Abstract
HIV-1 viral infectivity factor (Vif) is a viral accessory protein that is required for HIV-1 infection due largely to its role in recruiting antiretroviral factors of the APO-BEC3 (apolipoprotein B editing catalytic subunit-like 3) family to an E3 ubiquitin ligase complex for polyubiquitylation and proteasomal degradation. The crystal structure of the (near) full-length Vif protein in complex with Elongin (Elo)B/C, core-binding factor (CBF)β and Cullin (Cul)5 revealed that Vif has a novel structural fold. In our opinion the structural data revealed not only the protein–protein interaction sites that determine Vif stability and interaction with cellular proteins, but also motifs driving Vif homodimerization, which are essential in Vif functionality and HIV-1 infection. Vif-mediated protein–protein interactions are excellent targets for a new class of antiretroviral therapeutics to combat AIDS.
Keywords: viral infectivity factor, core-binding factor β, cullin 5, Elongin B/C, APOBEC
Introduction
The structural complexity of HIV-1 Vif has been studied for the better part of 3 decades [1] but now has been brought to light through the crystal structure of the (near) full-length protein recently published by the Huang group (PDB ID: 4N9F, [2]) (Figure 1). As revealed by its unique fold, Vif is unlike any other known protein to date, and its only structural homology with a cellular protein comprises just a small stretch of amino acids, termed the ‘BC-box’, found in SOCS-box (Suppressor of cytokine signaling) type E3 ubiquitin ligase substrate receptors. Vif is absolutely required for sustained HIV-1 infection and, although it has been implicated in a variety of viral functions, its main role is to recruit several human antiretroviral factors of the APOBEC3 family to an E3 ubiquitin ligase complex for polyubiquitylation, a signal for subsequent proteasomal degradation (Box 1).
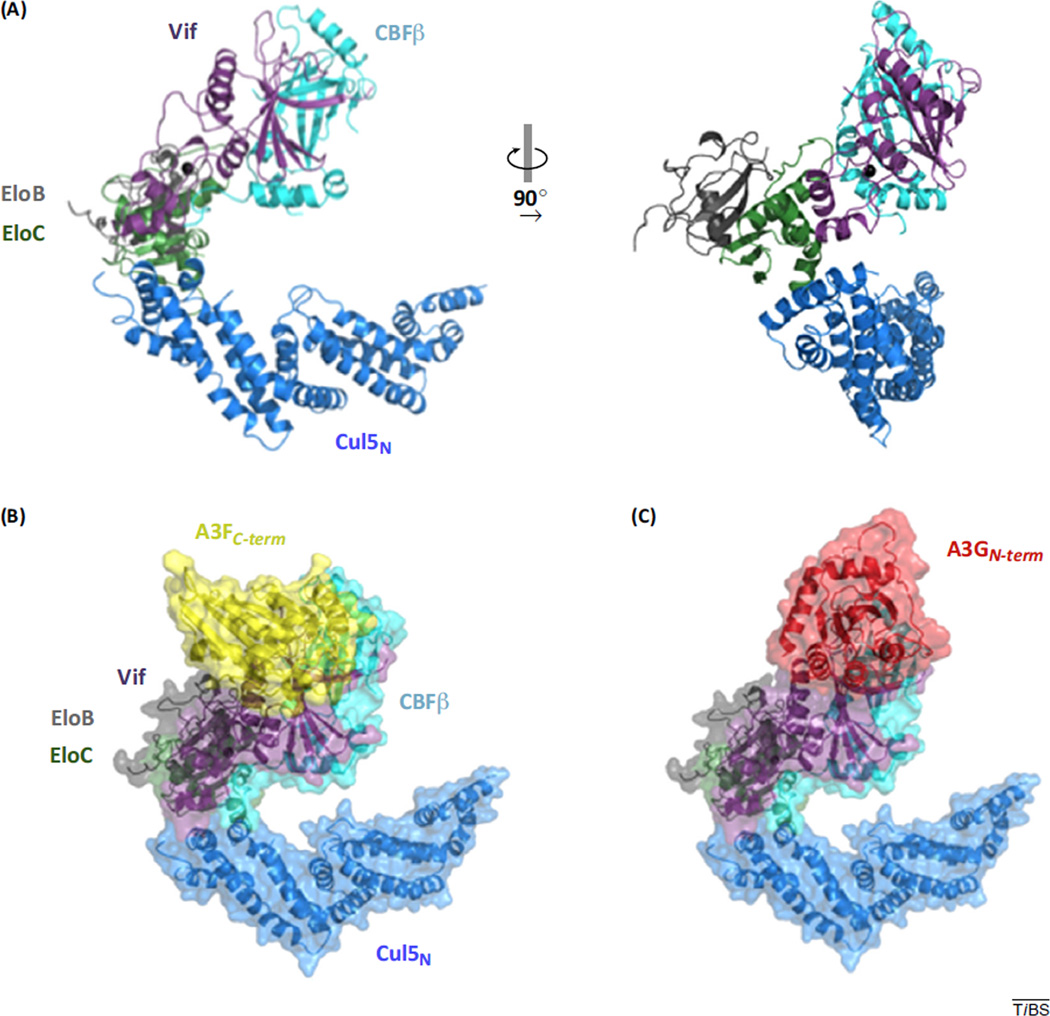
Figure 1.
Crystal structure of Vif/CBFβ/EloB/EloC/Cul5 and models of the E3 ubiquitin ligase complex docked with A3 substrates. (A) Cartoon representation of the E3 ubiquitin ligase complex comprising HIV-1 Vif (1–173) and human proteins CBFβ, EloB, EloC, and the N-terminal half (residues 1–386) of Cul5 (PDB 4N9F). Vif interacts with EloC and Cul5 through its C-terminal α domain and forms extensive interactions with CBFβ through its N-terminal α/β domain as well as the α domain. Coordination of a zinc ion (charcoal sphere) bridges the N and C-terminal domains of Vif. Top scoring ClusPro models of A3F (B) and A3G (C) docked with Vif in the context of the E3 ligase complex reveal interfaces formed between Vif and A3F differ from those between Vif and A3G. Abbreviations: A3F, APOBEC3F; A3G, APOBEC3G; APOBEC3, apolipoprotein B editing catalytic subunit-like 3; CBFβ, core-binding factor β subunit; Cul, Cullin; Elo, Elongin; Vif, viral infectivity factor.
HIV-1 Vif is an E3 ubiquitin-ligase substrate receptor that targets human A3 antiretroviral proteins for proteasomal degradation.
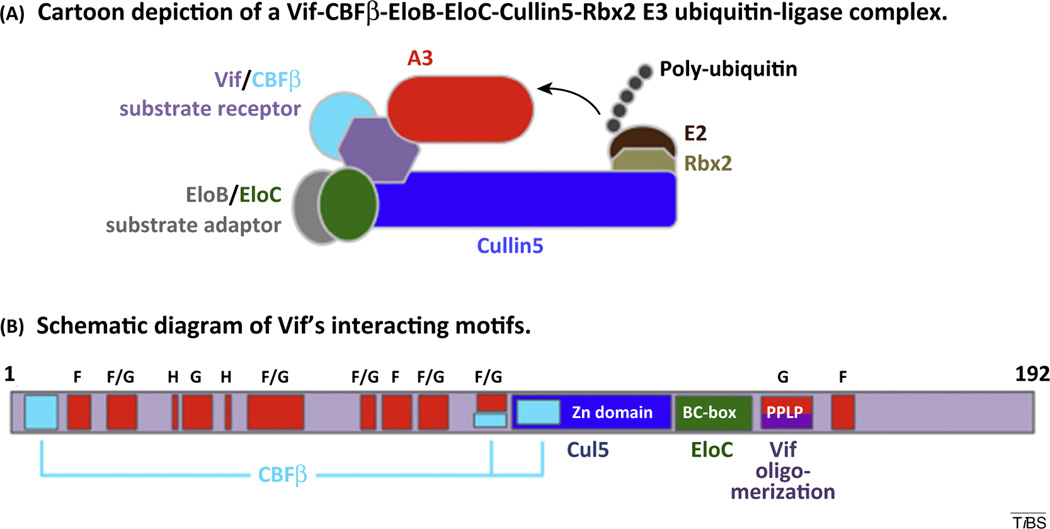
HIV-1 Vif is an auxiliary protein that is expressed during the late phase of viral infectivity and is essential to maintain a spreading infection in CD4+ T cells. The primary function of Vif is to prevent the accumulation of the antiretroviral A3 proteins (Box 2) in budding virions during viral infection. Vif acts as a SOCS-box type substrate receptor for a Cullin-RING E3 ubiquitin-ligase complex comprising the scaffold protein, Cul5, substrate adaptor heterodimer, EloBC, and the RING protein, Rbx2. Vif specifically binds and recruits substrate APOBEC3 proteins (A3C, D, F, G, and H) to the E3 ubiquitin ligase for covalent modification with polyubiquitin chains, which act as a target for proteasomal degradation (Figure 1A). Unlike cellular SOCS-box type substrate receptors, Vif requires an additional interaction with the cellular CBFβ. CBFβ acts as a chaperone that stabilizes the required conformation of Vif to interact with Cul5 and potentially regulates interaction with substrate proteins. As such, Vif forms specific protein–protein interactions with EloC through a BC-box motif and with Cul5 through a novel HCCH zinc-binding domain, both located in the C-terminal half of Vif (Figure 1A,B). CBFβ forms three critical interactions with the Vif N-terminal half and zinc-binding domain, burying a large amount of surface area. These structural interactions are described in greater detail by the Huang group [2]. Vif is predicted to interact with substrate proteins (A3C, D, F, G, and H) utilizing a variety of conserved sequence motifs primarily located throughout its N-terminal half (Box 3 and Figure 2 main text).
Figure I. HIV-1 Vif and its interactions with the E3 ubiquitin-ligase complex. (A) Schematic description of Vif-mediated E3 ubiquitin-ligase complex. Substrate A3 proteins are recruited to the N-terminal half of Cul5 through specific interaction with Vif substrate receptor. EloBC substrate adaptor bound to the Vif BC-box facilitates a combined interaction between the zinc-binding domain of Vif and Cul5. CBFβ stabilizes interactions of Vif with Cul5 and A3 substrates. The C-terminal half of Cul5 bound to Rbx2 recruits an E2 ubiquitin conjugation enzyme for transfer of polyubiquitin to substrate A3 proteins. (B) Functional motifs of Vif involved in protein–protein interactions of the E3 ubiquitin ligase complex. Regions of Vif that interact with CBFβ, Cul5, EloC, are colored cyan, blue, and green respectively. Regions implicated in binding A3 proteins are colored red and are labeled with F (for A3F), G (for A3G), H (for A3H), and F/G (for A3F and A3G).
In the past 12 years, numerous conserved residues and sequence motifs have been identified throughout the 192 amino acid length of Vif (Box 1). These amino acids contribute to the role of Vif in preserving the integrity of the HIV genome by suppression or destruction of the antiretroviral HIV-mutator family of human cytidine deaminase APOBEC3 proteins (Box 2). The requirement of these amino acids for Vif function can now be related to their structure and understood in the context of a Vif-mediated E3 ubiquitin ligase complex.
Antiretroviral activity of A3 proteins.
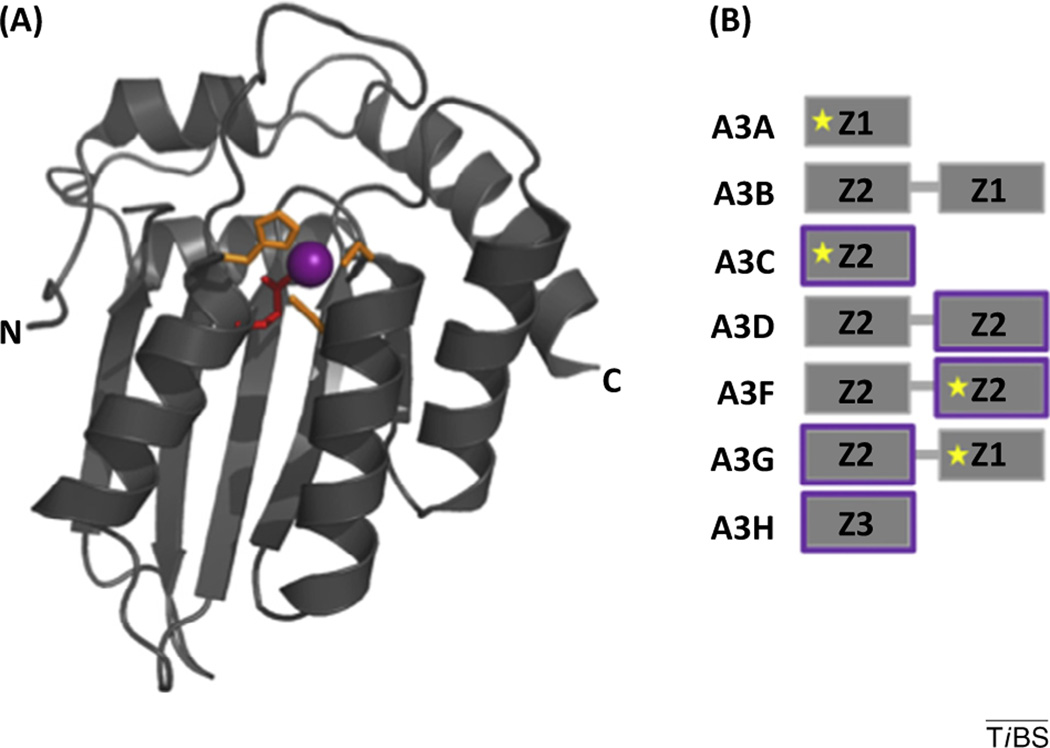
The seven members of the human APOBEC3 family (A3A, B, C, D, F, G, and H) are a group of evolutionarily related cytidine deaminases, forming part of the innate immune system, that actively restrict both endogenous and exogenous retroviral elements, including HIV-1, through both deaminase-dependent and -independent mechanisms. The structure of the A3 family of cytidine deaminases is shown (Figure 1). APOBEC evolutionary expansion, expression phenotypes, and activity against specific retroelements has been reviewed elsewhere [32]. Restriction of HIV-1 begins with the packaging of A3 proteins into budding HIV particles. Upon infection of subsequent cells, A3 binds and deaminates deoxycytidine (dC) to deoxyuridine (dU) of the single-stranded proviral DNA intermediate during reverse transcription after RNaseH-digestion of the genomic RNA strand. The dC-to-dU deamination of the negative strand proviral DNA generates deoxyguanine (dG) to deoxyadenosine (dA) mutations on the positive (coding) strand of HIV DNA. Lethal mutagenesis of the HIV DNA by the A3 family abrogates the spreading infection. This antiretroviral activity is countered by HIV-1 Vif, which is expressed in virion-producing cells and targets A3 proteins for ubiquitylation and subsequent proteasomal degradation (Box 1). The evolutionary pressure between A3 proteins and HIV-1, other lentiviruses, and endogenous retroelements likely led to the expansion of the A3 gene family in humans and other primates. It is now clear that at least A3D, A3F, A3G, and several haplotypes of A3H are bona fide restrictors of HIV-1 in CD4+ T cells.
Figure I. General structural features of the A3 family of antiretroviral cytidine deaminases. The seven APOBEC3 proteins (A, B, C, D, F, G, and H) are composed of either one or two Z domains (also known as cytidine deaminase domains). (A) The general fold (tertiary structure) of a canonical Z domain is exemplified by the C-terminal Z-domain of A3F (PDB ID 4IOU) [9]. The Z domain comprises a five-stranded mixed β sheet flanked by six α helices. The catalytic center of each Z domain comprises a zinc ion (purple sphere) that is coordinated by three conserved histidines or cysteines (side chains shown as orange sticks) and an activated water molecule (not shown). The glutamic acid (red) activates a catalytic water molecule and is involved in proton shuttling during deamination of deoxycytidine to deoxyuridine. (B) The Z domains are shown schematically (gray boxes) for each A3 member. Z domains are divided into three subtypes, Z1, Z2, and Z3, based on the presence or absence of several conserved residues that are located near the catalytic center [33,34]. Vif targets five A3 proteins (C, D, F, G, and H) for proteasomal degradation and the CDA domains that directly interact with Vif are highlighted in purple. The Z domains for which structural information (X-ray crystallographic structure or solution NMR structure) is available are so indicated with a five-pointed yellow star.
Here, we examine the implications of the crystal structure of Vif in complex with E3 ubiquitin ligase components for the role of Vif in APOBEC3 degradation and for the potential significance of the PPLP motif in Vif homodimerization. Our evaluation of the published data leads us to conclude that structural information can be leveraged not only to model the interaction of Vif with APOBEC3 proteins and the ubiquitylation machinery, but also to identify small molecules targeting Vif that will disrupt protein–protein interactions as a novel drug-discovery effort for HIV therapeutics and eradication.
Location of Vif residues implicated in binding of APOBEC3 proteins
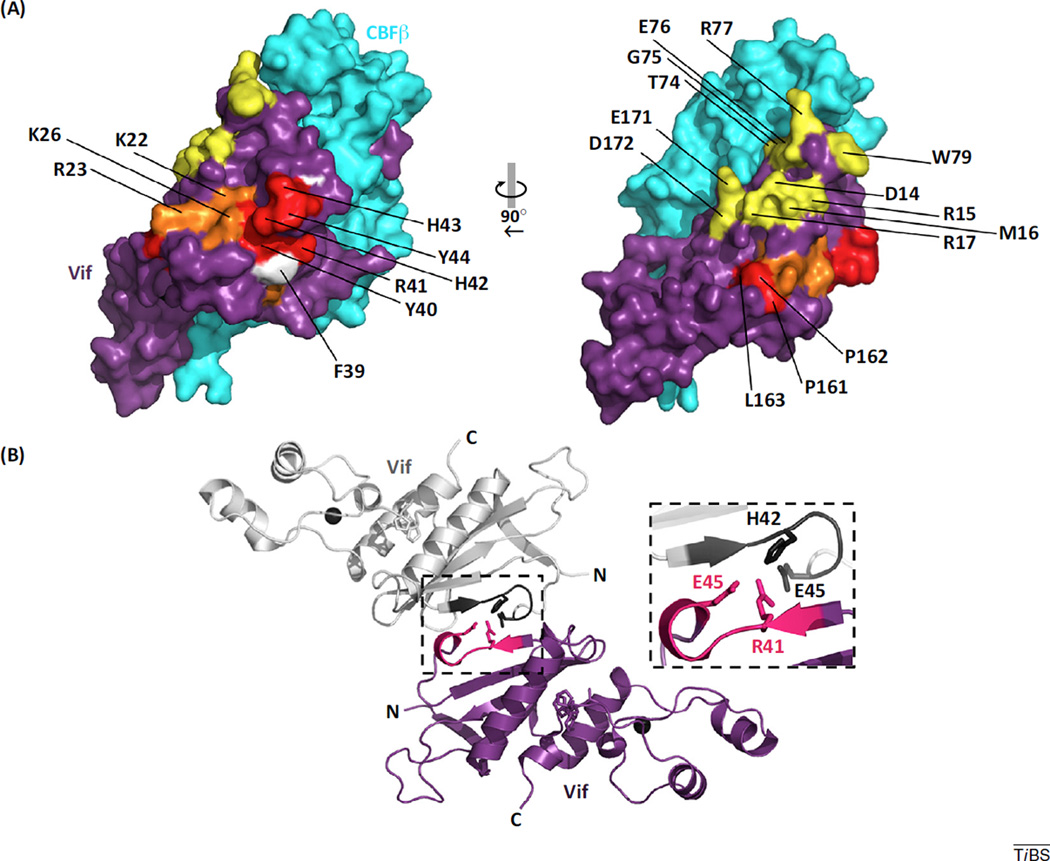
The role of a substrate receptor in an E3 ligase complex is to recruit a particular protein for polyubiquitylation through specific binding interactions. Unlike most cellular SOCS-box-type substrate receptors, which target a single protein, Vif targets several, albeit related, proteins of the APOBEC3 family. Specifically, Vif-dependent degradation of APOBEC3C, D, F, G and H (haplotype II), collectively referred to as the A3s, has been shown [3]. APOBEC3 proteins share a common core fold (Box 2) but positive evolutionary pressures to adapt to endogenous and exogenous retroelements led to expansion and diversification of the family. Ultimately, protein sequence variations are manifested structurally as subtle differences in shape, surface electrostatics, hydrophilicity, and quaternary arrangement (oligomerization). Thus, the APOBEC3 family does not have a single conserved surface to which Vif can bind and, as such, it is thought that Vif has evolved to recognize multiple surfaces presented by APOBEC3 family members. This is evident through functional mutational analyses of highly conserved Vif residues over the past decade. Such analyses have resulted in a large list of Vif sequence motifs implicated in binding specific APOBEC3 (F, G, and H) proteins (Table 1). Some sequence motifs have been shown to be important for binding multiple APOBEC3 proteins, whereas others are implicated in binding only a single APOBEC3 member. Some of the conserved motifs implicated in binding A3G and A3F are either buried within the core fold of Vif or buried at the interface with CBFβ. This suggests that these residues are likely not involved in direct interaction with A3 substrates, unless CBFβ dissociates upon A3 substrate binding, which as discussed below, may occur for A3F. Many of these residues map to the surface of Vif and are solvent exposed in the context of the E3 ubiquitin ligase complex structure, suggesting a direct role in binding of particular A3 proteins (Figure 2A). These solvent-exposed, surface-mapped residues implicated in binding A3F and A3G form two distinct and contiguous, though partially overlapping, regions [4].
Table 1.
Sequence motifs implicated in Vif–APOBEC3 interactions
| Vif sequence motifs implicated in binding specific APOBEC3 proteins |
APOBEC3 residues implicated in binding Vif |
||
|---|---|---|---|
| A3G | A3C | ||
| 40YRHHY44 | [35] | L72, F75, C76, I79, L80, | |
| 161PPLP164 | [23] | S81, Y86, E106, F107 | [8] |
| A3F | A3D | ||
| 14DRMR17 | [35] | L268, F271, C272, I275, L276, | |
| 74TGERxW79 | [36] | S277, Y282, E302, F303, H307 | [8] |
| 171EDRW174 | [37] | A3F | |
| A3G/F | L255, F258, C259, I262, L263, | ||
| 21WKSLVK26 | [38,39] | S264, Y269, E289, F290, H294 | [8] |
| 55VxIPLx4–5L64 | [36] | E286, E289, E316, E324 | [40] |
| 69YxxL72a | [41] | E324 | [42] |
| 81LGxGxxIxW89a | [37] | E289–H294 | [43] |
| 96TQx(D/E)PxxADxLI107a | [44] | A3G | |
| A3H | 128DPD130 | [45] | |
| F39, H48 | [46] | A3H (haplotype II) | |
| E121 | [47] | ||
Indicates the sequence motif is located within the core of Vif or buried at the interface with CBFβ, thus rendered inaccessible for a direct interaction with APOBEC3 proteins.
Figure 2.
Vif surface residues predicted to form important biological interfaces. (A) Vif residues implicated in binding A3 proteins form contiguous surface regions. Vif (purple) and CBFβ (cyan) from PDB 4N9F are depicted in surface representation. Coordinated zinc ions are depicted as charcoal spheres. Conserved residues implicated in binding A3G, A3F, or A3H (Table 1) are colored red, yellow, and white, respectively. Residues implicated in binding both A3G and A3F are colored orange. (B) Crystal contacts of the pentameric complex reveal a potential Vif oligomeric interface. Packing of the Vif/CBFβ/EloB/EloC/Cul5 complex (PDB ID: 4N9F) produces a Vif–Vif interface between adjacent pentamers in the asymmetric unit of the crystal. A single representative interface between two Vif molecules is depicted, which involves an interaction between the conserved 40YRHHY44 motif (hot pink and dark gray). Side chains of selected interface residues are shown in stick form. The PPLP motif is shown in stick form (magenta) for perspective. Abbreviations: A3F, APOBEC3F; A3G, APOBEC3G; A3H, APOBEC3H; APOBEC3, apolipoprotein B editing catalytic subunit-like 3; CBFβ, core-binding factor β subunit; Cul, Cullin; Elo, Elongin; Vif, viral infectivity factor.
Some Vif residues implicated in binding of A3F (and for A3G) are also partially buried at the interface with CBFβ, suggesting an overlap between the binding interfaces of Vif for A3F and CBFβ. As such, the Xiong group [5] showed that binding of A3F by a Vif/CBFβ/EloB/EloC complex displaces CBFβ, reinforcing the notion that binding of CBFβ and A3F may be mutually exclusive. If true, one is left to wonder if involvement of CBFβ in the E3 ligase is perhaps transient. Conversely, the initial work from the Yu group demonstrated that CBFβ binding to Vif did not prevent an interaction with A3G, suggesting the two interactions are not mutually exclusive, which supports an E3 ligase model with CBFβ as a permanent subunit [6].
The crystal structure of Vif has provided a physical means to interpret the myriad data regarding the importance of specific Vif residues for directly binding substrate A3s. As such, the structural model of Vif provides a road-map for implementing structure assisted drug design. Similarly, structural models of the A3 family lend insight into how Vif ‘sees’ each protein it targets for proteasomal degradation.
Vif-targeted interfaces on APOBEC3 family members
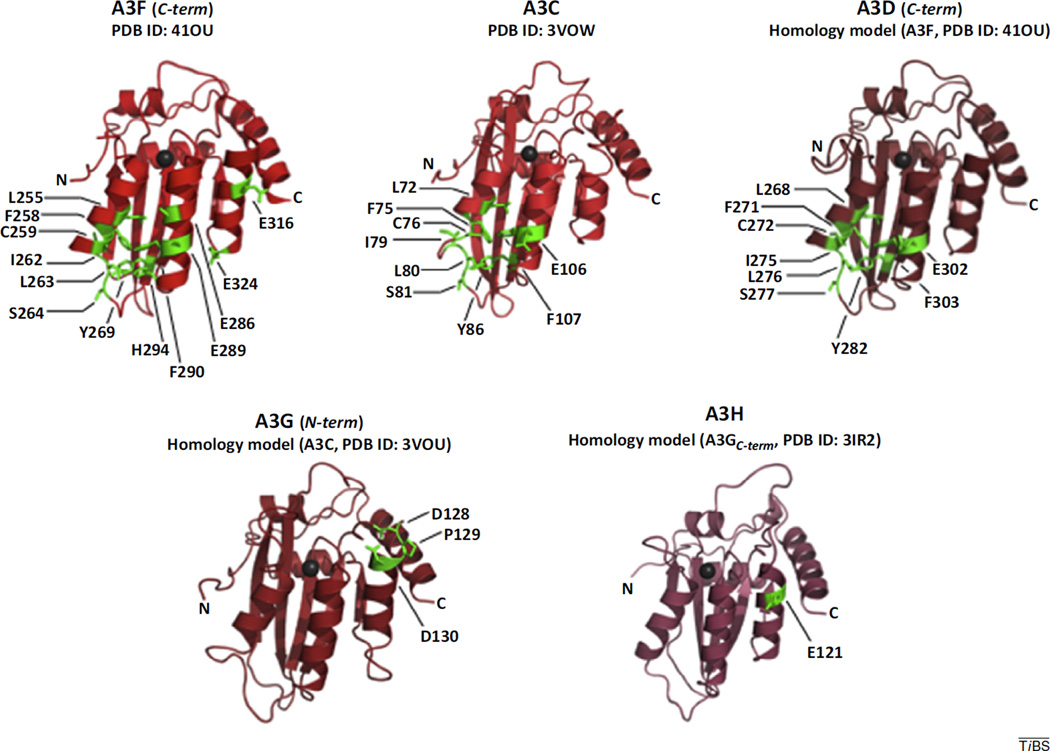
It has been suggested that Vif directly interacts with substrate APOBEC3 proteins to recruit them to the E3 ligase [7]. Of the seven A3 proteins expressed in humans, A3D, A3F, A3G, and A3H have been shown to possess true antiretroviral activity against HIV-1 [3]. Thus, it is not surprising that HIV-1 Vif has evolved to target each of these proteins for degradation. Indeed, specific residues from A3C, D, F, G, and H (haplotype II) have been shown to mediate binding with Vif (Table 1). Mapping these residues onto the available crystal structures of the Vif-binding domains for A3C [8] and A3F [9] and onto homology models of the Vif-binding domains of A3D, G and H, reveals conserved surface regions that are likely involved in direct interaction with Vif (Figure 3). As recently reviewed by Aydin et al. [4], the surface residues involved in Vif-binding for A3C, D, and F form a contiguous area, comprising both a hydrophobic and a negatively charged region. These surface properties are complementary to the hydrophobicity and net positive electrostatics associated with Vif. We suggest that although A3C can be bound and targeted for degradation by Vif, this recognition is likely driven by the sequence similarity with A3F alone. Given that A3C, unlike A3D, F, G, and H, is not packaged into virions [3], Vif targeting of A3C may or may not be required as an antagonist of antiretroviral activity.
Figure 3.
A3 protein residues implicated in Vif binding. The crystal structures of Vif-binding cytidine deaminase domains of (A) A3F (C-terminal domain) [9], and (B) A3C [8] are depicted as cartoon diagrams. Homology models of (C) A3D (C-terminal domain), (D) A3G (N-terminal domain), and (E) A3H haplotype II were generated with SWISS Model [48] from template structures A3F (C-terminal half) (PDB 4IOU), A3C (PDB ID 3VOW), and A3G (C-terminal half) (PDB ID 3IR2), respectively. Residues from each A3 protein that have been shown to modulate binding to Vif (Table 1) are shown in green with side chains depicted in stick representation. Zinc ions are depicted as charcoal spheres. Abbreviations: A3C, APOBEC3C; A3D, APOBEC3D; A3F, APOBEC3F; A3G, APOBEC3G; A3H, APOBEC3H; APOBEC3, apolipoprotein B editing catalytic subunit-like 3; CBFβ, core-binding factor β subunit; Vif, viral infectivity factor.
The surface charge properties of A3G and A3H differ widely from those of A3C, D, and F [4]. However, Vif-binding domains on the surface of A3G and A3H have not been well characterized and are likely incomplete (Table 1). Likewise, the conserved hydrophobic regions found on A3C, D, and F are not found on A3G and A3H [4]. We propose that there are likely three different types of interfaces that Vif forms to recruit APOBEC3 family members to the E3 ubiquitin ligase complex. This is realized by the identification of multiple APOBEC3-binding interfaces on the solvent-exposed surface of the Vif crystal structure. Considering the interactions at this surface, antiretroviral therapeutics that target Vif at this interface may have the potential to enable the innate antiretroviral activity of four different A3 effectors of viral mutation.
These are early days in the analysis, but the crystal structure of Vif and the Vif-binding C-terminal half of A3F (and the homology model of the A3G N-terminal half) enables modeling of the protein–protein interactions. Web-based rigid-body docking programs such as ClusPro (v 2.0) [10,11] can be used to construct models of Vif bound to A3F C-terminal half (PDB ID 4IOU) [9] and the A3G N-terminal homology model (as described in Figure 3) in the context of the E3 ligase complex structure. Top scoring docked models are shown in Figure 1B. Notably, the favorable locations of A3F and A3G docking to Vif superposes with residues that are predicted to bind A3F and A3G, respectively (Figure 2A). Although a precise determination of how Vif interacts with different members of the A3 family awaits structural studies on purified Vif-A3 complexes, structural models of individual Vif-binding A3 domains sheds light on the subtle differences and similarities among surfaces residues within the A3 family.
The PPLP motif and oligomerization of Vif
Oligomerization of Vif was first described in 2001 [12] through the use of phage display technology. Disruption of Vif oligomers with proline-rich peptidomimetics implicated the involvement of the short, yet completely conserved, 161PPLP164 sequence of Vif [13,14] in multimerization. This PPLP motif is not conserved in the less viremic HIV-2, but is conserved in some simian immunodeficiency virus (SIV) Vif proteins, including SIV that infect some chimpanzees, such as Pan troglodytes [15]. Likewise, SIV infecting gorillas (SIVgor) has a conserved PPFP sequence in Vif. The conservation of these short proline-rich motifs in Vif corroborates their importance to lentiviral infections. Importantly, the disruption of Vif oligomers was shown to increase the amount of packaged A3G in budding virions, which was associated with a marked reduction in HIV-1 infectivity [14]. The contribution of the PPLP motif to Vif oligomerization is supported by an abundance of biophysical analyses, including size-exclusion chromatography [16], dynamic light scattering [16], analytical ultracentrifugation [17], small-angle X-ray scattering [18], and chemical crosslinking mass spectrometry [19]. Furthermore, PPLP-dependent oligomerization of Vif was also shown in the context of living cells by fluorescence lifetime imaging microscopy (FLIM) [20]. In addition to mediating oligomerization, the PPLP motif is required for proper formation of the E3 ubiquitin ligase complex [21,22] and is involved in A3G binding and degradation [23]; however, as with its contribution to oligomerization, the mechanistic underpinnings of the role of the PPLP motif in these functions remain unclear. Furthermore, the aforementioned results describing PPLP-mediated oligomerization of Vif were obtained without the presence of CBFβ.
The new crystal structure reveals the PPLP motif (sequentially near the C terminus) is packed against αH1 of the N-terminal α/β A3-binding domain of Vif. Its location does not support the hypothesis that Vif oligomerizes through direct interaction between PPLP motifs of adjacent Vif moieties. In fact, mutation of PPLP-to-AALA reduced, but did not obliterate oligomerization [16,17]. Vif oligomerization mediated through regions other than the PPLP motif have been reported [16], including the zinc-binding domain [24], the N-terminal half, and the intrinsically disordered C-terminal end of Vif [19]. Mass spectrometry of chemically crosslinked Vif revealed extensive interactions among N- and C-terminal halves of isolated dimers and trimers of Vif [19]. As expected for a solvent-exposed surface, hydrogen–deuterium exchange mass spectrometry showed that the region encompassing the PPLP motif indeed underwent proton–deuterium exchange [25]. We therefore support the hypothesis that the role of the PPLP motif in oligomerization is to act as a conformational hinge to orient other regions of Vif for oligomerization [25].
However, a Vif–Vif interface does partly mediate interactions between individual pentameric complexes of the asymmetric unit of the Vif–CBFβ–EloBC–Cul5 crystal structure. This Vif–Vif interaction buries 352 Å2 of surface area. It is primarily formed by residues 39–47 and intermolecular salt bridge pairs HIS42–Glu45 and Glu45–Arg41 that are present six times in the asymmetric unit of the crystal structure (Figure 2B). This amount of buried surface area is smaller than that which typically governs protein–protein interactions. Most protein–protein inter-actions bury a larger amount of surface area (with an average range from 1200 to 2000 Å2), primarily formed through shape and electrostatic complementarity, and burial of hydrophobic regions [26]. This is exemplified well by the interaction between Vif and CBFβ, which buries a large amount (~4800 Å2) of surface area [2]. Still, it is intriguing to contemplate whether this small Vif–Vif crystallographic interface could represent part or all of a functional Vif–Vif oligomeric interaction, as described in the experimental data mentioned earlier. Notably, the residues at this interface include the A3G-binding motif, 40YRHHY44 (Table 1). It has been shown that binding of A3G by Vif decreased the oligomeric state of Vif [20]. Thus, A3G binding to Vif involving the 40YRHHY44 motif may be competitive with Vif oligomeric interactions. Specifically, we propose that this conserved Vif motif (40YRHHY44) participates in two mutually exclusive functionalities of Vif: oligomerization and A3G recognition. We suggest undertaking a mutational analysis of the residues at this crystal contact to determine the specific effect of each residue on Vif multimerization.
Although the aforementioned PPLP motif is implicated in oligomerization, the direct interactions involved may occur elsewhere on the Vif surface, as supported by the new crystal structure. As such, the inhibition of both Vif-dependent A3G degradation and HIV infectivity by PLPP peptidometics may be allosteric. Taken together, these data suggest a potentially intricate and as yet, not fully understood, role for the PPLP motif in Vif oligomerization. Nonetheless, given the importance of the PPLP motif to Vif function, A3G degradation, and the HIV-1 life cycle, and the fact that it is solvent exposed, we suggest that this motif represents a high priority target for development of antiretroviral therapeutics.
Vif protein–protein interactions are sites for therapeutic targeting
We propose that the crystal structure of Vif in the context of its E3 ligase complex may provide an excellent template for in silico screening methods [27]. Vif likely forms at least seven potentially druggable interfaces including that with CBFβ, EloC, Cul5, itself (dimerization domain), and the APOBECs (A3D, F, G, and H) (Box 1 Figure I). Although disrupting any of these protein–protein interactions involving Vif or any of the other interactions in the context of the E3 ubiquitin ligase would theoretically abrogate A3 degradation, the most prudent approach is to target Vif directly to avoid unwanted off-target interactions that may lead to cytotoxicity.
Specifically, disrupting interaction of Vif with CBFβ could unleash the natural host antiretroviral activity of each of the A3 proteins simultaneously, while mitigating the potential for nonspecific target effects on other cellular SOCS-box type E3 ubiquitin ligase complexes that function as a part of normal cell metabolism. CBFβ is thought to be required for degradation of each targeted A3 protein by Vif. In addition, CBFβ is not believed to be a component of cellular E3 ubiquitin ligase complexes, making its interaction with Vif unique. The interaction between Vif and CBFβ is large (~4800 Å2 of surface area is buried at the interface). Disruption of this interaction by a drug-like molecule that is smaller than the suggested upper size limit (500 Da) [28] may seem insurmountable. However, recent evidence suggests that much of the free energy of binding is confined to smaller sections of interacting protein surfaces, deemed ‘hotspots’, thus negating the notion that such a large PPI is undruggable [29].
It is becoming clear that Vif may interact with A3 proteins (D, F, G, and H) through unique and different interfaces (Figure 3). Developing a single small molecule drug to disrupt all Vif–A3 interactions seems unlikely. Thus, the Vif interaction with each A3 protein may have to be targeted separately. We suggest targeting interaction of Vif with A3G as a top priority because A3G has the most potent antiretroviral activity. Therapeutic inhibition of Vif-mediated degradation of A3G may facilitate A3G in-corporation into budding virions and subsequent A3G-mediated lethal mutagenesis of the HIV-1 single-stranded DNA during reverse transcription.
Likewise, targeting disruption of Vif oligomerization could potentiate the antiretroviral activity of A3s. Although the mechanism of action for Vif oligomerization is unknown, reduction of Vif oligomerization with peptidomimetics increased A3G incorporation into nascent virions [14]. We suggest first testing the residues at the Vif–Vif crystallographic interface (Figure 2B) for an effect on Vif oligomerization, followed by the in silico screening of small molecules that could target the interface. Conversely, targeting interaction of Vif with either Cul5 or EloC directly, while still preventing A3 degradation, would have a higher risk for off-target interactions and perhaps cytotoxic effects. The BC-box backbone from HIV-1 Vif and cellular SOCS-box proteins are nearly isomorphous, suggesting that therapeutic intervention targeting disruption of the Vif–EloC interface could also disrupt the interaction between cellular SOCS-box proteins and EloC. Thus, the Vif–EloC interface is a more tenuous and lower-priority target for small molecule disruption. In conclusion, we consider targeting interactions of Vif with CBFβ, A3G, and itself (oligomerization) top priorities for developing a new class of antiretroviral therapeutics.
Concluding remarks
Vif was described as a novel open reading frame ‘A’ (Sor) in 1985 and soon thereafter was appreciated for its critical role in HIV replication. It was not until 2002 that Michael Malim’s research group made clear that the mechanistic basis for this requirement was the degradation of APOBEC host defense factors [30]. The availability of a crystal structure of Vif in complex with components of the ubiquitylation machinery will usher in a new era of structure-based investigation and rational design for drug discovery. The past 30 years of exploration and discovery have laid a foundation of understanding from which we appreciate that HIV-1 defense against A3 restriction is paramount for viral replication. Unlike currently drug-targeted HIV proteins (protease, reverse transcriptase, and integrase), Vif itself is not an enzyme, functioning instead through protein–protein interactions. Vif must bind to A3 proteins as well as to CBFβ, EloC and Cul5 through defined protein interaction domains. The amino acid composition of host cell protein domains required for Vif interaction are slow to change in human/primate evolution and therefore may constrain the evolution of Vif.
An important future goal is to understand the mutability of Vif within domains that affect interactions, functionality, and viral competence. If, as expected, the mutational variability of Vif is limited within the binding domains for host cell proteins, then therapeutic intervention at these sites may be less likely to select for the emergence of drug-resistant strains. Hotspots, or subsets of residues at a protein–protein interface, contribute most of the free energy of binding, effectively reducing the surface area that would have to be disrupted to reduce or eliminate interactions [31]. Furthermore, surface residues that form appropriate protein–protein interfaces have more conformational flexibility than once thought, undergoing small perturbations that open up transient pockets available for small-molecule binding. These characteristics suggest that protein–protein interactions are amenable to disruption with small molecules identified through in vitro high-throughput screening, and indeed, several recent successes have been described [27]. Therefore, in our opinion, development of drugs that bind Vif and thereby antagonize Vif oligomerization and host protein interactions may just be the Achilles heel so long sought for eradication of HIV. In conclusion, the crystal structure of Vif in complex with its binding partners of the E3 ubiquitin ligase complex has left the field with several important questions to be addressed (Box 3).
Outstanding questions.
How have the amino acids involved in Vif protein–protein interactions evolved across species, clades, and drug-resistant strains?
How is the structure of Vif, which is stabilized by CBFβ in the pentameric crystal structure, affected by interactions with APO-BEC3 proteins?
Are Vif pentameric complexes the first stable or functional form following Vif translation or are there intermediates or different structures of Vif found in Vif:Vif, Vif:APOBEC3, CBFβ:Vif, or CBFβ:Vif:APOBEC3 complexes?
Could the relatively small crystal packing interface between Vif molecules represent a biologically relevant Vif–Vif oligomerization interface?
Given that the PPLP domain is not at the Vif–Vif crystal packing interface, why do substitutions in this amino acid sequence or peptide mimics of PPLP impair Vif multimerization and function?
Can the protein–protein interfaces that determine Vif function be targeted for disruption by small molecules?
Acknowledgments
The authors thank Drs. Ryan Bennett, Kimberly Prohaska and Andrew Torelli for helpful suggestions in preparing this manuscript. The modeling and preparation of the manuscript were supported in part by a Public Health Services Grant NIH/NIGM R01GM110568.
Dr. Harold C. Smith is a full-time tenured professor at the University of Rochester School of Medicine and Dentistry. He is also the founder and CEO of the University of Rochester spinout company OyaGen, Inc. Consistent with the policy of the University, Dr. Smith spends 20% of his time and effort leading the drug discovery program at OyaGen. Dr. Jason D. Salter is a scientist employed full-time by OyaGen. Dr. Guillermo A. Morales is the Director of Scientific Services at Cogent Professionals, an independent contractor working with OyaGen on in silico drug design.
Footnotes
Conflict of interest statement
The company has a financial interest in the development of Vif antagonists.
References
- 1.Strebel K, et al. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y, et al. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature. 2014;505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 3.Hultquist JF, et al. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J. Virol. 2011;85:11220–11234. doi: 10.1128/JVI.05238-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aydin H, et al. Structure-guided analysis of the human APOBEC3-HIV restrictome. Structure. 2014;22:668–684. doi: 10.1016/j.str.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Fribourgh JL, et al. Core binding factor beta plays a critical role by facilitating the assembly of the Vif-cullin 5 E3 ubiquitin ligase. J. Virol. 2014;88:3309–3319. doi: 10.1128/JVI.03824-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang W, et al. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature. 2012;481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 7.Schrofelbauer B, et al. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl. Acad. Sci. U.S. A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitamura S, et al. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat. Struct. Mol. Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 9.Bohn MF, et al. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comeau SR, et al. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. (Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozakov D, et al. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 12.Yang S, et al. The multimerization of human immunodeficiency virus type I Vif protein: a requirement for Vif function in the viral life cycle. J. Biol. Chem. 2001;276:4889–4893. doi: 10.1074/jbc.M004895200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang B, et al. Potent suppression of viral infectivity by the peptides that inhibit multimerization of human immunodeficiency virus type 1 (HIV-1) Vif proteins. J. Biol. Chem. 2003;278:6596–6602. doi: 10.1074/jbc.M210164200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JH, et al. The dimerization domain of HIV-1 viral infectivity factor Vif is required to block virion incorporation of APOBEC3G. Retrovirology. 2007;4:81. doi: 10.1186/1742-4690-4-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barraud P, et al. Advances in the structural understanding of Vif proteins. HIV Res. 2008;6:91–99. doi: 10.2174/157016208783885056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernacchi S, et al. Importance of the proline-rich multimerization domain on the oligomerization and nucleic acid binding properties of HIV-1 Vif. Nucleic Acids Res. 2011;39:2404–2415. doi: 10.1093/nar/gkq979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Techtmann SM, et al. Hydrodynamic and functional analysis of HIV-1 Vif oligomerization. Biochemistry. 2012;51:2078–2086. doi: 10.1021/bi201738a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallerano D, et al. Biophysical characterization of recombinant HIV-1 subtype C virus infectivity factor. Amino Acids. 2011;40:981–989. doi: 10.1007/s00726-010-0725-x. [DOI] [PubMed] [Google Scholar]
- 19.Auclair JR, et al. Mass spectrometry analysis of HIV-1 Vif reveals an increase in ordered structure upon oligomerization in regions necessary for viral infectivity. Proteins. 2007;69:270–284. doi: 10.1002/prot.21471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Batisse J, et al. APOBEC3G impairs the multimerization of the HIV-1 Vif protein in living cells. J. Virol. 2013;87:6492–6506. doi: 10.1128/JVI.03494-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bergeron JR, et al. The SOCS-box of HIV-1 Vif interacts with Elongin BC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog. 2010;6:e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfe LS, et al. Dissection of HIV Vif interaction with the human E3 ubiquitin ligase. J. Virol. 2010;84:7135–7139. doi: 10.1128/JVI.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donahue JP, et al. The HIV-1 Vif PPLP motif is necessary for human APOBEC3G binding and degradation. Virology. 2008;377:49–53. doi: 10.1016/j.virol.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giri K, et al. Molecular structure and biochemical properties of the HCCH-Zn2+ site in HIV-1 Vif. Biochemistry. 2009;48:7969–7978. doi: 10.1021/bi900677w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marcsisin SR, et al. On the solution conformation and dynamics of the HIV-1 viral infectivity factor. J. Mol. Biol. 2011;410:1008–1022. doi: 10.1016/j.jmb.2011.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreira IS, et al. Hot spots – a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 27.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 28.Lipinski CA, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 29.Winter A, et al. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: applications in structure-guided drug discovery. Q. Rev. Biophys. 2012;45:383–426. doi: 10.1017/S0033583512000108. [DOI] [PubMed] [Google Scholar]
- 30.Sheehy AM, et al. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 31.Keskin O, et al. Hot regions in protein – protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 32.Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- 33.Conticello SG, et al. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 34.LaRue RS, et al. Guidelines for naming nonprimate APOBEC3 genes and proteins. J. Virol. 2009;83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell RA, Pathak VK. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Z, et al. Characterization of conserved motifs in HIV-1 Vif required for APOBEC3G and APOBEC3F interaction. J. Mol. Biol. 2008;381:1000–1011. doi: 10.1016/j.jmb.2008.06.061. [DOI] [PubMed] [Google Scholar]
- 37.Dang Y, et al. Identification of 81LGxGxxIxW89 and 171EDRW174 domains from human immunodeficiency virus type 1 virus Vif that regulate APOBEC3G and APOBEC3F neutralizing activity. J. Virol. 2010;84:5741–5750. doi: 10.1128/JVI.00079-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita T, et al. Identification of amino acid residues in HIV-1 Vif critical for binding and exclusion of APOBEC3G/F. Microbes Infect. 2008;10:1142–1149. doi: 10.1016/j.micinf.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 39.Dang Y, et al. Identification of a novel WxSLVK motif in the N terminus of human immunodeficiency virus and simian immunodeficiency virus Vif that is critical for APOBEC3G and APOBEC3F neutralization. J. Virol. 2009;83:8544–8552. doi: 10.1128/JVI.00651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siu KK, et al. Structural determinants of HIV-1 Vif susceptibility and DNA binding in APOBEC3F. Nat. Commun. 2013;4:2593. doi: 10.1038/ncomms3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pery E, et al. Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif. J. Virol. 2009;83:2374–2381. doi: 10.1128/JVI.01898-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albin JS, et al. A single amino acid in human APOBEC3F alters susceptibility to HIV-1 Vif. J. Biol. Chem. 2010;285:40785–40792. doi: 10.1074/jbc.M110.173161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith JL, Pathak VK. Identification of specific determinants of human APOBEC3F, APOBEC3C, and APOBEC3DE and African green monkey APOBEC3F that interact with HIV-1 Vif. J. Virol. 2010;84:12599–12608. doi: 10.1128/JVI.01437-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dang Y, et al. Identification of a critical T(Q/D/E)×5AD×2(I/L) motif from primate lentivirus Vif proteins that regulate APOBEC3G and APOBEC3F neutralizing activity. J. Virol. 2010;84:8561–8570. doi: 10.1128/JVI.00960-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Binka M, et al. The activity spectrum of Vif from multiple HIV-1 subtypes against APOBEC3G, APOBEC3F, and APOBEC3H. J. Virol. 2012;86:49–59. doi: 10.1128/JVI.06082-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhen A, et al. A single amino acid difference in human APOBEC3H variants determines HIV-1 Vif sensitivity. J. Virol. 2010;84:1902–1911. doi: 10.1128/JVI.01509-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnold K, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]