Abstract
The CEBPA gene is mutated in 9% of patients with acute myeloid leukemia (AML). Selective expression of a short 30 kDa C/EBPα translational isoform, termed p30, represents the most common type of CEBPA mutations in AML. The molecular mechanisms underlying p30-mediated transformation remain incompletely understood. We show that C/EBPα p30, but not the normal p42 isoform, preferentially interacts with Wdr5, a key component of SET/MLL histone-methyltransferase complexes. Accordingly, p30-bound genomic regions were enriched for MLL-dependent H3K4me3 marks. The p30-dependent increase in self-renewal and inhibition of myeloid differentiation required Wdr5, as its down-regulation inhibited proliferation and restored differentiation in p30-dependent AML models. OICR-9429 is a novel small-molecule antagonist of the Wdr5-MLL interaction. This compound selectively inhibited proliferation and induced differentiation in p30-expressing human AML cells. Our data reveal the mechanism of p30-dependent transformation and establish the essential p30-cofactor Wdr5 as a therapeutic target in CEBPA-mutant AML.
Introduction
Leukemia is characterized by de-regulation of hematopoietic progenitor cell function, leading to ectopic self-renewal and disruption of normal differentiation properties. As lineage-specific transcription factors are key regulators of hematopoietic homeostasis, their dysregulated activity is frequently associated with leukemia.
The basic region-leucine zipper transcription factor C/EBPα (CCAAT-enhancer binding protein-alpha) is a master regulator of myeloid gene expression programs in the hematopoietic system, linking terminal differentiation to growth arrest1. C/EBPα deficiency leads to a complete block of terminal myeloid differentiation at the pre-Granulocyte-Monocyte (preGM) cell stage2.
CEBPA mutations are present in 9% of patients presenting with acute myeloid leukemia (AML)3. The most prevalent type of mutation involves frameshifts in the N-terminal part of the C/EBPα coding sequence4. These mutations ablate expression of the full-length p42-isoform but still allow a shorter protein (termed p30) to be expressed from an AUG codon downstream of the mutated region. Under physiological circumstances, nutrient and growth factor availability decreases the p42/p30 ratio by increasing bypass of the upstream initiation codon, promoting the maintenance of an undifferentiated cellular state5. Mice engineered to express only the p30 variant of C/EBPα (Cebpap30/p30 genotype) develop AML with complete penetrance6. Cebpap30/p30 hematopoietic progenitors hyper-proliferate in vitro6, and over-expression of p30 in hematopoietic progenitor cell lines blocks myeloid differentiation5.
Various hypotheses have been put forward to explain the molecular basis for the transforming function of C/EBPα p30. N-terminal deletions strongly reduce transcriptional activation by C/EBPα7,8. Yet, certain properties that are exerted by the p42 isoform are still maintained by the p30 variant. These include the trans-activating potential and the recruitment of the SWI/SNF complex to regulate the expression of downstream target genes8,9. In addition, C/EBPα p42 can inhibit the activity of E2F proteins through direct, physical interaction10. This is critically required for C/EBPα-dependent induction of terminal myeloid differentiation11, with c-Myc as a critical E2F-regulated C/EBPα target12,13. The ability of C/EBPα to repress E2F target genes gets lost upon deletion of its N-terminus11,12, indicating that de-repression of E2F target genes is key to leukemogenesis upon p42 loss. Additionally, C/EBPα p42 and p30 isoforms might have variable affinities for consensus C/EBP binding sites in the genome14.
Finally, while a plethora of interaction partners has been described for C/EBPα15-19, it is not clear to which extent these co-factors differ between p42 and p30 isoforms and if any differences in the protein interactomes contribute to the differential effects of C/EBPα p42 versus p30.
We hypothesized that differences in the biochemical properties of C/EBPα isoforms could be reflected in a differential ability to engage the molecular machinery of the cell. The protein complexes associated with p42 or p30 should be able to account for differences in their biological activities, in particular with respect to altered regulation of self-renewal, proliferation and differentiation. Thus, the identification of any factor conferring differential and druggable vulnerability to p30-expressing cells would represent an attractive target for therapy. Here we identified Wdr5 as such a target, and demonstrate that a novel, first-in-class small molecule antagonist of the Wdr5-MLL interaction can selectively target AML cells with N-terminal CEBPA mutations.
Results
p30 interacts with Wdr5 and co-localizes with H3K4me3
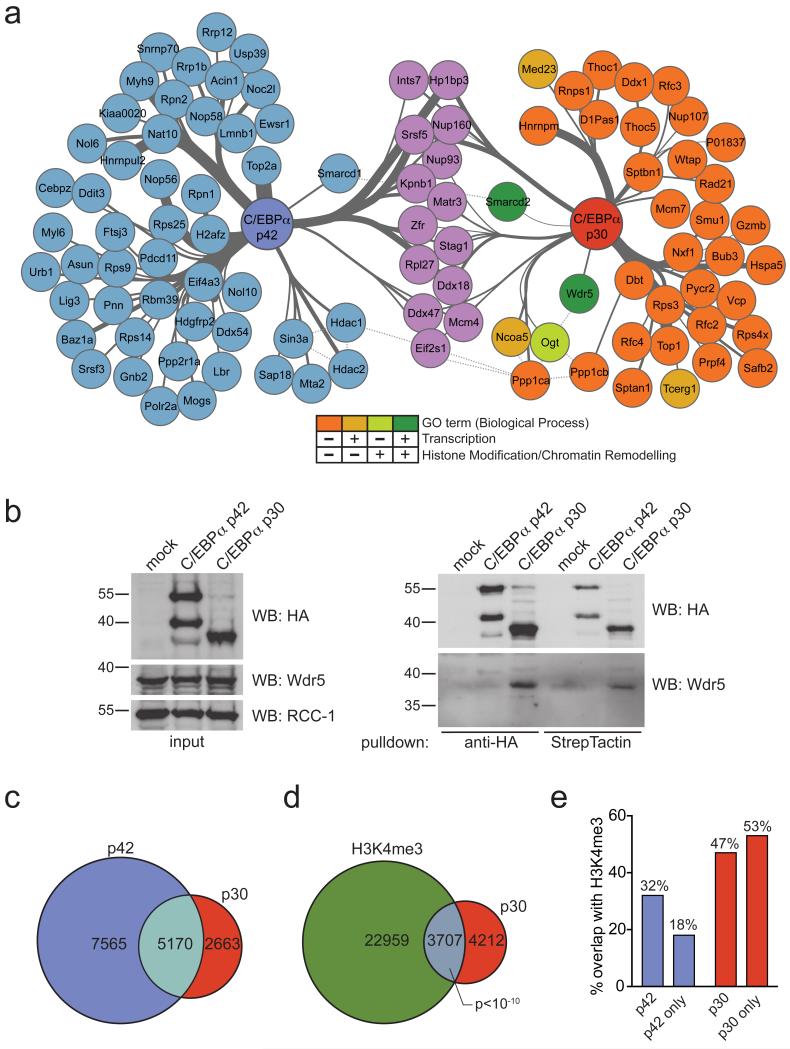
We expressed affinity-tagged variants of C/EBPα p42 and p30 in the murine myeloid progenitor cell line FDCP-1 (Supplementary Results, Supplementary Fig. 1a). While p42 over-expression in this system induced the down-regulation of the progenitor surface marker c-Kit as well as the acquisition of morphological signs of myeloid maturation, these features were blocked upon p30 over-expression (Supplementary Fig. 1b, c). Gene expression profiling revealed that p30 had profound effects on global gene expression patterns, including the up-regulation of genes important for cell growth and division and the induction of E2F target genes (Supplementary Fig. 1d, e). This confirms that our isogenic cell system could recapitulate principal aspects of C/EBPα-mediated terminal myeloid differentiation as well as p30-associated changes in gene expression. We affinity-purified protein complexes containing p42 and p30 from nuclear extracts of these cell lines and characterized them by liquid chromatography tandem mass spectrometry (LC-MSMS). Using stringent statistical evaluation, we found 64 proteins that reproducibly associated with p42, while 52 proteins co-purified with p30 (Fig. 1a, Supplementary Table 1, 2). We recapitulated several previously published interactions of C/EBPα p42 (e.g. C/EBPζ, Ddit3 and Hdac1/Hdac2) under the conditions used, supporting the relevance of our interactome data. Only 14 proteins interacted with both p42 and p30 (Fig. 1a), suggesting that p30-selective protein-protein interactions could be instrumental to leukemogenesis. Further, we hypothesized that any p30-specific characteristics would mostly manifest on the level of transcription and/or chromatin regulation. Among all p30-specific interactors, three proteins were annotated with the GO-term “transcription” and one protein with “histone modification/chromatin remodeling”. Two proteins were positive for both selected GO terms (Fig. 1a). Of those, Smarcd2 - a component of the SWI/SNF complex - has been previously shown to be associated with and regulate the activity of C/EBPα9. The other p30-specific interactor was Wdr5, a component of SET/MLL histone methyltransferase (HMT) complexes. MLL belongs to the Trithorax (trxG) family of proteins, which positively regulate gene expression through the catalysis of histone 3 lysine 4 (H3K4) methylation. The H3K4 methyltransferase activity of MLL critically depends on the presence of Wdr520.
Figure 1. C/EBPα p30 preferably interacts with Wdr5 and the SET/MLL HMT complex, leading to increased co-localization of p30 and H3K4me3 on chromatin.
(a) Network representation of proteins reproducibly identified in AP-MS experiments of tagged C/EBPα variants from FDCP-1 after background normalization. Proteins identified in the purifications of both C/EBPα p42 and p30 variants are shown in purple. p42-specific interactors are shown in blue, proteins specifically bound to p30 are color-coded according the scheme below the network. Thickness of edges represents strength of the interaction (based on spectral counts). (b) Left panel: nuclear extracts from 32D cells expressing tagged variants of p42 or p30 were analyzed by Western blot for expression of HA and Wdr5. RCC-1 was used as loading control. Right panel: Western blot analysis of HA and Wdr5 from anti-HA- or StrepTactin purifications of tagged C/EBPα variants from 32D cell lines stably expressing p42 and p30. Representative images of at least 2 replicate experiments are shown. (c) Venn diagram showing the overlap of ChIP-seq peaks between C/EBPα p42 and p30 (d) Venn diagram showing the overlap of ChIP-seq peaks between C/EBPα p30 and H3K4me3 in p30-epxressing cells (hypergeometric t-test, duplicate experiments) (e) Bar diagram showing percentages of overlap between p42/p42-only and p30/p30-only peaks and H3K4me3 ChIP-seq data in the respective cellular background.
As Wdr5 is part of large, multi-subunit protein complexes, we next tested whether p30 also displays enhanced association with reported Wdr5/MLL interaction partners. Out of 29 proteins previously reported to participate in Wdr5/MLL containing complexes, 23 proteins (79%) were present at greater or equal levels in purifications of p30, while only 6 proteins (21%) showed enrichment in the analyses of p42 (Supplementary Fig. 2a). This indicates that, globally, p30 has a higher preference to incorporate into Wdr5-containing protein complexes than p42, despite equal expression levels of Wdr5 in p42- vs. p30-expressing cells (Fig. 1b and Supplementary Fig. 2b). Importantly, the selective interaction of p30 with Wdr5 could be confirmed by co-immunoprecipitation experiments in FDCP-1 cells (Supplementary Fig. 2b) and was conserved in mouse 32D cells, a different isogenic cell line model engineered to express affinity tagged p42 and p30, respectively (Fig. 1b). While the specificity of the Wdr5-p30 interaction was not retained upon over-expression in HEK293 cells, we observed a strong interaction between endogenous Wdr5 and C/EBPα p30 proteins in a murine Cebpap30/p30 myeloid progenitor cell line (Supplementary Fig. 2c, d).
As the association of p30 with Wdr5 and components of the SET/MLL HMT complex suggested a specific role in the regulation of gene expression, we investigated the genome-wide distribution of p42 and p30. Both C/EBPα isoforms showed very similar distributions across genomic loci and displayed significantly enriched chromatin binding around annotated transcription start sites (TSS), consistent with functional roles in transcriptional regulation (Supplementary Fig. 2e, 3a, b). Furthermore, 66% (5,170) of all sites occupied by p30 also overlapped with p42 binding, while p30 was exclusively bound to 2,663 sites (Fig. 1c and Supplementary Fig. 3c).
As Wdr5 is required for efficient H3K4 methylation by SET/MLL20 and the H3K4me3 mark is strongly associated with actively transcribed gene promoters, we performed H3K4me3 ChIP-seq experiments in p42- or p30-expressing cells. Despite the smaller overall number of binding sites, p30-ChIP-seq peaks showed greater overlap with H3K4me3-marked genomic regions than p42-bound sites (47% vs. 32%, p<10−10, Fig. 1d, e). Only 18% of regions with only p42-, but no p30 binding were H3K4me3-positive, while 53% of regions exclusively associated with p30 were marked with H3K4me3 (Fig. 1d). This strong positive correlation of p30 binding and H3K4me3 marks on chromatin is in line with increased interaction of p30 with Wdr5 and the SET/MLL HMT complex.
Assignment of ChIP-seq peaks with overlapping p30-H3K4me3 occupancy to gene targets based on their proximity to the nearest annotated TSS yielded 2,606 actively transcribed p30-target genes. 268 of these genes showed significant p30-dependent changes in mRNA expression levels (FDR 0.01, p=3.3×10−7, Supplementary Fig. 4a). p30-bound and up-regulated genes were enriched for functions in cell cycle control and mitosis, while p30-dependent down-regulated genes were involved in the regulation of ER-stress and transcriptional suppression (Supplementary Fig. 4b).
Together, these data indicate that p30 preferably associates with Wdr5 and SET/MLL HMT complex components, leading to increased p30-H3K4me3 co-localization on chromatin.
The p30-mediated differentiation block is Wdr5-dependent
Next we tested the functional consequences of inducible Wdr5 loss on p30-dependent cellular features. In the system used by us, shRNA expression is coupled to the induction of a fluorescent reporter after doxycycline (Dox)-administration (Fig 2a, Supplementary Fig. 5a). Two independent Wdr5-targeting shRNAs efficiently silenced Wdr5 mRNA and protein expression in different cell systems (Supplementary Fig. 5b-e). Given the prominent p30-H3K4me3 co-localization on chromatin and the described role of Wdr5 in the SET/MLL HMT complex, we hypothesized that Wdr5 down-regulation might have a specific effect on H3K4me3 marks at promoters of p30-target genes. Indeed, H3K4me3 levels on promoters of the p30-target genes Cdk6, Etv6 and Kit were higher in p30- than in p42-expressing FDCP-1 cells (Supplementary Fig. 5f). Furthermore, we observed a significant reduction of H3K4me3 marks on these promoters upon shRNA-mediated depletion of Wdr5 in p30-expressing cells, while they remained unchanged in p42-expressing cells (Supplementary Fig. 5f).
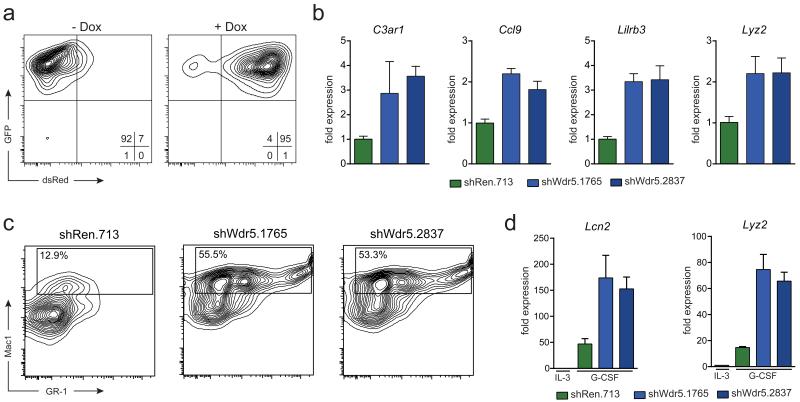
Figure 2. Loss of Wdr5 restores granulocytic differentiation potential in C/EBPα p30-expressing cells.
(a) Flow cytometric analysis of dsRed-reporter induction in 32D rtTA3 p30 cells upon Dox administration. (b) qRT-PCR analysis of the indicated genes in 32D rtTA3 p30 cells expressing indicated shRNA constructs after 48 h of Dox treatment. (c) Flow cytometry analysis for Mac-1 and GR-1 surface markers of 32D rtTA3 p30 cells expressing indicated shRNA constructs 96 h after Dox treatment and 48 h after exposure to G-CSF (10 ng/mL). Presented events are gated on the GFP+ dsRed+ population. (d) qRT-PCR analysis of Lcn2 and Lyz2 expression in 32D rtTA3 p30 cells transduced with indicated shRNA constructs after 96 h of Dox treatment followed by 48 h exposure to G-CSF. Data are presented as mean +/− standard deviation (SD) of triplicate experiments.
Gene expression profiling in p30-expressing FDCP-1 cells revealed that knockdown of Wdr5 led to increased expression of genes associated with terminal myeloid differentiation (Supplementary Fig. 5g). Importantly, the same up-regulation of myeloid genes was observed in p30-expressing 32D cells after Wdr5 knockdown (Fig. 2b), pointing to a conserved role of Wdr5 in preventing the expression of differentiation-associated genes in p30-mutated leukemia. To test this hypothesis in a mechanistically defined model, we turned to G-CSF-induced granulocytic maturation of 32D cells, which is characterized by up-regulation of myeloid surface markers and induction of myeloid gene expression. Expression of p30 in 32D cells completely blocked all signs of G-CSF-induced maturation, validating this model for monitoring of p30-dependent effects on myeloid differentiation (Supplementary Fig. 6a, b). If Wdr5 was indeed a key effector of p30 action, then its loss of function should overcome the p30-dependent differentiation block. As expected, expression of a control shRNA (shRen.713) did not alter the expression of myeloid surface markers or genes associated with terminal granulocytic maturation (Fig 2c, d). In contrast, suppression of Wdr5 using two independent shRNAs triggered robust expression of Gr-1 and Mac-1 surface markers in response to G-CSF, resulting in a 4-fold increase in the abundance of Mac-1-positive cells (Fig. 2c, Supplementary Fig. 7a). Furthermore, knockdown of Wdr5 allowed high expression of the granulocyte marker genes Lcn2 and Lyz2 in G-CSF-treated 32D-p30 cells (Fig. 2d). Importantly, the G-CSF-mediated differentiation-promoting effect was restricted to p30-expressing cells. Knockdown of Wdr5 in mock- or C/EBPα p42-transduced cells only slightly increased the abundance of Mac-1 positive cells and myeloid marker gene expression, as the basal levels of both parameters were elevated in these cell types (Supplementary Fig. 7a-e). Also, Wdr5 down-regulation did not have any effect in culture conditions that do not permit myeloid differentiation (Supplementary Fig. 7f). Finally, knockdown of WDR5 did not result in altered proliferation rates in human K562 and U-937 cells, further supporting the specificity of the observed effects in p30-expressing cells (Supplementary Fig. 8).
As Wdr5 was shown to be a critical mediator of MLL activity, we next tested the involvement of the SET-domain-containing MLL enzymes MLL1, MLL2, MLL3 and MLL4/Wbp7 in maintaining the p30-induced differentiation block of 32D cells. Only shRNA-mediated knockdown of Mll1, and to a lesser extent Mll3, could phenocopy the differentiation-inducing effect of Wdr5-downregulation in p30-expressing 32D cells (Supplementary Fig. 9a-c). This suggests differential preferences of Wdr5 in its functional interaction with different MLL family members.
Hence, Wdr5 is a critical effector of C/EBPα p30 action, as inactivation of a Wdr5-MLL1 axis reduces H3K4me3 levels on p30-target genes and leads to the up-regulation of myeloid marker genes culminating into a release of the p30-dependent block of myeloid differentiation.
p30-induced self-renewal and leukemogenesis require Wdr5
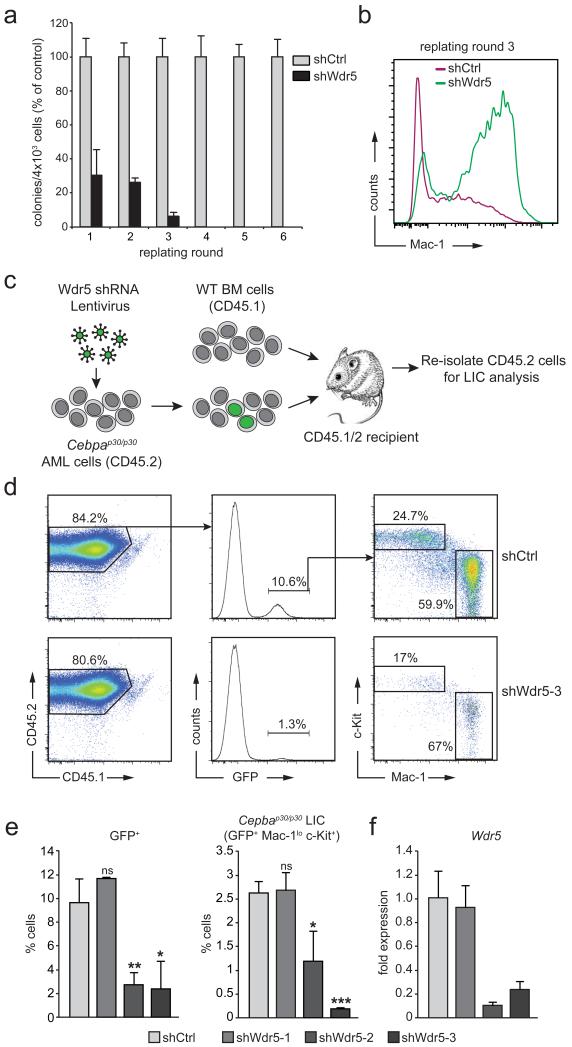
We next asked whether Wdr5 was required for the development of C/EBPα p30-dependent leukemia. As cells from Cebpap30/p30 animals show enhanced self-renewal capacity consistent with a pre-leukemic state, we tested whether Wdr5 was required for this phenotype. While hematopoietic stem/progenitor cells from Cebpap30/p30 mice expressing a control shRNA (shCtrl) showed sustained proliferation in a serial re-plating assay, the proliferative capacity of shWdr5-expressing cells gradually dropped until no immature cells could be isolated after the third plating round (Fig. 3a and Supplementary Fig. 10). Consistently, immuno-phenotypic analysis of these cells showed a high proportion of mature myeloid cells with strong Mac-1 staining in Cebpap30/p30 cultures with Wdr5 knockdown, while Mac-1 expression was mostly absent on shCtrl-expressing Cebpap30/p30 cells (Fig. 3b). We next determined the influence of Wdr5 on the maintenance of p30-dependent leukemia in vivo. We knocked down Wdr5 in freshly isolated Cebpap30/p30 leukemia cells using three different lentiviral constructs expressing GFP-coupled Wdr5-targeting shRNAs and transplanted the transduced cells into irradiated recipient mice (Fig. 3c). Using the same titer for all viral stocks, we observed robust GFP expression in the donor-derived fraction of bone marrow in mice transplanted with control shRNA-expressing lentivirus. In contrast, two shRNAs that efficiently down-regulate Wdr5 expression led to a significant reduction in the percentage of GFP-positive bone marrow cells (Fig. 3d, e). The same trend was observed when the Mac-1loc-Kit+ p30-leukemia initiating cell (LIC) fraction was analyzed (Fig. 3d, e). Importantly, the abundance of GFP-positive p30-LICs was correlated to Wdr5 expression levels, as a dysfunctional Wdr5-targeting shRNA (Wdr5-1) had no effect in this system (Fig. 3e, f).
Figure 3. Wdr5 is required to maintain C/EBPα p30-dependent self-renewal in vitro and in vivo.
(a) Serial re-plating assay of primary Cebpap30/p30 fetal liver cells after knockdown of-Wdr5. To prevent outgrowth of cells escaping retroviral shRNA expression after several rounds of re-plating, we used a lentiviral shRNA construct allowing for continuous selection of shRNA-expressing cells through puromycin addition. Colony numbers were normalized to cells expressing a control shRNA construct (shCtrl) in every round of re-plating. (b) Flow cytometry analysis of Mac-1 surface marker expression of cells from the experiment shown in (a) after round 3 of re-plating. (c) Schematic outline of the experimental setup of the transplantation experiment. (d) Flow cytometric analysis of Cebpap30/p30 LIC from the bone marrow of transplanted mice. Presented events are gated on the CD45.2+ Lineage- singlet population. Cebpap30/p30 LIC display a c-Kit+ Mac-1lo immunophenotype. Top, representative example of a mouse transplanted with Cebpap30/p30 AML cells transduced with a control shRNA-expressing lentivirus. Bottom, representative example of a mouse transplanted with Cebpap30/p30 AML cells transduced with shWdr5-3-expressing lentivirus. (e) Statistical representation of percentages of GFP+ cells (left) and LIC (GFP+ c-Kit+ Mac1lo cells, right) within the CD45.2+ population from the experiment described in (b). (f) qRT-PCR analysis of Wdr5 expression in WT fetal liver cells transduced with indicated shRNA constructs. Data are presented as mean +/− standard deviation (SD) of triplicate experiments. ns, non-significant; * p ≤ 0.05; ** p ≤ 0.01; * p ≤ 0.001 (t-test).
Together, this indicates that the maintenance of transformed Cebpap30/p30 leukemic cells in vitro and in vivo requires Wdr5 expression.
OICR-9429 occupies the MLL-binding pocket of Wdr5
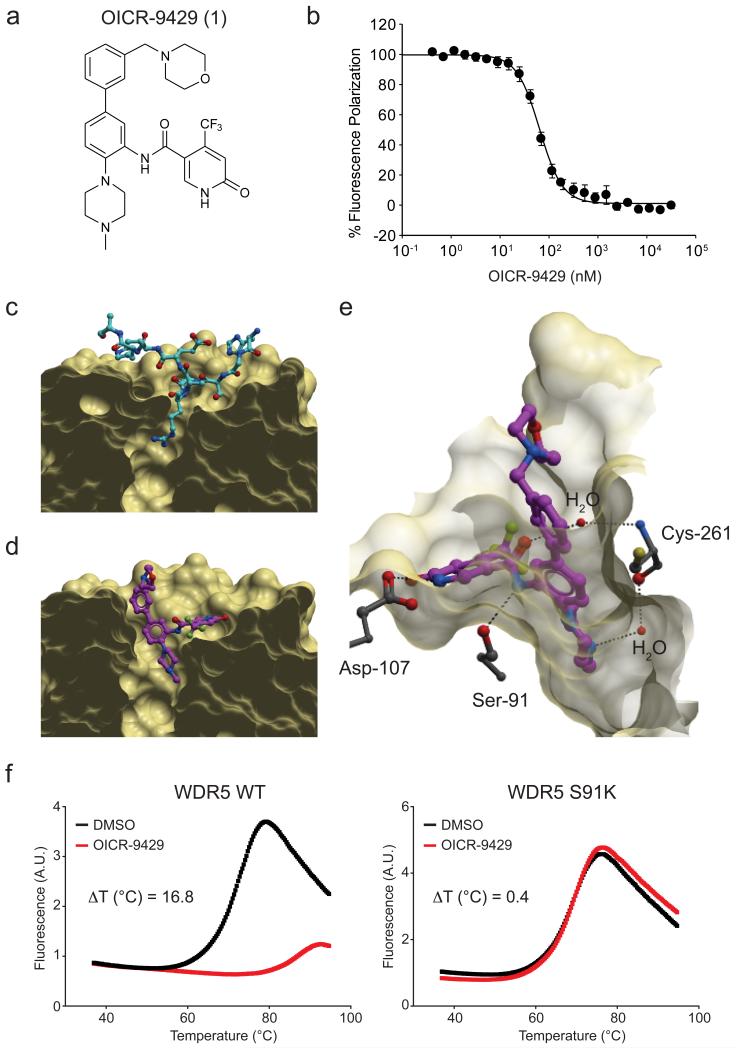
As our loss-of-function experiments suggest that Wdr5 is indeed an obligate and specific effector of p30 action, we reasoned that pharmacological antagonism of Wdr5 could selectively target p30-dependent cellular functions. We previously reported a series of compounds that bound to human WDR5 and antagonized its interaction with MLL complex components in biochemical assays21,22. Optimization of this series using structure-guided medicinal chemistry resulted in OICR-9429 (1) (Fig. 4a), which binds to WDR5 with high affinity (KD = 93 ± 28 nM, Supplementary Fig. 11) and competitively disrupts its interaction with a high-affinity Wdr5-INteracting (WIN) peptide of MLL (Kdisp = 64 ± 4 nM, Fig. 4b). OICR-9429 is highly selective for WDR5, as it showed no significant binding to or inhibition of 22 human methyltransferases, 9 different WD40- and histone reader domains and a panel of over 250 human kinase, GPCR, ion channel, and transporter drug targets (Supplementary Table 3, 4).
Figure 4. OICR-9429 binds WDR5 in the MLL WIN motif-binding pocket.
(a) Chemical structure of OICR-9429 (N-(4-(4-methylpiperazin-1-yl)-3′-(morpholinomethyl)-[1,1′-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide). (b) Peptide displacement assay monitoring the decrease in fluorescence polarization (FP) signal of a fluorescently labeled MLL peptide upon OICR-9429-induced dissociation from WDR5. (c) Crystal structure of WDR5 in complex with a WIN peptide (cyan; PDB code 4ESG). (d) Structure of WDR5 bound to OICR-9429 (purple, PDB code 4QL1). (e) Key direct and water-mediated hydrogen-bonds between OICR-9429 and WDR5 are shown as dashed lines. Extensive hydrophobic contacts and other interactions are shown in Supplementary Fig. 12. (f) Differential Scanning Fluorimetry (DSF) of WDR5 WT (left panel) and an S91K mutant protein (right panel) incubated with DMSO or OICR-9429 (50 μM).
To obtain insight into the detailed mode of interaction, we determined the crystal structure of WDR5 in complex with OICR-9429 at 1.5 Å resolution (Supplementary Table 5). As expected from our molecular design, OICR-9429 bound in the MLL WIN peptide-binding pocket of WDR5, thereby preventing its interaction with MLL via the same protein surface (Fig. 4c, d). Thus, OICR-9429 occupies the arginine-binding pocket exploited by the WDR5-binding peptides of SET1 methyltransferases23 and KANSL124. Conformational rearrangements critical for OICR-9429 binding were observed (i) at the bottom of the cavity (Phe-263) to accommodate the N-methyl piperazine moiety in lieu of the WIN peptide guanidinium group, and (ii) at the rim (Phe-133, Phe-149) to accommodate the ‘northern’ substituent of the OICR-9429 compound (Supplementary Fig. 12). The piperazine, amide linker and pyridone groups of OICR-9429 form direct or water-mediated hydrogen bonds with surrounding residues (Cys-261, Ser-91, Asp-107, Fig. 4e), while the northern fragment interacts with a hydrophobic patch in WDR5 (Phe-133, Tyr-191, Phe-149, Pro-173, Supplementary Fig. 12). Ser-91 in WDR5 is critical for compound binding, as a S91K mutant of WDR5 did not bind to OICR-9429 (Fig. 4f).
OICR-9429 is a potent and selective Wdr5 antagonist
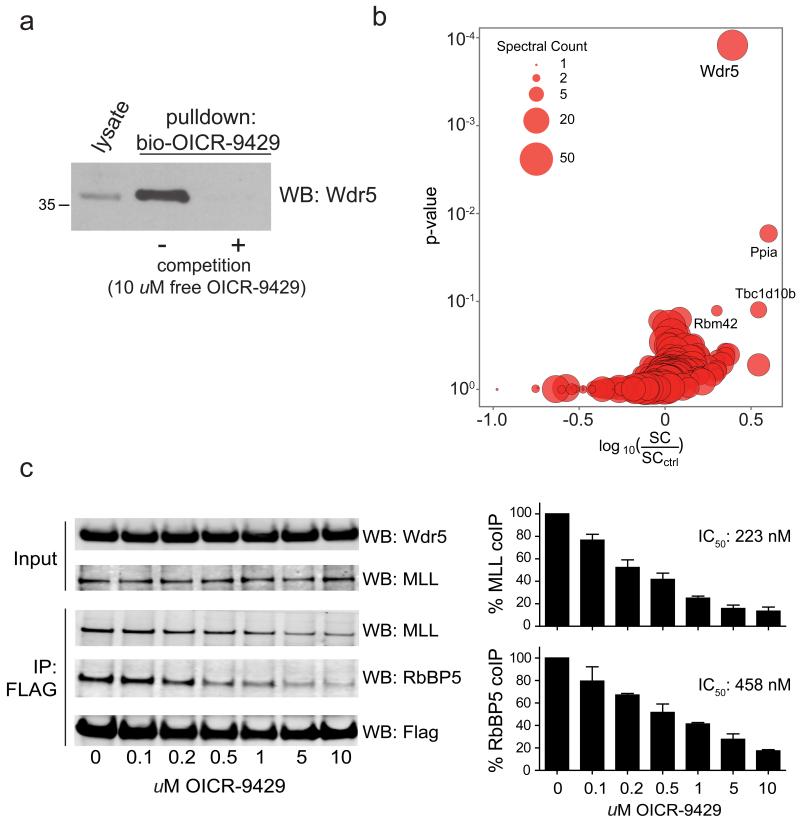
First we tested if OICR-9429 was able to disrupt the C/EBPα p30-Wdr5 interaction. Wdr5 was readily detected in C/EBPα -immunoprecipitates from lysates of Cebpap30/p30 cells in the presence of OICR-9429, suggesting that the interaction of Wdr5 with MLL did not influence p30 binding (Supplementary Fig. 13). We next tested the effect of OICR-9429 on Wdr5-dependent protein-protein interactions in cells using a biotinylated variant of the compound in a chemical proteomics experiment. While we were able to efficiently isolate Wdr5 using the biotinylated variant of OICR-9429, this enrichment was lost upon competition with excess unmodified OICR-9429 (Fig 5a). Bioinformatic analysis of LC-MSMS data revealed that Wdr5 was the primary target protein of OICR-9429 in cells (Fig 5b). Notably, our analysis did not identify any other components of SET/MLL HMT complexes, indicating that OICR-9429 disrupts integral protein-protein interactions between Wdr5 and its binding partners. Indeed, OICR-9429 reduced the amount of endogenous MLL and RBBP5 that co-immunoprecipitated with exogenously expressed FLAG-tagged WDR5 in a dose-dependent manner (Fig. 5c).
Figure 5. OICR-9429 is a small molecule antagonist of the Wdr5-MLL interaction.
(a) Western blot analysis of drug-affinity purification experiments using a biotinylated variant of OICR-9429 in Cepbap30/p30 cells. (b) Cellular target profile of OICR-9429. Log ratio of total spectral counts in normal drug pulldown experiments vs. competition experiments (X axis) and corresponding binding specificity p-value as calculated by Decontaminator58 (Y axis). (c) Left, HEK293 cells were transfected with FLAG-tagged WDR5 and treated with vehicle alone (DMSO) or indicated doses of OICR-9429. FLAG-immunoprecipitates were blotted for MLL and RBBP5. Right, Histogram representation of quantified data from immunoprecipitation experiments. Data are presented as mean +/− standard deviation (SD) of triplicate experiments.
Gene expression profiling of OICR-9429-treated Cebpap30/p30 cells showed that Wdr5 antagonism led to the up-regulation of myeloid-specific transcripts (Supplementary Fig. 14a). Importantly, Gene Set Enrichment Analysis25 showed a close correlation between OICR-9429-induced genes and genes that were up-regulated after Wdr5 knockdown. Furthermore, the gene signature of Cebpap30/p30 LICs6 was down-regulated upon Wdr5 antagonism by OICR-9429 (Supplementary Fig. 14b).
Taken together, these data demonstrate that OICR-9429 is a potent, selective and cell-active antagonist of the Wdr5-MLL interaction, which is able to elicit a profound disruption of the protein-protein interaction network around Wdr5 and the SET/ MLL HMT complex.
p30-expressing cells are sensitive to Wdr5 antagonism
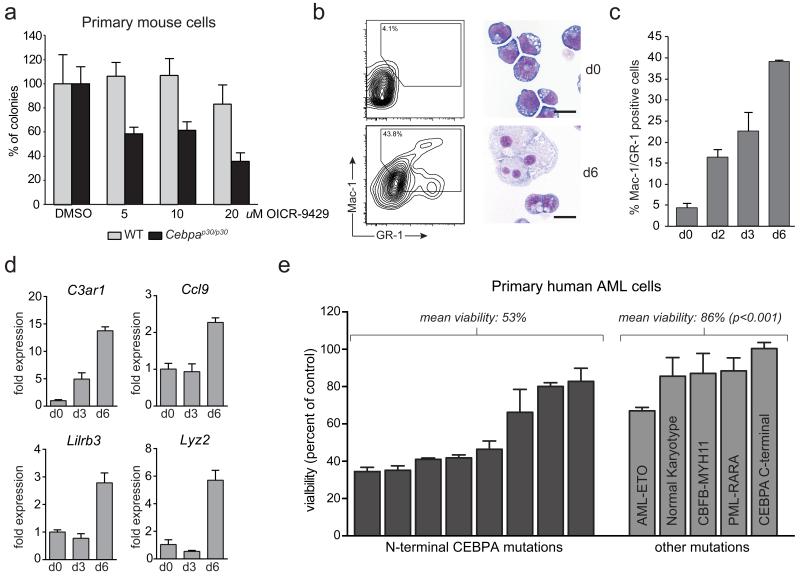
Next we tested whether OICR-9429 would have a selective inhibitory effect on cells expressing C/EBPα p30. Primary fetal liver cells from Cebpap30/p30 animals showed a markedly increased sensitivity towards OICR-9429 treatment compared to wild-type fetal liver cells in colony formation assays (Fig. 6a). Similarly, the sensitivity to OICR-9429 was dramatically increased in a Cebpap30/p30 cell line as compared to HoxA9/Meis1-transformed murine cells and different human leukemia cell lines (Supplementary Fig. 15a, b). Also, OICR-9429 did not affect the number and distribution of murine bone marrow CFUs (Supplementary Fig. 15c). The specificity of OICR-9429 is further illustrated by the fact that the closely related derivative OICR-0547 (2), an inactive control compound that no longer binds to WDR5, did not show any effect on the viability of OICR-9429-sensitive or -resistant cells at relevant concentrations (Supplementary Fig. 16). Prolonged exposure of Cebpap30/p30 cells to OICR-9429 caused up-regulation of surface markers associated with myeloid differentiation and loss of progenitor morphology (Fig. 6b, c). In fact, OICR-9429 induced the up-regulation of myeloid marker genes in Cebpap30/p30 cells, such as C3ar1, Ccl9, Lcn2, Lilrb3, and Lyz2 (Fig. 6d), indicating that Wdr5 antagonism is able to overcome the differentiation block of Cebpap30/p30 cells.
Figure 6. Pharmacological antagonism of the WDR5-MLL interaction by OICR-9429 selectively affects p30-expressing cells.
(a) Colony formation assay of primary Cebpap30/p30 and WT fetal liver cells in response to OICR-9429 at indicated concentrations. Numbers were normalized to colonies generated in response to DMSO for each genotype. (b) Cebpap30/p30 cells were left untreated (top panels) or treated with 20 μM OICR-9429 (bottom panels) for 6 days. Left, flow cytometry analysis of Mac-1/Gr-1 surface expression. Right, representative micrographs of stained cytospins. Scale bar, 10 μm. (c) Quantification of Mac-1/Gr-1 double positive cells at indicated time points after exposure to OICR-9429 (20 μM). (d) qRT-PCR analysis of the indicated genes in Cebpap30/p30 cells treated with 20 μM OICR-9429 for the indicated periods. (e) Cell viability of primary human AML cells with the indicated mutational status after 3 days of exposure to 5 μM of OICR-9429. In samples with N-terminal CEBPA mutations, these are either isolated or occur in conjunction with a C-terminal mutation on the other allele. Numbers were normalized values obtained in response to DMSO for each patient sample. Data are presented as mean +/− standard deviation (SD) of triplicate experiments. p-values were calculated using the t-test.
Finally, we tested whether primary human AML cells with N-terminal C/EBPα mutations would be sensitive to WDR5 antagonism. At 5 μM, OICR-9429 caused a significant decrease in viability in the majority of patient cells with mutations in the N-terminal part of the CEBPA gene (mean viability; 53%, n=8, Fig. 6e). Strikingly, the same concentration of OICR-9429 had little effect on viability in AML patient cells with other mutations (mean viability: 86%, n=5).
In summary, our data indicate that p30-recruited WDR5-containing protein complexes represent a functional vulnerability that can be pharmacologically exploited to trigger differentiation and growth arrest in C/EBPα-mutant AML.
Discussion
Here we demonstrate that significant differences in protein-protein interactions as well as protein-DNA interactions of p42 vs. p30 isoforms of C/EBPα underlie their distinct biological functions. Comparative AP-MS experiments showed that the leukemogenic p30 variant of C/EBPα, but not the wild-type p42 isoform, preferably interacted with Wdr5, a critical component of mammalian SET/MLL HMT complexes. These complexes are composed of the SET/MLL histone methyltransferases and three catalytically inactive core subunits (Wdr5, Ash2l and Rbbp5)26. Among those, Wdr5 mediates MLL-dependent H3K4 methylation by physically interacting with both histone H3 and the MLL protein27-30. Thus, the maintenance of MLL-dependent global H3K4me3 levels and gene expression requires Wdr520,26. Wdr5 expression was required for ES cell pluripotency31, suggesting that Wdr5 is a key factor in linking chromatin remodeling and differentiation potential.
As deposition of the H3K4me3 mark is mediated by the SET/MLL HMT complex, we used the distribution of H3K4me3 ChIP-seq as a proxy for the presence and activity of Wdr5 on chromatin. Co-localization of H3K4me3 with p30 was enriched over p42 and was predominantly found in proximity to genes that showed p30-dependent expression changes. Formally, our data do not exclude the possibility that the p30-Wdr5 interaction occurs outside of the SET/MLL complex. However, Wdr5 down-regulation led to a selective reduction of H3K4me3 levels on promoters of p30-target genes, indicating a functional consequence of the p30-Wdr5 interaction in the SET/MLL-dependent regulation of gene expression. These data not only cross-validate our AP-MS results of the specific association of p30 with Wdr5, they also extend the functional aspect of the p30-Wdr5 interaction to the chromatin level.
Wdr5 was not the only p30-specific interactor in our AP-MS survey. Indeed, C/EBPα p42 and p30 were associated with a surprisingly different subset of the molecular machinery of the cell. The observation that a smaller isoform of a protein can have specific binding partners that are not shared with the longer variant seems counterintuitive. It is possible that different biophysical properties of C/EBPα variants may account for changes in sub-nuclear localization and thus also manifest in differential protein interaction patterns. This could be reflected by the differences in chromatin association between p42 and p30, which are likely to be influenced by specific binding proteins, as was previously shown to be important for transcription factor function32.
As Wdr5 could also interact with C/EBPα p42 upon overexpression in HEK293 cells, we speculate that the preferred interaction of p30 and Wdr5 in myeloid cells is likely to be indirect and mediated through other proteins whose expression is restricted to the hematopoietic system and which are differentially regulated by p30.
Wdr5, and also MLL1 were critical to sustain p30-dependent functions, including inhibition of myeloid differentiation, enhanced self-renewal and leukemogenesis. Importantly, this effect was restricted to the p30 isoform of C/EBPα, as depletion of Wdr5 had little effect on wild-type cells or on cells over-expressing p42. In line with this, shRNA-mediated down-regulation of Wdr5 had no effects in fibroblasts or myoblasts31. We were able to phenocopy the effect of Wdr5 down-regulation by using a novel pharmacological antagonist of the Wdr5-MLL interaction. OICR-9429 has all the essential properties of a high-quality chemical probe. Its on-target, dose-dependent activity in cells directly reflects its in vitro biochemical activity. Furthermore, OICR-9429 directly engages the endogenous cellular target at pharmacologically relevant concentrations and displays a disease-related activity that is related to the target hypothesis. OICR-9429 displays exquisite cellular selectivity and specificity in disrupting critical protein-protein interactions between WDR5 and other MLL complex members, thereby compromising MLL activity.
C/EBPα p42 was shown to sequester E2F proteins through direct protein-protein interactions, leading to repression of E2F transcriptional activity, cell cycle arrest and differentiation5. Reduction of the E2F-C/EBPα interaction in the p30 variant has been postulated to contribute to leukemogenic effects of p30. Our identification of Wdr5 as a specific interactor of p30 allows speculation over another mechanism: Wdr5 has been reported to interact with E2F633, and another reported binding partner of Wdr5, Hcf-1, was shown to mediate recruitment of the SET/MLL HMT complex to E2F target genes by physical interaction with E2F family members34. Thus, p30 could actively induce the expression of E2F target genes via the specific recruitment of the SET/MLL HMT complex.
It has been reported that LICs from Cebpap30/p30 mice show gene expression patterns that are related to AML with MLL-rearrangements6. Consistently, a peptide-based antagonist of the Wdr5-MLL interaction was recently shown to selectively target MLL-leukemia cells35.
In summary, our comparative integrated analyses of C/EBPα variants provide evidence for the existence of gain-of-function features of the C/EBPα p30 protein. We reveal a novel and specific functional connection between the p30 variant of the C/EBPα transcription factor and Wdr5 that controls the transforming properties of p30. We describe OICR-9429 as a novel compound that disrupts the interaction of Wdr5 with MLL in cells, thereby selectively triggering a differentiation program in p30-expressing leukemia cells. Thus, our data provide an attractive alternative by targeting the MLL complex in C/EBPα-mutated AML. As direct pharmacological targeting of transcription factors remains difficult, targeting of critical co-factors such as the MLL complex could represent an attractive strategy to target cancer entities that depend on oncogenic transcription factor mutations.
Online Methods
Constructs
The coding regions of C/EBPα p42 and p308 were cloned into a modified pMSCV-IRES-GFP retroviral vector with a C-terminal Strep-HA tag. A two-component miR-30-based retroviral expression system was used for inducible shRNA studies36. A DNA fragment encoding human WDR5 (residues 24-334) was amplified by PCR and sub-cloned into pET28-MHL vector, downstream of the poly-histidine coding region.
Cell culture
Primary murine fetal liver cells were cultivated in DMEM plus 10% FCS plus SCF, IL-6, and IL-3. c-kit-positive Cebpap30/p30 mutant bone marrow cells were enriched using magnetic beads (Miltenyi). Cells were cultured in IMDM medium supplemented with IL-6, IL-3, SCF, polybrene and β-mercaptoethanol. FDCP-1 and 32D cell lines and immortalized Cebpap30/p30 fetal liver cells were maintained in RPMI 1640 medium supplemented with 10% FCS and IL-3. Plat-E and HEK293T cells were maintained in DMEM medium with 10% FCS. For colony assays, 1×104 cells were plated in complete methylcellulose medium (MethoCult M3434) and colonies were scored 7 days later. For re-plating assays, colonies were scored in 7 day-intervals and 1×104 cells were replated. The Cebpap30/p30 cell line was established by picking single cell clones after the 6th round of re-plating and continuous liquid culture in the presence of SCF, IL-3 and IL-6 for 4 weeks. The HoxA9/Meis1-transformed cell line was a kind gift from Dr. R. Slany. Ecotropic retroviral particles were generated using the Plat-E packaging cell line. Lentiviruses were produced by transient transfection of HEK293T cells. Human leukemic blast cells were derived from diagnosed AML patients and isolated on Ficoll-Hypaque gradients. Cells were cultured in RPMI 1640 supplemented with 20% FCS plus human IL-3, IL-6, G-CSF, GM-CSF and SCF. Patient recruitment and sample processing were performed according to protocols from the Dutch-Belgian Hematology/Oncology Cooperative Group (HOVON trials). All studies were approved through institutional human ethics review board, and all patients provided written informed consent in accordance with the Declaration of Helsinki.
Bone marrow transplantation
Cells were transplanted in lethally irradiated CD45.1/2 8-10 week old recipients along with 1×106 whole BM competitor cells (CD45.1) via tail vein injection. 6 mice were transplanted for every candidate shRNA in two independent experiments. Mice were sacrificed 28 days post transplantation and subjected to LIC analysis. All mice were bred and maintained at University of Oxford in accordance with Institutional and UK Home Office guidelines.
Affinity Purification of C/EBPα protein complexes
All steps described in the protocol were carried out at 4°C. Purifications were performed from 1×109 freshly harvested cells. After washing with PBS, cells were incubated in buffer N (300 mM sucrose, 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1 mM DTT, 0.75 mM spermidine, 0.15 mM spermine, 0.1% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, protease inhibitors) for 5 min on ice to lyse the cytoplasm. Nuclei were collected by centrifugation (500×g for 5 min), and the supernatant was removed. The nuclear pellet was washed with buffer N. Nuclei were resuspended in buffer C420 (20 mM HEPES pH 7.9, 420 mM NaCl, 25% glycerol, 1 mM EDTA, 1 mM EGTA, 0.1 mM DTT, 50 mM NaF, 1 mM Na3VO4, protease inhibitors), shaken vigorously for 30 min. Nuclear extracts were cleared by centrifugation for 1 h at 100,000×g. Prior to purification, 15 mg of extracts were adjusted to 2 mg/mL and 150 mM NaCl with 20 mM HEPES, 50 mM NaF, 1 mM Na3VO4, protease inhibitors. After 20 min of pre-clearing with RNAse A, benzonase and avidin, nuclear extracts were incubated with 200 μL StrepTactin sepharose beads for 2 h on a rotating wheel at 4°C. Beads were washed 3× with TNN-HS buffer, and 2× using TNN-HS buffer without detergent and inhibitors. Bound proteins were eluted by incubation with 100 μL 2.5 mM biotin in TNN-HS buffer 10 min at 4°C, followed by centrifugation for 3 min at 300×g. Samples were alkylated with iodoacetamide and separated by 1D SDS-PAGE on a 4 – 12% bis-Tris gel (NuPAGE, Invitrogen). Proteins were visualized by silver staining and the entire gel lane was excised and 20 slices digested in situ with modified porcine trypsin (Promega) as previously described37. Peptides were pooled into 10 samples and analysed by online LC-MSMS (1D-gel-MS).
Chemical proteomics
Per experiment, 100 μL UltraLink streptavidin bead slurry (Pierce) was centrifuged, and the supernatant removed. After washing with lysis buffer (50 mM Tris–HCl, 100 mM NaCl, 0.2% NP-40, 5% glycerol, 1.5 mM MgCl2, 25 mM NaF, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol (DTT), 10 μg/mL TLCK, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 10 μg/mL soybean trypsin inhibitor (Sigma), pH 7.5), biotin-conjugated OICR-9429 derivative (0.05 μmol) was added and incubated on a roto-shaker for 30 min at 4 °C. After centrifugation and one additional washing step, beads were re-suspended in cell lysates (10 mg total protein per pull-down) and incubated on a roto-shaker for 2 h at 4 °C. For competition experiments, lysates were pre-incubated with 10 μM of unmodified OICR-9429, for 20 min at 4 °C. After centrifugation, beads were transferred to spin columns (MoBiTec), washed with lysis buffer and HEPES, followed by elution of bound proteins with elution buffer (50% 6 M urea, 50% 100 mM formic acid).
Mass spectrometry
All affinity purifications were analyzed on a hybrid linear trap quadrupole (LTQ) Orbitrap Velos mass spectrometer (ThermoFisher Scientific) coupled to a 1200 series high-performance liquid chromatography system (Agilent Technologies) via a nanoelectrospray ion source using liquid junction (Proxeon). Solvents for HPLC separation of peptides were as follows: solvent A consisted of 0.4% formic acid in water and solvent B consisted of 0.4% formic acid in 70% methanol and 20% isopropanol. 8 μL of the tryptic peptide mixture were automatically loaded onto a trap column (Zorbax 300SB-C18 5 μm, 5×0.3 mm, Agilent Biotechnologies) with a binary pump at a flow rate of 45 μL/min. 0.1% trifluoroacetic acid was used for loading and washing the precolumn. After washing, the peptides were eluted by back-flushing onto a 16 cm fused silica analytical column with an inner diameter of 50 μm packed with C18 reversed phase material (ReproSil-Pur 120 C18-AQ, 3 μm, Dr. Maisch GmbH) with a 27 min gradient ranging from 3 to 30% solvent B, followed by a 25 min gradient from 30 to 70% solvent B and, finally, a 7 min gradient from 70 to 100% solvent B at a constant flow rate of 100 nL/min38. Analyses were performed in a data-dependent acquisition mode and dynamic exclusion for selected ions was 60 s. A top 15 collision-induced dissociation (CID) method was used, and a single lock mass at m/z 445.120024 (Si(CH3)2O)6 was employed.39 Maximal ion accumulation time allowed in CID mode was 50 ms for MSn in the LTQ and 500 ms in the C-trap. Automatic gain control was used to prevent overfilling of the ion traps and was set to 5,000 in MSn mode for the LTQ and 106 ions for a full FTMS scan. Intact peptides were detected in the Orbitrap Velos at 60,000 resolution at m/z 400.
Protein identification
Peak list information was extracted from the RAW MS files and converted into an MGF format with the msconvert tool (ProteoWizard Library v2.1.2708). MGF files were searched against the mouse component of the UniProtKB/SwissProt database (www.uniprot.org), including all protein isoforms plus rat C/EBPα and known contaminant sequences. An initial search was performed with Mascot (www.matrixscience.com, version 2.3.02). Mass error tolerances on the precursor and fragment ions were ±10 ppm and ±0.6 Da, respectively. Only fully-tryptic peptides were considered with a maximum of one missed cleavage site, and carbamidomethyl cysteine and methionine oxidation set as fixed and variable modifications, respectively. The Mascot peptide ion score threshold was equal to 30, and at least 3 peptide identifications per protein were required.
For both the precursor and fragment ion data, linear recalibration transformations that minimise the mean square deviation of the measured from theoretical values were deduced from initial identifications. Re-calibrated files were searched against the same protein database with Mascot and Phenyx (GeneBio) using narrower mass tolerances (±4 ppm and ±0.3 Da)40. All other search parameters were identical to the initial first pass search. Mascot and Phenyx output files were processed by internally-developed parsers to filter and integrate protein identifications. The following peptide score thresholds were used: T1=14, T2=40 and T3=10; and T1=4.2, T2=4.75 and T3=3.5, respectively (p<10−3). Proteins with at least two unique peptides above score T1, or with a single peptide above T2 were selected as unambiguous identifications.
Additional peptides from these validated proteins with a score >T3 were appended to the final result. The validated identifications from both algorithms were merged, spectral conflicts discarded, and protein groups defined according to shared peptides. A false discovery rate (FDR) of <0.01 for protein identifications and <0.001 for peptides (including peptides exported with lower scores) was estimated by applying the same filtering procedure against a database of reversed protein sequences. All affinity purifications were analyzed based on protein spectral counts. For each purification strategy, proteins identified in the control cells were subtracted from proteins identified from the corresponding C/EBPα affinity purifications. Moreover, contaminants such as keratin, spectrin, and plectin were removed from the list as known non-specific binders frequently observed in AP-MS.
To estimate confident drug targets in chemical proteomics experiments, the drug pull-downs were compared with the competition experiments using the Decontaminator algorithm modified to take advantage of using multiple protein search engines (p-values were calculated separately for spectral counts and both Mascot and Phenyx protein scores as described in and then combined into single p-value using the Fisher method). The p-value cutoff for the confident hits was set to 0.001.
Expression and purification of human WDR5
WDR5 was over-expressed in E. coli BL21 by addition of 1 mM IPTG overnight at 15°C. cells were resuspended in 50 mM HEPES buffer, pH 7.4 containing 250 mM NaCl, 5 mM imidazol, 2 mM β-mercaptoethanol, 5% glycerol and lysed using a microfluidizer (Microfluidics Corporation, 20,000 psi). The clarified lysate was loaded onto a Ni2+-charged HiTrap Chelating column (GE Healthcare). After washing with 10 column volumes of 20 mM HEPES, pH 7.4, 250 mM NaCl, 50 mM imidazole, 5% glycerol, the protein was eluted with elution buffer (20 mM HEPES pH 7.4, 250 mM NaCl, 250 mM imidazole, 5% glycerol) and loaded on a Superdex200 column (GE Healthcare) equilibrated with 20 mM PIPES buffer, pH 6.5, and 250 mM NaCl. TEV protease was added to combined WDR5-containing fractions to remove the His-tag. The protein was further purified to homogeneity by ion-exchange chromatography.
Crystallization of the WDR5-OICR-9429 complex
Purified WDR5 protein (10 mg/mL) was mixed with OICR-9429 at 1:5 molar ratio of protein:compound and crystallized using the sitting drop vapor diffusion method by mixing 1 μL of protein solution with 1 μL of the reservoir solution containing 25% PEG 3350, 0.2 M ammonium acetate, 0.1 M BisTris, pH 6.5. Crystals were soaked in the corresponding mother liquor supplemented with 20% ethylene glycol as cryoprotectant before freezing in liquid nitrogen.
Crystallographic Data Collection and Structure Determination
X-ray diffraction data for WDR5 structure in complex with OICR-9429 was collected at 100K at beam-line 08ID-1 of CLS, Canadian Light Source. Data sets were processed using the HKL-3000 suite41. The structures of WDR5 in complex with OICR-9429 inhibitor was solved by molecular replacement using PHASER42 with PDB entry 4IA9 as search template. PRODRG was used to generate geometry restraints for the compound OICR-9429 refinement43. REFMAC was used for structure refinement44. The graphics program COOT was used for model building and visualization45. MOLPROBITY was used for structure validation46.
Fluorescence polarization assays
The 9-Ala-FAM ((Ac)-ARAEVHLRK-(Ahx-Ahx)-K(5,6-FAM)) peptide for WDR5 was synthesized, c-terminally labelled with FAM and purified by Peptide 2.0 (Chantilly). Peptide displacement assays were performed in 125 μL reactions, at a constant labelled peptide concentration of 5 nM and WDR5 concentration of 50 nM in 80 mM sodium phosphate pH 6.5, 20 mM KCl and 0.008% Triton X-100 using 96-well Microfluor 2 plates (Thermo Scientific). Fluorescence polarization was measured using a ViewLux imager (Perkin Elmer) at excitation wavelength of 480 nm and an emission wavelength of 540 nm.
OICR-9429 Selectivity Assays
OICR-9429 was evaluated at concentrations up to 50 μM for potential inhibition of the following human protein methyltransferases using a radioactivity based catalytic assay as previously reported22,47,48: SMYD2, G9a, EHMT1, SUV39H2, SETDB1, SETD7, SETD8, SUV420H1, SUV420H2, PRMT1, PRMT3, PRMT5-MEP50 complex, PRMT6, PRMT8, PRMD9, EZH1, EZH2, NSD1, NSD2, NSD3, SETD2, DOT1L, DNMT1. The following chromatin histone binding ‘reader domains’ were tested for binding to OICR-9429 (200 μM) using differential scanning fluorimetry (DSF) and differential static light scattering (DSLS) as previously described49: TDRD3, SND1, L3MBTL1, L3MBTL3, UHRF1, 53BP1, STRAP, EED. Additional selectivity screening of >200 enzymes, receptors and transporters was performed by Cerep.
Transient transfection and immunoprecipitation of WDR5
293 cells were transfected with 1 μg of FLAG-tagged human WDR5 plasmid (a kind gift from Dr. Wysocka) using GeneJuice (EMD) and 24 h later the media was replaced with fresh media containing OICR-9429, and the cells incubated for 5 h. Cells were collected and lysed in 0.5 mL of cold 0.3 M KCl, 20 mM TrisHCl pH 8, 1 mM EDTA, 0.1% NP40, 10% glycerol, protease inhibitors (Roche) for 10 min on ice. The lysate was centrifuged for 5 min at 14000 rpm and the supernatant was collected. Supernatants were used for immunoprecipitation with 1 μg Flag M2 (Sigma) and 30 μL protein G dynabeads (Invitrogen) overnight at 4°C. Beads were washed 3 times with lysis buffer containing 0.15 M KCl before SDS-PAGE loading buffer was added.
Chromatin Immunoprecipitation
1×108 cells were crosslinked with 10% formaldehyde for 10 minutes, quenched with glycine for 5 min and then harvested. Cells were resuspended in LB1 buffer (50 mM Hepes pH 7.6, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) to lyse the cytoplasm. Nuclei were washed once in LB2 buffer (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). before lysis in LB3 buffer (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% NaDeoxycholate, 0.5% N-lauroylsarcosine, EDTA, 1 mM EGTA, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4 and protease inhibitors). The released chromatin was sonicated to obtain fragments of 150 bp using a COVARIS sonicator. 0.5% Triton X-100 was added to the samples immediately after sonication to aid solubilization of the sheared DNA. Samples were spun at 10,000×g for 10 min. Half of the supernatant was incubated with 5μg anti-HA antibody (Abcam) or anti-H3K4me3-antibody (Millipore) at 4°C overnight. Antibody-bound material was pulled down using Dynal protein G magnetic beads (Invitrogen), washed 5 times and released using elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) at 65°C. DNA-protein crosslinks were reverted by incubating the samples overnight at 65°C. The DNA was treated with RNaseA and proteinase K and purified through phenol-chloroform extraction. 10 ng of precipitated material was used for sequencing on a Genome Analyzer IIx (Illumina).
Deep Sequencing and data analysis
Short read sequences were aligned to the Genome Reference Consortium Mouse Build 38 (GCA_000001635.2, GRCm38, UCSC mm10) using Bowtie2, version 2.1.050 in end-to-end, gapped alignment mode. Peaks were called with the Model-based Analysis of ChIP-Seq (MACS) algorithm, version 1.4.251 with parameter settings suggested by the authors52. The resulting peak lists were annotated with regard to transcription start sites using the Bioconductor package ChIPpeakAnno (http://www.bioconductor.org/packages/release/bioc/html/ChIPpeakAnno.html) and PeakAnnotator53 and post-processed with custom R scripts utilizing the ggplot2 plotting system (http://ggplot2.org/).
Microarray Analysis
Total RNA (200 ng) was used for GeneChip analysis. Preparation of terminal-labeled cDNA, hybridization to genome-wide murine Gene Level 1.0 ST GeneChips (Affymetrix) and scanning of the arrays were performed as described previously54 and according to manufacturer’s protocols. Signal extraction, normalization using the RMA algorithm and probe filtering was performed using R/bioconductor as described previously55,56.
Western blotting
Western blotting was performed according to standard laboratory protocols. Antibodies used were: peroxidase labeled anti-HA (HA-7, Sigma), anti-HA (HA-11, Covance), anti-Wdr5 (Abcam), anti-C/EBPα (14AA, Santa Cruz), anti-tubulin (Abcam), anti-RCC-1 (E-6, Santa Cruz), anti-MLL (N-terminal, Santa Cruz), anti-FLAG (Sigma), anti-RbBP5 (Abcam). For Figure 5c, Secondary IR600- and IR700-conjugated antibodies (LiCor) were used to visualize the bands on an Odyssey scanner (LiCor). The band intensity was quantified using LiCor software, with data normalized to the WDR5-IP signal and plotted as percent of control and standard errors representing SEMs. Experiments were performed 3-4 independent times with compound incubated on cells for 5-6 h.
Real time PCR analysis
Total RNA was isolated using RNeasy Mini Kit (Qiagen). 300 ng RNA was reverse transcribed using oligo(dT) primers using RevertAid Reverse Transcriptase (Fermentas). Quantitative PCR was carried out on a Qiagen - RotorGene RG-600 PCR machine using the SensiMix SYBR kit (Bioline). Results were quantified using the 2−ddC(t) method57.
FACS
Cells were stained with fluorescence-labelled antibodies against c-Kit, Mac-1, GR-1 after incubation with anti-mouse CD16/CD32 to block Fc-receptors. Data were collected on a FACS Fortessa Instrument (BD Biosciences). For the analysis of the LIC fraction in the bone marrow of transplanted mice, bone marrow cells were stained with a mix of antibodies against CD4, CD5, CD8 and Ter119 (lineage cocktail) and antibodies against c-Kit, Sca-1, Mac1, CD45.2 and CD45.1 following red blood cell lysis. 7-Amino-Actinomycin D (7-AAD) was added to exclude dead cells. Cells were analyzed on a LSR Fortessa SORP analyzer. Data were analyzed with FlowJo software (Treestar).
Cytospin analysis
Cells were cytocentrifuged onto glass slides and stained with Rapid-Chrome Kwik-Diff Staining System (Thermo Scientific) prior to microscopic analysis.
Cell viability measurements
20,000 viable, actively proliferating primary human AML cells/well were seeded in 96 well-plates in triplicates and treated with 0.05% DMSO or OICR-9429 at indicated concentrations. Cell viability was measured using the Cell Titer-Glo luminescent cell viability assay (Promega) on a VICTOR X4 luminometer (Perkin Elmer) after 72 h.
Statistical analysis
Two-tailed Student’s t-tests were used for statistical analysis if not stated otherwise.
Supplementary Material
Acknowledgements
We thank Manuela Gridling, Melanie Planyavsky, Dieter Printz and Andreas Spittler for experimental and Kumaran Kandasamy, and Michael Schuster for bioinformatic help. Next generation sequencing was performed at the CSF NGS Unit (www.csf.ac.at). F.G. and R.G. were funded by the Austrian Science Fund (FWF grant P22282-B11). The Superti-Furga laboratory is supported by the Austrian Academy of Sciences and by ERC grant ERC-2009-AdG-250179-i-FIVE. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer, Boehringer Ingelheim, Genome Canada through the Ontario Genomics Institute [OGI-055], GlaxoSmithKline, Janssen, Lilly Canada, the Novartis Research Foundation, the Ontario Ministry of Economic Development and Innovation, Pfizer, Takeda, and the Wellcome Trust [092809/Z/10/Z]. The Ontario Institute for Cancer Research is funded by the Government of Ontario. Funding was also provided by the Leukemia and Lymphoma Society of Canada.
Footnotes
Competing financial interests
The authors do not have any competing financial interests.
Accession codes
PDB 4QL1
shRNA- and qPCR primer sequences are provided in Supplementary Tables 6 and 7.
Samples of OICR-9429 for research purposes are available upon request at http://www.thesgc.org/node/9204
References
- 1.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J. Clin. Oncol. 2009;27:619–28. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang P, et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–63. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Leroy H, et al. CEBPA point mutations in hematological malignancies. Leukemia. 2005;19:329–34. doi: 10.1038/sj.leu.2403614. [DOI] [PubMed] [Google Scholar]
- 4.Fasan A, et al. The role of different genetic subtypes of CEBPA mutated AML. Leukemia. 2014;28:794–803. doi: 10.1038/leu.2013.273. [DOI] [PubMed] [Google Scholar]
- 5.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat. Rev. Cancer. 2004;4:394–400. doi: 10.1038/nrc1363. [DOI] [PubMed] [Google Scholar]
- 6.Kirstetter P, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13:299–310. doi: 10.1016/j.ccr.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Friedman A, McKnight S. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 8.Nerlov C, Ziff EB. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 2001;15:3208–16. doi: 10.1101/gad.209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slomiany BA, Arigo KLD, Kelly MM, Kurtz DT. C / EBP α Inhibits Cell Growth via Direct Repression of E2F-DP-Mediated Transcription. Mol. Cell. Biol. 2000;20:5986–5997. doi: 10.1128/mcb.20.16.5986-5997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porse BT, et al. E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell. 2001;107:247–58. doi: 10.1016/s0092-8674(01)00516-5. [DOI] [PubMed] [Google Scholar]
- 12.D’Alo’ F, et al. The amino terminal and E2F interaction domains are critical for C/EBP alpha-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003;102:3163–71. doi: 10.1182/blood-2003-02-0479. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q-F, Cleaves R, Kummalue T, Nerlov C, Friedman AD. Cell cycle inhibition mediated by the outer surface of the C/EBPalpha basic region is required but not sufficient for granulopoiesis. Oncogene. 2003;22:2548–57. doi: 10.1038/sj.onc.1206360. [DOI] [PubMed] [Google Scholar]
- 14.Cleaves R, Wang Q, Friedman AD. C/EBPalpha p30, a myeloid leukemia oncoprotein, limits G-CSF receptor expression but not terminal granulopoiesis via site-selective inhibition of C/EBP DNA binding. Oncogene. 2004;23:716–25. doi: 10.1038/sj.onc.1207172. [DOI] [PubMed] [Google Scholar]
- 15.Zada A, et al. Proteomic discovery of Max as a novel interacting partner of C/EBPalpha: a Myc/Max/Mad link. Leukemia. 2006;20:2137–46. doi: 10.1038/sj.leu.2404438. [DOI] [PubMed] [Google Scholar]
- 16.Trivedi A, et al. Proteomic identification of C/EBP-DBD multiprotein complex: JNK1 activates stem cell regulator C/EBPalpha by inhibiting its ubiquitination. Oncogene. 2007;26:1789–801. doi: 10.1038/sj.onc.1209964. [DOI] [PubMed] [Google Scholar]
- 17.Bararia D, et al. Proteomic identification of the MYST domain histone acetyltransferase TIP60 (HTATIP) as a co-activator of the myeloid transcription factor C/EBPalpha. Leukemia. 2008;22:800–7. doi: 10.1038/sj.leu.2405101. [DOI] [PubMed] [Google Scholar]
- 18.Koleva RI, et al. C/EBPα and DEK coordinately regulate myeloid differentiation. Blood. 2012;119:4878–88. doi: 10.1182/blood-2011-10-383083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimoto T, Anderson K, Jacobsen SEW, Nishikawa S-I, Nerlov C. Cdk6 blocks myeloid differentiation by interfering with Runx1 DNA binding and Runx1-C/EBPalpha interaction. EMBO J. 2007;26:2361–70. doi: 10.1038/sj.emboj.7601675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 21.Bolshan Y, et al. Synthesis, Optimization, and Evaluation of Novel Small Molecules as Antagonists of WDR5-MLL Interaction. ACS Med. Chem. Lett. 2013;4:353–357. doi: 10.1021/ml300467n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senisterra G, et al. Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5. Biochem. J. 2013;449:151–9. doi: 10.1042/BJ20121280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Migliori V, et al. Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance. Nat. Struct. Mol. Biol. 2012;19:136–44. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- 24.Dias J, et al. Structural analysis of the KANSL1/WDR5/KANSL2 complex reveals that WDR5 is required for efficient assembly and chromatin targeting of the NSL complex. Genes Dev. 2014;28:929–942. doi: 10.1101/gad.240200.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Gene set enrichment analysis : A knowledge-based approach for interpreting genome-wide. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–9. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 27.Couture J-F, Collazo E, Trievel RC. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- 28.Song J-J, Kingston RE. WDR5 interacts with mixed lineage leukemia (MLL) protein via the histone H3-binding pocket. J. Biol. Chem. 2008;283:35258–64. doi: 10.1074/jbc.M806900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A, Dharmarajan V, Cosgrove MS. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 2008;283:32158–61. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 30.Ruthenburg AJ, et al. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–12. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ang Y-S, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–97. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell. 2009;36:682–95. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou Y, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 34.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 35.Cao F, et al. Targeting MLL1 H3K4 Methyltransferase Activity in Mixed-Lineage Leukemia. Mol. Cell. 2014:1–15. doi: 10.1016/j.molcel.2013.12.001. doi:10.1016/j.molcel.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuber J, et al. Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol. 2011;29:79–83. doi: 10.1038/nbt.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shevchenko A, Wilm M, Vorm O, Mann M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 38.Bennett KL, et al. Proteomic analysis of human cataract aqueous humour : Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ ® -labelled specimens. J. Proteomics. 2010;74:151–166. doi: 10.1016/j.jprot.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 39.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics. 2005;4:2010–21. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Colinge J, Masselot A, Giron M, Dessigny T, Magnin J. OLAV : Towards high-throughput tandem mass spectrometry data identification. Proteomics. 2003;3:1454–1463. doi: 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W, Carter Charles W. Macromol. Crystallogr. Part A. Vol. 276. Academic Press; 1997. pp. 307–326. J. B. T.-M. in E. [Google Scholar]
- 42.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuttelkopf AW, van Aalten DMF. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin a a, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siarheyeva A, et al. An allosteric inhibitor of protein arginine methyltransferase 3. Structure. 2012;20:1425–35. doi: 10.1016/j.str.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Yu W, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat. Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 49.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2007;2:2212–21. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 50.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–40. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 2010;11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Astapova I, et al. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci U S A. 2008;105:19544–9. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irizarry R. a, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 56.Bilban M, et al. Deregulated expression of fat and muscle genes in B-cell chronic lymphocytic leukemia with high lipoprotein lipase expression. Leukemia. 2006;20:1080–8. doi: 10.1038/sj.leu.2404220. [DOI] [PubMed] [Google Scholar]
- 57.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 58.Lavallée-Adam M, Cloutier P, Coulombe B, Blanchette M. Modeling contaminants in AP-MS/MS experiments. J. Proteome Res. 2011;10:886–95. doi: 10.1021/pr100795z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.