that defined architectural compartments within the cell nucleus can regulate the transcriptional activity of chromosomal domains within their vicinity. However, it is generally unclear how these compartments are formed. The nuclear periphery has received a great deal of attention as a repressive compartment that is implicated in many cellular functions during development and disease. The inner nuclear membrane, the nuclear lamina, and associated proteins compose the nuclear periphery and together they interact with proximal chromatin creating a repressive environment. A new study by Harr et al. identifies specific protein:DNA interactions and epigenetic states necessary to re-position chromatin to the nuclear periphery in a cell-type specific manner. Here, we review concepts in gene positioning within the nucleus and current accepted models of dynamic gene repositioning within the nucleus during differentiation. This study highlights that myriad pathways lead to nuclear organization.
Keywords: epigenetics, lamina-associated domains, nuclear lamina, nuclear organization
Introduction
DNA is not randomly dispersed throughout the nucleus. Rather, eukaryotic cells carefully package their DNA in complex physical structures forming higher-order chromatin states. During interphase, specific chromosomal domains occupy defined areas within the nucleus [1]. This physical organization has been shown to ensure proper cellular events by positioning chromosomal domains in proximity to regulatory bodies in the nucleus [2, 3]. For example, highly active genes localize to subnuclear foci that engender proximity to the transcriptional machinery [3]. Similarly, inactive genes tend to cluster together near transcriptionally repressive areas of the nucleus. In vertebrates, gene-rich and transcriptionally active DNA tends to locate towards the nuclear interior while heterochromatic regions that are gene-poor, less transcribed, and late replicating cluster towards the nuclear periphery [4]. This organization is almost universal; exceptions to this organization exist in specialized conditions such as rod photoreceptor cells in nocturnal mammals [5] and during oncogene induced senescence [6]
Nuclear organization appears to play a key role in regulating many developmental genes [7]. For example, as embryonic stem cells (ESCs) differentiate to various cell types, chromatin is dynamically rearranged, correlating with different gene expression programs being activated or repressed [8, 9, 10, 11, 12]. Three main mechanisms work in unison to control developmentally regulated gene expression patterns: availability of transcription factors, epigenetics, and higher-order chromatin organization. While the former two have been extensively studied, much less is known about higher-order chromatin organization and its role in controlling gene expression [3, 13, 14]. Gene regulation can be manipulated by creating higher-order chromatin loops, supporting the critical role that chromatin-organization plays in gene regulation [15, 16]. Additionally, targeting of ectopic genes to the nuclear periphery leads to a general (albeit gene specific) repressive state [16, 17]. Together, these artificial manipulations suggest that nuclear organization is important for gene regulation and development. It is critical to understand the endogenous mechanisms driving gene rearrangement in the nucleus as it offers insights on the process of regulating gene activity and function. In this paper, we highlight recent advances in understanding genetic and epigenetic factors driving chromosomal association with the nuclear periphery.
The nuclear periphery: A familiar landmark of nuclear organization
The nucleus is encased by an outer and inner nuclear membrane interrupted by pores spanning both of the membranes that control traffic into and out of the nucleus. Early cytogenetic studies of mammalian nuclei observed an accumulation of heterochromatin near the inner nuclear membrane (INM), forming an easily recognizable transcriptionally repressed compartment [18]. Animals possess a 15-20 nm layer of protein meshwork lining the nucleoplasmic side of the INM called the nuclear lamina, which is a polymerized structure composed of intermediate filament proteins known as lamins. This dense underlying mesh network of lamins fortifies the INM and mechanically supports the nucleus [19]. The nuclear lamina is also characterized by myriad lamina-associated proteins that interact with a large portion of the mammalian genome creating repressive compartments along the nuclear periphery. Many of these compartments are comprised of heterochromatic DNA regions and have been shown to physically interact with the nuclear lamina at the periphery of the nucleus. Furthermore, mutations in lamin proteins have been associated with a host of heritable human diseases such as dilated cardiomyopathy with variable muscular dystrophy and those associated with extreme premature aging [20] underscoring the pleiotropic importance of this nuclear compartment.
In order to understand how chromatin is regulated at the periphery, one series of experiments characterized the segments of chromosomes that reside at the periphery through the use of DNA adenine methyltransferase identification (DamID) techniques [21]. DamID works by fusing the E. coli Dam methylase to a protein of interest, directing adenine methylation to GATC sequences that come into close proximity to the protein of interest [21]. In one case, DamID was fused to the lamin protein, lamin B1, and identified 1,300 regions of DNA known as lamina associated domains (LADs) ranging from 0.1-10 Mb in cultured human lung fibroblasts [22]. Interestingly, LAD borders, marking the junctions between LAD and interiorly-localized chromatin termed inter-LADs, show enrichment for specific chromatin proteins such as CCCTC-binding factor (CTFC) as well as conserved histone modifications, which suggest possible mechanisms for separating or “insulating” inter-LAD chromatin from the repressive effects of the lamina [11, 22].
LADs are characterized by low gene expression levels and their interaction with the lamina can be developmentally regulated [11, 22]. Hence, it has been speculated that cell-type specific interaction with the nuclear lamina is involved in the repression of developmental genes. In fact, DamID studies of differentiating mouse ESCs demonstrate a strong correlation between movement of chromatin to or away from the lamina and transcriptional repression or upregulation, respectively [11]. These developmentally regulated LADs, known as “variable LADs” (vLADs), have been shown to occur by shifting LAD border regions [11], suggesting that a dissection of LAD borders will enable a greater understanding of the dynamic regulation of their genomic organization.
Targeting genes to the nuclear lamina generally represses their expression
The studies described above identified a correlation between localization at the nuclear lamina and transcriptional repression. However, such correlations do not assess whether gene repression is a direct consequence of targeting to the periphery. Methods of molecular tethering have been developed to overcome these pitfalls [17, 23, 24]. These systems work to position genes to the periphery in an inducible fashion using a two-component inducible system. In the absence of an allosteric inhibitor, a fusion of the LacI protein with any of several proteins that are localized to the nuclear periphery (e.g. Lap2β, emerin, laminB1) recruits target genomic loci bearing an array of lacO repeats to the periphery. With such systems, it has been shown that physically tethering genes to the nuclear periphery can reversibly repress gene expression in human cells, suggesting that nuclear reorganization during development also has the potential to modulate gene expression [17, 24, 25].
As suggested by the correlative studies, not all genes are silenced at the periphery. In one of these studies, recruitment to the nuclear periphery had no effect on the transcriptional activity of a reporter gene under the control of a potent artificial transcriptional activating domain [23]. Finlan et al. repositioned sites on two different human chromosomes (4 and 11) to the periphery and found a silencing of some of the endogenous genes but not others [17]. Finally, Zuleger et. al. identified novel cellular nuclear envelope proteins that could promote peripheral localization when artificially targeted to specific chromosomal locations, with similarly variable effects on transcription of the re-positioned genes [26]. Although the three studies used separately developed gene targeting systems as well as different test genes, the simplest conclusion is that targeting to the nuclear periphery is generally repressive to gene activity, but this repression can be overcome by certain promoters. These studies also demonstrate that transcription is compatible with peripheral localization. In apparent contradiction to this conclusion, a recent study found that targeting multiple copies of the same transcriptional activator used by Kumaran and Spector to a promoter drove chromatin off of the periphery [27], but it was found that the chromatin decondensation activity of this activator, not its ability to induce transcription was responsible for repositioning.
LADs contain lamina associated sequences that drive peripheral targeting
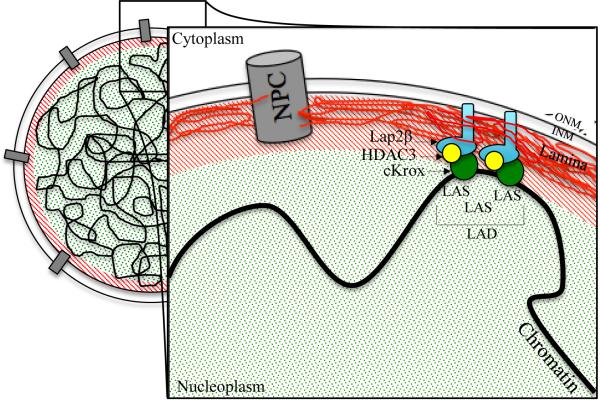
The artificial systems described above demonstrate that targeting to the periphery is sufficient to repress transcription of certain classes of genes, but understanding the dynamic reorganization of the genome during differentiation requires the identification of the endogenous mechanisms that mediate this targeting. Various molecular mechanisms have been proposed to explain how endogenous chromatin domains become targeted to the periphery [28]. Recent studies have begun to identify short sequences (4-6 kb) that can target to the periphery [29]. In this study, the researchers used developmentally regulated loci including IgH and Cyp3a, which are contained within vLADs associated with the nuclear lamina and are repressed in fibroblasts. Regions of LADs within the IgH and Cyp3a carried on bacterial artificial chromosomes (BACs) were assayed for their ability to target to the lamina when randomly inserted into the genome of mouse fibroblasts. By 3D-immunoFISH, the authors observed that BACs containing LAD-derived regions of DNA associated with the lamina independent of where the BACs were inserted. In contrast, control BACs comprised of inter-LAD DNA localized to the nuclear interior. Control inter-LAD BACs that did not contain active genes also localized to the nuclear interior demonstrating the inter-LAD control BACs were not simply positioned to the interior because of the activity of genes transcribed from the BACs. Furthermore, vLAD-derived DNA was shown to reduce the synthesis of a fluorescent reporter protein. This strongly suggests that specific DNA sequences, rather than the absence of transcriptional activity, target chromatin to a transcriptionally repressive environment at the periphery. The authors then used their random integration assay followed by 3D-immunoFISH to identify several sub-fragments within both vLAD-containing BACs that were sufficient to promote chromatin association with the nuclear lamina, which they referred to as lamina associated sequences (LASs). The authors then searched for DNA sequence motifs within the LASs, revealing enrichment for an extended GAGA motif, which is the recognition site for the transcription factor Zbtb7b (also known as THPOK, cKrox, or ZFP67). Furthermore, Zbtb7b was found in a complex with HDAC3 and Lap2β proteins and knockdown of either Zbtb7b or HDAC3 eliminated peripheral targeting. These results reveal the first glimpses of mechanisms targeting chromatin domains to the nuclear periphery, coordinated with the control of gene activity (Fig. 1).
Figure 1. Model depicting the interaction of LASs within a LAD with the nuclear periphery.
Depicted above is a view of the nucleus with nuclear pore complexes (NPCs) spanning the inner and outer nuclear membranes (INM/ONM). Red and green shading corresponds to repressive and activating nuclear compartments respectively. The transcription factor cKrox binds to GAGA motifs enriched in LASs. Since cKrox has been shown to interact with both HDAC3 and Lap2β, it is hypothesized that the LAS:cKrox complex then mediates interaction with HDAC3 and the inner nuclear membrane protein Lap2β [29].
Epigenetics work in conjunction with LASs in nuclear periphery association
In a recent issue of the Journal of Cell Biology, Harr et al. reasoned that there are likely to be many different mechanisms targeting chromatin to the periphery during development and set out to identify others [29]. First, the group applied the DamID method described above to identify that vLADs are at the periphery in fibroblasts but are interiorly located in pro-B cells and contain key B-cell specific developmental genes. Because vLADs occur by shifting LAD border regions between cell types, the group focused their analysis on border regions. BACs containing genomic regions from vLAD borders were used to test the sufficiency of each sequence to associate with the periphery in fibroblasts, using the random integration assay described above. Of three independent BACs tested derived from vLAD borders near the IgH, Bcl11a or Ikzf1 genes, all three showed strong association with the nuclear periphery. A control BAC constructed from an inter-LAD domain localized to the interior. As in the Zullo et al. paper, subfragments of the vLAD border containing BACs were also tested for targeting ability, identifying several LAS-containing subfragments. However, since random integration generally leads to several tandem copies of the inserted sequence, the authors wanted to test whether a single copy of a LAS-containing element would be sufficient to target genes to the periphery. To do so, they employed a previously developed system that uses site-specific recombination to target a single copy of any DNA sequence to the same specific chromosomal locus [30]. This also controlled for any effects of the integration site. Using this strategy, they found that the probability of targeting with a single LAS was the same as their multiple-insert counterparts.
Similar to the Zullo et al. study, the group next sought to decipher what genetic elements within the LASs enable protein binding and assist in the molecular tethering. The group identified CTCF, previously shown to be enriched at LAD borders [22], and Zbtb7b binding sites previously shown to be enriched within LASs [28]. Additionally, they identified strong enrichment for binding sites for a protein known as Ying-Yang 1 (YY1), which is speculated to participate in many facets of genome regulation including gene activation and repression. To determine whether each of these proteins was necessary for targeting to the periphery, the group employed a targeted small hairpin RNA (shRNA)-mediated knockdown strategy to test the involvement of a host of proteins, including known lamina-associated proteins (Zbtb7b, HDAC3, and Lap2β), lamina subunits LMNA and LMNC and the proteins binding to the novel motifs discovered in this study (CTFC, YY1, and EZH2). All knockdowns with the exception of LMNA revealed a loss of peripheral association of the targeting fragments (although knock down of Lap2β and HDAC3 resulted in excessive cell death and could not be analyzed). Therefore, CTFC, YY1, EZH2, Zbtb7b and LMNC are all necessary to maintain lamina-proximal association of the identified LASs. Interestingly, Zbtb7b-mediated repression of Runx is required for generation of CD4+CD8- thymocytes in mice. When Zbtb7b is mutant, Runx expression in progenitors of these thymoctes redirects their differentiation toward the CD8+ lineage [31]. It is enticing to speculate that LAS peripheral localization may be part of the mechanism by which Zbtb7b represses Runx in MHC class II– selected thymocytes. It should also be noted that a recent study examining genome-wide assoication of chromatin with the nuclear periphery using an emering-DamID-fusion found that, in mouse ESCs, Lamins A, B and C are dispensible for peripheral localization, suggesting that cell-type specific differences may exist [32].
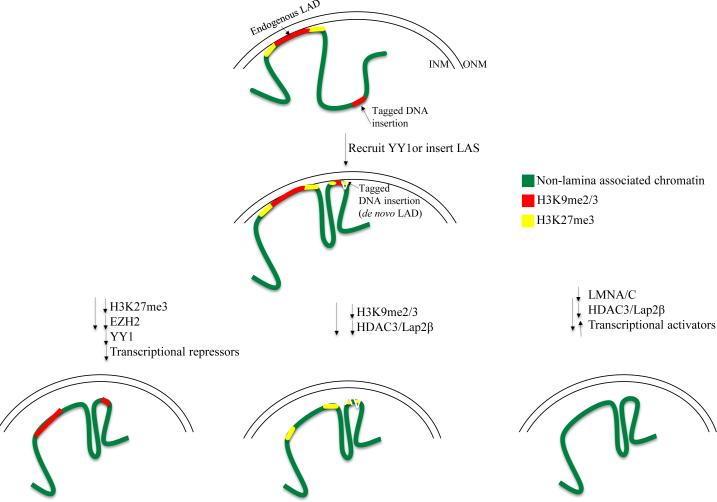
Next, the group assayed whether either a YY1- or a Zbtb7b-GFP-LacI fusion protein was sufficient to target an integrated lacO array to the nuclear periphery. They found that YY1, but not Zbtb7b was sufficient to target to the periphery. They also found that the histone modification H3K27me3, which is enriched at natural LAD borders [22], was also enriched at the lacO-array containing sites to which YY1 was recruited. They then showed that chemical inhibition of H3K27 methylation prevented YY1-mediated recruitment to the periphery, suggesting that H3K27 methylation is necessary in conjunction with YY1 binding. In fact, there is evidence that YY1 can recruit PRC2 to mediate H3K27 methylation [33]. The groups also noticed that the repressive H3K9me2/3 modifications were constitutively present at the lacO array site, before recruitment to the periphery. Since a recent study showed that this histone modification plays a key role in the repositioning of ectopic sites to the nuclear lamina [34, 35], Harr et al. also treated cells with a chemical inhibitor of H3K9me2/3 methylation, and found that this treatment also prevented YY1-mediated targeting to the periphery. Together, these results suggest cooperation of YY1 with both pre-existing and induced epigenetic modifications (Figure 2).
Figure 2. Many factors are necessary and/or sufficient for peripheral targeting.
vLADs are depicted as regions with H3K9me2/3 (red) bordered by H3K27me3 (yellow) after they are recruited to the periphery, either by endogenous sequences, or when targeted by artificial recruitment of YY1. Perturbation of H3K27me3, H3K9me2/3, or LMNA/C leads to loss of association of endogenous or ectopic LADs with the nuclear periphery. This image is from [29], and is displayed is under the terms of a Creative Commons License (Attribution-Noncommercial-Share Alike 3.0 Unported license, as described at http://creativecommons.org/licenses/by-nc-sa/3.0/).
Conclusions: Moving towards the full picture of gene interaction with the nuclear periphery
We have come a long way from the initial finding that genes occupy conserved pre-defined locations within the interphase nucleus. We are getting closer to understanding the complexity of the nucleus and its immense role in gene regulation and development. Of all the subnuclear compartments, the nuclear periphery is arguably one of the most important in transcriptional repression. Several models have emerged as to the cis and trans elemental control of the tethering of chromosomal domains to the inner-nuclear membrane and lamina. Most recently, focus has been directed toward understanding how large segments of chromosomes are repositioned during differentiation to engender proper gene expression. The experiments described in Harr et al. demonstrate that a combination of DNA sequence, binding proteins, and chromatin post-translational modifications work in concert to reposition genes to the nuclear periphery. This paints a much more complex picture than was initially laid forth by the pioneering studies of Zullo et al. Clearly these findings suggest that there are many avenues of research remaining to achieve a complete understanding of how nuclear organization regulates genome function and its disruption in disease.
Acknowledgements
Our research is supported by NIH grants GM083337 and GM085354 to DMG. We would like to extend a special thank you to the members of the Gilbert lab for thoughtful conversation on this review.
Abbreviations
- (v)LAD
(variable) lamina associated domain
- LAS
lamina associated sequence
References
- 1.Okeefe RT, Henderson SC, Spector DL. Dynamic Organization of Dna-Replication in Mammalian-Cell Nuclei - Spatially and Temporally Defined Replication of Chromosome-Specific Alpha-Satellite Dna-Sequences. J. Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. ST – DYNAMIC ORGANIZATION OF DNA–REPLIC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Bortle K, Corces VG. Nuclear Organization and Genome Function. Annu. Rev. Cell Dev. Biol. 2012;28:163–87. doi: 10.1146/annurev-cellbio-101011-155824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambeyron S, Bickmore W a. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–30. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Croft JA, Bridger JM, Boyle S, Perry P, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–31. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solovei I, Kreysing M, Lanctôt C, Kösem S, et al. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell. 2009;137:356–68. doi: 10.1016/j.cell.2009.01.052. [DOI] [PubMed] [Google Scholar]
- 6.Chandra T, Ewels PA, Schoenfelder S, Furlan-Magaril M, et al. Global Reorganization of the Nuclear Landscape in Senescent Cells. Cell Rep. 2015;10:471–83. doi: 10.1016/j.celrep.2014.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat. Rev. Genet. 2007;8:507–17. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 8.Hiratani I, Ryba T, Itoh M, Yokochi T, et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:2220–36. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M, et al. Chromatin Fibers Are Formed by Heterogeneous Groups of Nucleosomes In Vivo. Cell. 2015;160:1145–58. doi: 10.1016/j.cell.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 10.Williams RR, Azuara V, Perry P, Sauer S, Dvorkina, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J. Cell Sci. 2015;119:132–40. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 11.Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SWM, et al. Molecular Maps of the Reorganization of Genome-Nuclear Lamina Interactions during Differentiation. Mol. Cell. 2010;38:603–13. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, Jung I, Selvaraj S, Shen Y, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–6. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson I, Berlivet S, Eskeland R, Boyl S, et al. Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev. 2014;28:2778–91. doi: 10.1101/gad.251694.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 2015;16:245–57. doi: 10.1038/nrm3965. [DOI] [PubMed] [Google Scholar]
- 15.Deng W, Lee J, Wang H, Miller J, et al. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–44. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng W, Rupon JW, Krivega I, Breda L, et al. Reactivation of Developmentally Silenced Globin Genes by Forced Chromatin Looping. Cell. 2014;158:849–60. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finlan LE, Sproul D, Thomson I, Boyle S, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heun P, Laroche T, Shimada K, Furrer P, et al. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–6. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- 19.Schirmer EC, Foisner R. Proteins that associate with lamins: Many faces, many functions. Exp. Cell Res. 2007;313:2167–79. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 20.Worman HJ, Courvalin JC. Nuclear envelope, nuclear lamina, and inherited disease. Int. Rev. Cytol. 2005;246:231–79. doi: 10.1016/S0074-7696(05)46006-4. [DOI] [PubMed] [Google Scholar]
- 21.Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–78. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 22.Guelen L, Pagie L, Brasset E, Meuleman W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 23.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 25.Kind J, van Steensel B. Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol. 2010;22:320–5. doi: 10.1016/j.ceb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Zuleger N, Boyle S, Kelly DA, de las Heras JI, et al. Specific nuclear envelope transmembrane proteins can promote the location of chromosomes to and from the nuclear periphery. Genome Biol. 2013;14:R14. doi: 10.1186/gb-2013-14-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therizols P, Illingworth RS, Courilleau C, Boyle S, et al. Chromatin decondensation is sufficient to alter nuclear organization in embryonic stem cells. Science (80-. ) 2014;346:1238–42. doi: 10.1126/science.1259587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zullo JM, Demarco I a, Piqué-Regi R, Gaffney DJ, et al. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–87. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 29.Harr JC, Luperchio TR, Wong X, Cohen E, et al. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng YQ, Seibler J, Alami R, Eisen a, et al. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–85. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 31.Egawa T, Littman DR. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx- mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 2008;9:1131–9. doi: 10.1038/ni.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amendola M, van Steensel B. Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 2015;16:610–7. doi: 10.15252/embr.201439789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 34.Bian Q, Khanna N, Alvikas J, Belmont AS. ??-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J. Cell Biol. 2013;203:767–83. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tobin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-Wise Methylation of Histone H3K9 Positions Heterochromati at the Nuclear Periphery. Cell. 2012;105:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]