Abstract
Particles in aquatic environments host distinct communities of microbes, yet the evolution of particle-specialized taxa and the extent to which specialized microbial metabolism is associated with particles is largely unexplored. Here, we investigate the hypothesis that a widely distributed and uncultivated microbial group—the marine group II euryarchaea (MGII)—interacts with living and detrital particulate organic matter (POM) in the euphotic zone of the central California Current System. Using fluorescent in situ hybridization, we verified the association of euryarchaea with POM. We further quantified the abundance and distribution of MGII 16 S ribosomal RNA genes in size-fractionated seawater samples and compared MGII functional capacity in metagenomes from the same fractions. The abundance of MGII in free-living and >3 μm fractions decreased with increasing distance from the coast, whereas MGII abundance in the 0.8–3 μm fraction remained constant. At several offshore sites, MGII abundance was highest in particle fractions, indicating that particle-attached MGII can outnumber free-living MGII under oligotrophic conditions. Compared with free-living MGII, the genome content of MGII in particle-associated fractions exhibits an increased capacity for surface adhesion, transcriptional regulation and catabolism of high molecular weight substrates. Moreover, MGII populations in POM fractions are phylogenetically distinct from and more diverse than free-living MGII. Eukaryotic phytoplankton additions stimulated MGII growth in bottle incubations, providing the first MGII net growth rate measurements. These ranged from 0.47 to 0.54 d−1. However, MGII were not recovered in whole-genome amplifications of flow-sorted picoeukaryotic phytoplankton and heterotrophic nanoflagellates, suggesting that MGII in particle fractions are not physically attached to living POM. Collectively, our results support a linkage between MGII ecophysiology and POM, implying that marine archaea have a role in elemental cycling through interactions with particles.
Introduction
Archaea are ubiquitous in marine-water columns, but their roles in the biogeochemical cycles of carbon (Pearson et al., 2001; Herndl et al., 2005; Ingalls et al., 2006) and nitrogen (Wuchter et al., 2006; Santoro et al., 2011) are only beginning to be understood. Three groups of archaea are abundant in marine-water columns: marine Group I Thaumarchaeota (MG I), Group II Euryarchaeota (MGII) and Group III Euryarchaeota (MGIII) (Delong, 1992; Fuhrman et al., 1992; Fuhrman and Davis, 1997). Of these groups, cultivated representatives exist only from the MGI (Konneke et al., 2005; Santoro and Casciotti, 2011; Park et al., 2014; Qin et al., 2014), limiting our ability to understand the contribution of other archaea to elemental cycling in the ocean.
A central biogeochemical role of microbes in the marine carbon cycle is the degradation of particulate organic matter (POM), which regulates oceanic carbon sequestration through the biological pump (for example, Charette et al., 1999; Buesseler et al., 2008). Microbes have a critical role in the degradation and remineralization of both sinking (Cho and Azam, 1988; Steinberg et al., 2008) and suspended POM (Karl et al., 1988; Giering et al., 2014), but genetic and physiological differences between POM-attached and free-living microbes are still poorly understood (Herndl and Reinthaler, 2013; Ganesh et al., 2014). Particle attachment likely has a role in driving bacterial metabolic diversification within Vibrio species (Hunt et al., 2008; Shapiro et al., 2012). It remains unknown, however, whether similar diversification (on and off of particles) has occurred within marine archaea, and the contribution of archaea to particle degradation in the ocean is completely unknown.
Metagenomic and environmental data suggest a potential role for the MGII in the degradation of POM. An MGII genome assembled from a coastal metagenome (Iverson et al., 2012) contained multiple adhesion domains and genes involved in the degradation of proteins and fatty acids, similar to their deep-sea relatives (Reysenbach et al., 2006; Reysenbach and Flores, 2008). Other metagenomic evidence suggests that MGII may be capable of anaerobic respiration with dimethylsulphoxide as a terminal electron acceptor (Martin-Cuadrado et al., 2008), further suggesting a potential association with low-oxygen microenvironments within particles. MGII are relatively more abundant in particle-rich surface waters (Massana et al., 2000; Herndl et al., 2005; DeLong et al., 2006) compared with deeper waters and archaeal abundance at the domain level has been found to correlate with particle concentrations (Wells et al., 2006). Furthermore, a positive relationship between MGII abundance and phytoplankton has been observed (Galand et al., 2010), and MGII have been detected on POM (Crump and Baross, 2000), in particle-rich waters (Galand et al., 2008) and in size-fractionated metagenomes (Smith et al., 2013; Ganesh et al., 2014). The effects, however, of MGII–POM interactions on MGII physiology, metabolism and evolution are unknown, as is the extent to which particle-attachment is obligate or facultative. The MGII Euryarchaeota are a diverse group with multiple clades defined based on 16 S ribosomal RNA (rRNA) gene sequences (Massana et al., 2000; Hugoni et al., 2013), the multiple clades may correspond to differential adaptation to different environments or microenvironments in the water column, such as those created by POM.
We investigated interactions between uncultivated MGII Euryarchaeota and living and detrital POM in the euphotic zone of the central California Current System (CCS). This region exhibits a large range in primary production (Collins et al., 2003) and supports abundant and diverse prokaryote and eukaryotic microbial populations, including MGII (Suzuki et al., 2000; Rich et al., 2008; Preston et al., 2009; Ottesen et al., 2013). To better understand linkages between MGII ecophysiology and POM, we present the first detailed analysis of MGII abundance, diversity and metabolic capacity in size-fractionated samples from a transect covering different oceanographic regimes. Our analyses suggest the existence of genetically and physiologically distinct particle-attached MGII ecotypes, which can outnumber free-living MGII under oligotrophic conditions. We also present the first MGII-specific net growth rates, indicating that their growth is stimulated in the presence of phytoplankton, and use flow cytometry to investigate whether or not free-living and POM-attached MGII are physically associated with phytoplankton cells. Our findings suggest that interactions with particles have shaped the life history and diversification of MGII, and imply that MGII have a role in POM transformations in the ocean.
Materials and methods
Sampling and environmental contextual data
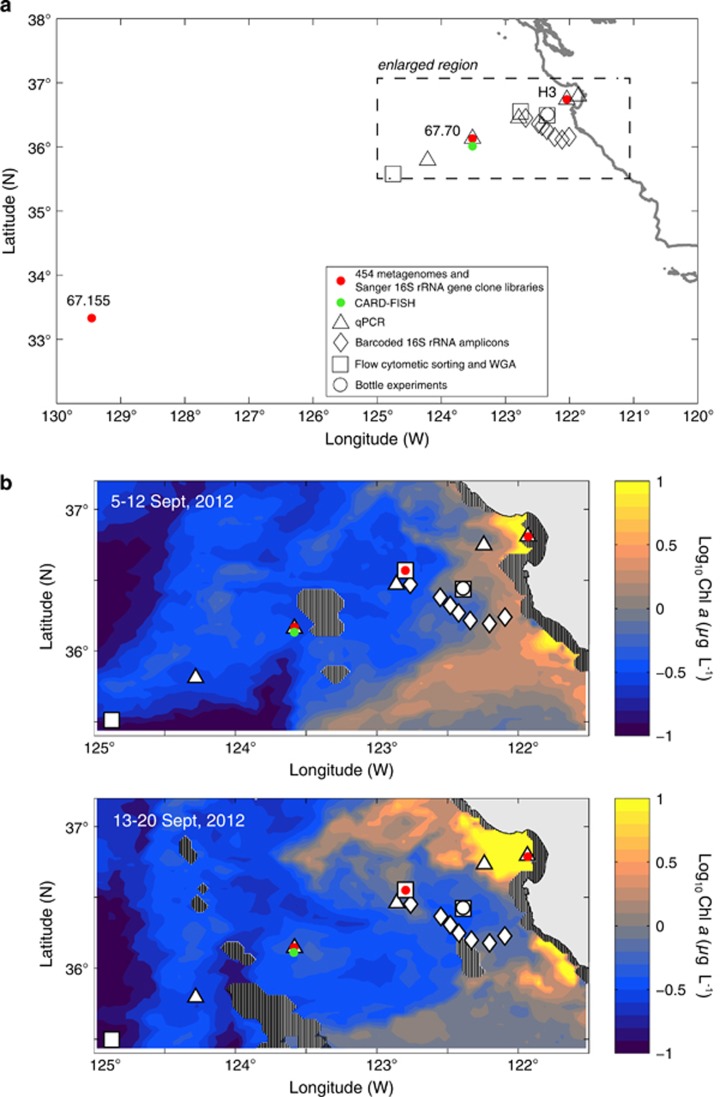
Seawater was collected from established stations along, and in the vicinity of, the California Cooperative Fisheries Investigations Line 67 during three cruises aboard the R/V Western Flyer: October 2007 (cruise CN207), September 2012 (cruise C0912) and October 2013 (cruise CN13ID) (Figure 1, Supplementary Table S1). From the CN207 cruise, broad survey samples were collected and analyzed, specifically metagenomic samples sequenced as part of another study (Monier et al., 2012) and the 16 S rRNA gene clone libraries generated here. During C0912, experiments and targeted samples building on the survey results were collected for quantitative polymerase chain reaction (qPCR), flow cytometric cell sorting, incubation experiments and high-throughput Illumina-based 16 S rRNA gene barcoded sequencing. During CN13ID, a sample was taken at station 67–70 from a depth of 20 m for microscopy and catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH). For all cruises, sampling and hydrographic profiling were conducted using a conductivity-temperature-depth rosette sampler (Sea-Bird Electronics) equipped with (12) 10 l Niskin-type sample bottles. In addition to standard conductivity-temperature-depth measurements, the sensor package also included a C-Star transmissometer (WET Labs) and a WETstar fluorometer (WET Labs).
Figure 1.
Sampling locations for the different data types analyzed in this study for (a) all sampling sites and data collected, and (b) enlarged maps of selected sample sites and data collected from the 2012 cruise, superimposed on 8-day composite satellite ocean color data (MODIS-Aqua Level 3 data, 4-km resolution, obtained from oceancolor.gsfc.nasa.gov) from the dates immediately preceding (September 5–12) and during (September 13–20) the cruise. See Supplementary Table S1 for number of sequences obtained from the different samples and size fractions. Cross-hatching represents missing data due to cloud cover.
Metagenomic samples from the CN207 cruise were collected as described previously (Monier et al., 2012). In brief, ∼ 200 l of water were filtered first through a 20-μm mesh, and then filtered sequentially via pressure filtration through a 293-mm diameter, 3-μm-pore-size filter (Pall Versapor-3000T, Cortland, NY, USA), 0.8-μm-pore-size filter (Pall Supor-800), and finally a 0.1-μm-pore-size filter (Pall Supor-100). For qPCR analysis, seawater samples (1–2 l) from the C0912 cruise were serially size fractionated via pressure filtration through 10-μm, 3-μm, 0.8-μm and 0.2-μm-pore-size filters (25-mm diameter) by in-line filtration through Supor (Pall, 0.2-μm and 0.8-μm-pore-size) or polyester (GE Osmonics, Watertown, MA, USA, 3-μm and 10-μm-pore-size) membrane filters housed polypropylene filter holders (Swin-Lok, Whatman). Samples for high-throughput 16 S rRNA gene V4 Illumina-based barcoded sequencing were prefiltered through a 10-μm-pore-size polyester prefilter (GE Osmonics) prior to collection on a 0.2 μm Supor (Pall) filter. Filters were immediately frozen at −80 °C until further processing.
Discrete measurements of macronutrient concentrations (NO3−, NO2−, SiO44−, PO43−) were determined on whole seawater samples, frozen at sea and brought ashore for subsequent measurement by segmented flow analysis (Sakamoto et al., 1990). NH4+ concentrations in whole seawater samples were determined at sea by fluorometric detection following treatment with borate-phthaldialdehyde-sulfite buffer (Holmes et al., 1999). Chlorophyll a (Chl a) pigment concentrations were determined by fluorometry using established protocols (Pennington and Chavez, 2000).
Flow cytometry, whole-genome amplification and rRNA gene screening
Populations of chlorophyll-containing picoeukaryotes were sorted at sea on the basis of forward scatter and red fluorescence (692 nm) intensity using a flow cytometer equipped with a 200 mW, 488-nm laser (Becton Dickson Influx, Franklin Lakes, NJ, USA). Three populations of picoeukaryotic phytoplankton were sorted on the basis of gates drawn on forward scatter and red fluorescence plots. Heterotrophic picoeukaryotes were stained using Lysotracker Green (Life Technologies Molecular Probes, Grand Island, NY, USA), prior to sorting on the basis of gates drawn on red (692 nm) and green fluorescence (531 nm). For staining, whole seawater samples were 10 × concentrated via gravity filtration through a 47 mm, 0.8-μm-pore-size Supor filter (Pall). Concentrated cells were stained for 10 min in the dark at a final concentration of 100 nM Lysotracker Green.
For each chlorophyll-containing eukaryote population, 50–300 cells were sorted into 2 ml sterile cryotubes (six replicate tubes per population) and frozen at −80 °C. For cells stained with Lysotracker Green, 300 cells were sorted (six replicate tubes). Multiple displacement amplification (MDA) was performed using the RepliG Midi kit (Qiagen) according to the manufacturer's instructions. To reduce founder effects, the reaction was split into five equal aliquots prior to amplification, and pooled after amplification. Two negative controls (no template DNA added) were run with every amplification reaction. In addition, a contamination control for each sort was performed that contained ‘sorted' sheath fluid as template for the MDA reaction. Amplifications were monitored with 0.5 μl SYBR green (1:50 dilution) for 3–12 h on a CFX96 real-time PCR machine (Bio-Rad, Hercules, CA, USA) and terminated at the first sign of fluorescence increase in the negative control. No sheath fluid samples or negative controls exhibited MDA amplification after 16 h of amplification at 30 °C. The MDA success rate was 64% (n=25). MDA products were screened for the presence of archaeal 16 S rRNA genes using three different primer sets: 21F (5′-TTCCGGTTGATCCYCCCGGA- 3′)/958R (5′-YCCGGCGTTGAMTCCAATT) (Delong, 1992), 21F/1391R (5′-GACGGGCGGTGTGTRCA-3′) (Delong, 1992; Reysenbach and Pace 1995) and 554F (5′-GTCGMTTTTATTGGGCCTAA-3′)/1391R (Reysenbach and Pace 1995; Massana et al., 1997).
Bottle experiments
Water was collected from 10 m depth at station 67–60 (36.74° N, 122.02° W) on the C0912 cruise and sampled from Niskin bottles directly into acid-washed 4 l polycarbonate bottles. Two additional 2 l samples were taken directly from the Niskin and filtered as described above as time zero samples. Duplicate treatments were prepared for controls (no addition) and experimental treatments (addition of axenic Micromonas pusilla CCMP 1545, final concentration ∼103; ml−1). Bottles were incubated in dark for 72 h in an on-deck incubator with flowing surface seawater. At the conclusion of the experiment, size-fractionated samples were collected for DNA by in-line pressure filtration through 3-μm, 0.8-μm and 0.2-μm–pore-size filters (as described above). DNA extraction and quantification of MGII 16S rRNA genes using qPCR was performed as described below. Growth rates for free-living and particle-attached MGII were calculated using the semi-log equation from Button (1985).
Catalyzed reporter deposition fluorescence in situ hybridization
We utilized the Eury806 horseradish peroxidase-labeled oligonucleotide probe originally reported by Teira et al. (2004), which targets marine Euryarchaeota. The CARD-FISH protocol generally followed that of Teira et al (2004). In brief, seawater was fixed with paraformaldehyde at a final concentration of 2%. A fixed sample (15 ml) was filtered onto 0.2 μm polycarbonate filters (Millipore, Temecula, CA, USA) and washed once with molecular grade water before freezing at −80 °C. An enrichment culture of thaumarchaea (Santoro and Casciotti, 2011) was used as a negative control for the Eury806 probe, and a successful CARD-FISH protocol was confirmed by using the Arch915 probe (Stahl and Amann, 1991) with the same culture. After thawing filters, thin slices were cut out with a sterile scalpel and overlain with ∼10 μl low melting point agarose (SeaPlaque, Lonza, Basel, Switzerland). For hybridizations, a hybridization buffer with 20% formamide was used for a 12 h hybridization at 35 °C. Amplification using Alexa488 (Life Technologies Molecular Probes) conjugated tyramides was carried out at 37 °C for 1 h. After amplification, filter sections were then washed with a mixture of 1 × phosphate-buffered saline solution amended with 0.05% Triton X-100 (PBS-T), followed by two rinses with molecular grade water. Filters were washed for 30 min in 100% ethanol prior to drying, counterstaining with 4,6-diamidino-2-phenylindole, and mounting for microscopy. Hybridized filter sections were imaged with a Zeiss AxioImager 2 epifluorescence microscope equipped with an Apotome.2. Images were captured with a Photometrics CoolSNAP HQ2 CCD camera.
DNA extraction
DNA for Sanger 16 S rRNA gene amplification and 454 metagenomic sequencing was extracted from samples collected during CN207 as previously described (Monier et al., 2012). For qPCR, DNA from size-fractionated filters was extracted according to a protocol previously described (Santoro et al., 2010). In brief, 850 μl of a sucrose ethylenediaminetetraacetic lysis buffer (0.75 M sucrose, 0.05 M Tris-Base, 0.02 M ethylenediaminetetraacetic, 0.4 M NaCl, pH 9.0) and 100 μl of 10% sodium dodecyl sulfate were ultraviolet treated for 30 min and then added to 2 ml gasketed bead-beating tubes containing the filters and 0.1 mm sterile glass beads (Biospec). Bead beating was performed for 1 min and the samples were subsequently heated at 99 °C for 2 min. After heating, 25 μl of 20 mg ml−1 proteinase K was added and tubes were incubated at 55 °C overnight. DNA was extracted and purified from the lysate using the DNeasy Blood and Tissue Kit (Qiagen). Extracts for 16 S rRNA gene-barcoded amplicon sequencing were processed in a similar manner with a slightly modified lysis protocol (Smith et al., 2014).
16 S rRNA gene amplification, cloning and Sanger sequencing
A region of the bacterial and archaeal 16 S rRNA gene was amplified using the universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 1391R (5′-GACGGGCGGTGTGTRGA-3′) and cloned into the pCR2.1 vector using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA, USA). Approximately 5000 clones were picked and sequenced bi-directionally using Big Dye Terminator chemistry on an AB3730xl sequencer (Applied Biosystems, Waltham, MA, USA) using the M13F and M13R sequencing primers.
Sanger-based 16 S rRNA gene sequences were clustered into operational taxonomic units (OTUs) using the Quantitative Insights Into Microbial Ecology software package (Caporaso et al., 2010) at 97% sequence identity using UCLUST (Edgar, 2010). OTUs were identified as MGII through Basic Local Alignment Search Tool for nucleotides (BLASTn) searches against the SILVA database (Pruesse et al., 2007). Alignments of MGII 16S rRNA genes using OTU sequence representatives were performed in ARB (Ludwig et al., 2004) using the FastAligner function, followed by manual curation of the alignment. Phylogenetic analyses were performed in SeaView (Gouy et al., 2010) and Mr Bayes (Huelsenbeck and Ronquist, 2001). PhyML and Bayesian analyses of 120 MGII 16 S rRNA gene sequences were constructed using an alignment of 719 unambiguous positions under the GTR+I+Gamma model of sequence evolution. For PhyML trees, bootstrapping was performed on 1000 replicates. The Bayesian analysis consisted of two independent runs with five million generations. Trees were sampled every 1000 generations with 25% discarded as burn-in. Topologies of maximum likelihood and Bayesian trees were compared and the tree with the best log likelihood was chosen for presentation. The Sanger-sequenced 16 S rRNA gene sequences have been deposited in GenBank under accession numbers KJ608270–KJ608349.
Quantitative PCR
MGII 16 S rRNA gene abundance was quantified using a modified assay from a previously published TaqMan qPCR assay for MGII (Suzuki et al., 2000). We compared the 120 Sanger-sequenced MGII 16S rRNA gene sequences obtained from the CCS in this study (see above) to the previously published primers using the probe design and probe match tools in ARB (Ludwig et al., 2004). The original MGII333F primer reported by Suzuki et al. (2000) was found to be biased against the MGII.D clade in our CCS data set, and we were able to increase the coverage of this clade by adding a degeneracy position (in bold) to the 5th base (ARCHGII333Fmod: 5′-GAGAYGGATTCTGAGACACGAA-3′). The original reverse primer reported by Suzuki et al. (ARCHGII554R: 5′-TTAGGCCCAATAAAAKCGAC-3′) was modified with a degeneracy position at the 4th base (in bold) (ARCHGII554R: 5′-TTASGCCCAATAAAAKCGAC-3′). ARCHGII554Rmod was used together with ARCHGII333Fmod with the original TM519AR TaqMan probe reported by Suzuki et al.
qPCR reactions were carried out in 25 μl volumes on a CFX96 real-time PCR machine (Bio-Rad). Prior to qPCR, the concentration of DNA template in each sample was quantified using Picogreen dye (Life Technologies Molecular Probes) and a fluorometric plate reader (Bio-Tek Synergy HT, Winooski, VT, USA). The DNA in each sample was then diluted to equimolar concentrations prior to qPCR. The reaction chemistry for each qPCR reaction was as follows: 12.5 μl TaqMan Environmental Master Mix 2.0 (Applied Biosystems), 400 nM of each primer, 320 nM of probe and ∼5 ng of DNA template. A gel-purified amplicon was quantified in triplicate using QuantiT BR reagent (Life Technologies) and used as a standard. All reactions were run in triplicate in 96-well plates with a standard curve spanning ∼101–105 templates. Two negative control reactions were run for each plate, which did not exhibit amplification. Standard curves were run and analyzed in duplicate for each amplification run. The thermal cycling profile for the reaction consisted of 35 cycles of 95 °C for 15 s and 60 °C for 30 s, with a plate read step at 68 °C for 10 s. The average qPCR efficiency was 91%, and the R2 values of the standard curves were >0.99. All qPCR reactions were prepared using an epMotion automated liquid handler (Eppendorf, Hamburg, Germany). Correlations of MGII 16S rRNA gene abundance with distance from shore were performed in R (http://vegan.r-forge.r-project.org/) and Chl a data were plotted and contoured using Ocean Data View version 4.5.6 (Schlitzer, 2013) using weighted-average gridding.
Analysis of MGII distribution in barcoded amplicon libraries
Barcoded 16 S rRNA gene PCR amplification and library construction (including quality control) for Illumina (San Diego, CA, USA), MiSeq sequencing were done at the Joint Genome Institute using standard protocols, as reported by (Lindemann et al., 2013) using the universal 16 S primers 515F and 806R, targeting the V4 region. In brief, paired-end reads were assembled using FLASH (Magoc and Salzberg, 2011). Using a sliding window of 10 bp all sequences with a quality score of <33 were removed, as well as any sequence containing an ambiguous nucleotide position. Sequences were checked for chimeras using UCHIME (Edgar et al., 2011) and removed prior to further processing of the data set. Gene sequences of 16 S rRNA passing quality control (n=8,343,259) were clustered into OTUs (n=13,089) in Quantitative Insights Into Microbial Ecology (Caporaso et al., 2010) at 97% sequence identity using UCLUST (Edgar, 2010). OTUs were annotated against the SILVA database (Pruesse et al., 2007) through Basic Local Alignment Search Tool for nucleotides (BLASTn) searches. MGII OTU abundance in barcoded amplicon libraries was rarified across each sample (to the sample with the least number of MGII sequences: site 67.155 80 m (n=208)) by rarifying the OTU table and matrices of rarified MGII OTU abundance in the size-fractionated samples were used as input for the Non-Metric Multidimensional Scaling and Analysis of Similarity (ANOSIM) analyses. ANOSIM was performed with 999 permutations, and both Non-Metric Multidimensional Scaling and ANOSIM were performed using a Bray-Curtis distance metric. The 95% confidence intervals for Non-Metric Multidimensional Scaling were calculated and plotted using the ‘ordiellipse' command in R using the vegan package (http://vegan.r-forge.r-project.org/). MiSeq data are deposited in the NCBI Short Read Archive (BioProject ID PRJNA237060).
Metagenomic analyses
Library preparation and sequencing methods for the 454-FLX metagenomic data sets were reported previously (Monier et al., 2012). Reads from size-fractionated metagenomes were searched for homology to genes in a closed MGII genome originated from Puget Sound (Iverson et al., 2012) through BLASTx searches (e−5,>60% amino-acid identity). However, because MGII reference genomes are limited to this single genome (which was assembled from a metagenome) some MGII diversity was likely missed by this metagenomic approach. To account for biases in MGII representation as a result of different MGII population sizes in the different size fractions, the data were normalized against the average abundance of a suite of universally conserved single copy genes (SCG) (Kawai et al., 2014). The number of reads mapping to each open-reading frame in the MGII reference genome were normalized to the average abundance of homologs to 35 universal SCG in each sample. Significant differences in open-reading frame recruitment between size fractions were calculated in R using a Kruskal–Wallis significance test with a Bray-Curtis distance metric using a significance threshold of P<0.05. To further increase the stringency of our significance threshold, we only considered genes with a P-value<0.05 to be significant if they had at least a twofold increase in relative coverage. Fold increase in POM-attached MGII gene coverage was calculated as the ratio of the average SCG-normalized coverage in each POM fraction to the average SCG-normalized coverage in free-living fractions. Fold increase in free-living MGII recruitment was calculated as the ratio of the average normalized coverage of free-living MGII homologs to their average normalized coverage in POM fractions. Metagenomic reads with homology to euryarchaeal fosmids (other than the closed assembly reported by Iverson et al.) were identified via BLASTx (e−5, >60% amino-acid identity) searches against the M5nr database (Wilke et al., 2012). These fosmids derive from previous water column metagenomic studies (Beja et al., 2000a, 2000b; Moreira et al., 2004; Frigaard et al., 2006; Martin-Cuadrado et al., 2008; Ghai et al., 2010). Annotated reads were then subjected to BLASTx searches against the Clusters of Orthologous Genes database. ANOSIM was performed with 999 permutations using a Bray-Curtis distance metric.
Proteorhodopsin gene assembly and phylogenetic analysis
Metagenomic reads with homology to proteorhodopsin were identified via BLASTx searches against a custom database of proteorhodopsin protein sequences. Homologous reads were assembled into contigs, using their closest relative's DNA sequence as scaffolds in CLC Genomics Workbench 6.0. Chimeric assemblies of proteorhodopsin genes were checked by BLASTx searches of random sections of the contig, to make sure that the same subject was obtained for each section (query) of the contig. Consensus scaffolds were translated into amino-acid sequences using a six reading frame translation and aligned against their closest homologs in SeaView (Gouy et al., 2010). The alignment was manually curated for accuracy, and only conserved portions of the alignment that could be reliably aligned were used for the phylogenetic analysis. Phylogenies were performed using BioNJ (neighbor-joining distance) in SeaView with 1000 bootstrap replicates.
Results and discussion
Defining free-living and particle-attached MGII
Here, we define ‘free-living' MGII as those found in the<0.8-μm-pore-size fraction, and ‘attached' MGII as those found in fractions>0.8 μm. To validate assignment of the>0.8 μm fractions as POM-attached and not an artifact of a reduction in the effective pore size of the larger pore size filters, we examined size-fractionated distributions of known free-living bacterial groups, specifically members of the SAR11 and SAR86 clades, (Giovannoni et al., 1990; DeLong et al., 1993) using 16 S rRNA gene clone libraries (Supplementary Figure S1). As expected, SAR11 OTUs were markedly more abundant in the free-living fraction (Supplementary Figure S1A). SAR86 OTUs were also more abundant in the free-living fraction (Supplementary Figure S1B). In contrast, the majority of MGII OTUs were overrepresented in the >0.8-μm fractions (Supplementary Figure S1C). The different distributions of SAR11, SAR86 and MGII across the three fractions suggest that free-living cells<0.8 μm in size are not significantly represented in >0.8-μm fractions due to filter clogging. We acknowledge, however, that some presumably free-living prokaryote taxa may also be detected within the >0.8-μm fractions (indeed some SAR86 and SAR11 OTUs were detected there), including larger cells such as the picocyanobacterium Synechococcus (Olson et al., 1990; Sudek et al., 2015).
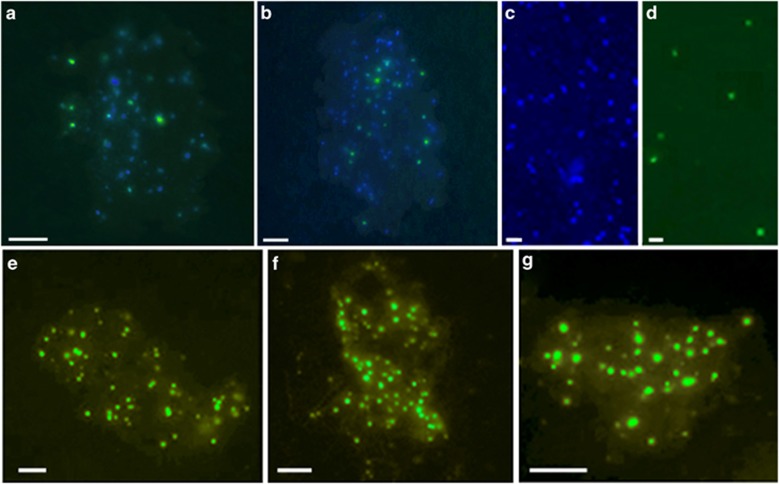
The presence of euryarchaeal cells physically attached to POM in the CCS was also confirmed by CARD-FISH and microscopy (Figure 2a, b and e–g). All free-living and particle-attached euryarchaea observed were <0.8 μm in diameter (Figure 2a–g), and dense populations of euryarchaea were observed on particles (2–75 μm in size) (Figure 2e–g). This provides additional evidence beyond size-fractionated filtration that MGII are found in physical association with POM.
Figure 2.
Epifluorescence microscopy of marine euryarchaeal cells (Station 67–70, 20 m water depth) hybridized with the Eury806 CARD-FISH probe after amplification with tyramide-Alexa488. (a, b) Euryarchaeal cells (green) and DAPI-stained bacteria (blue) physically attached to particles (2-D overlay of same image). (c, d) Free-living microbial cells stained with DAPI (c), and the same field of view showing hybridization of the Eury806 CARD-FISH probe to MGII cells (d). (e–g) Dense populations of MGII cells (green) attached to detrital particles (brown). Images were captured in black and white. Green color was assigned to pixels with grey-values >6000 on images captured with the green fluorescence filter set (FITC). Brown color (particles) was assigned to pixels with grey-values in the range of 4000–2000 on images captured with the FITC filter set (e.g., lower than the cells and higher than the background). Scale bars=a, b: 5 μm, c, d: 1 μm, e–g: 4 μm.
Quantitative distribution of MGII in size-fractionated samples
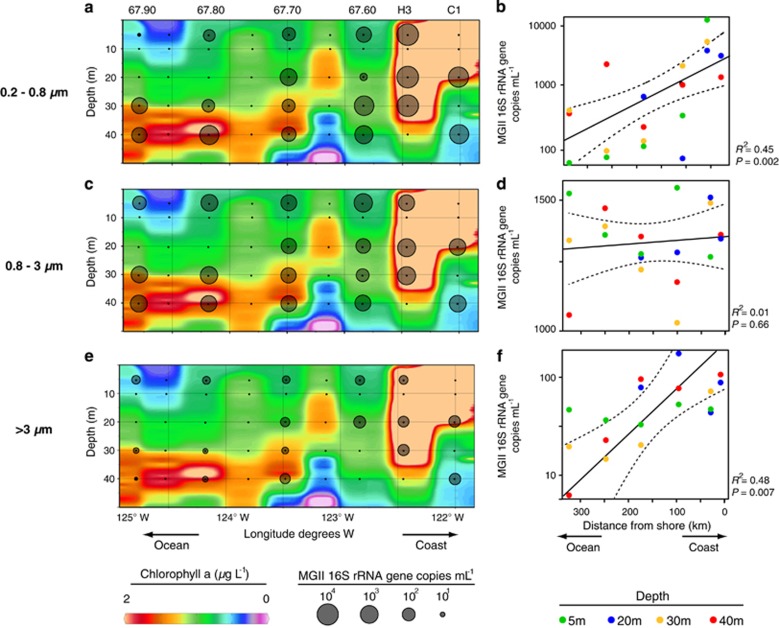
We used qPCR to quantify the abundance of MGII in size-fractionated plankton samples from the CCS. The qPCR assay modifications described above (see Materials and methods) increased the number of MGII captured based on comparisons with both our CCS 16 S rRNA gene data set and the SILVA 16 S rRNA database. In the CCS data, the percent of MGII captured by the qPCR assay increased from 82 to 90%, and increased from 78 to 83% for MGII in the SILVA database. Using the modified primers, MGII were detected at all stations, depths and size-fractions, underscoring their ubiquitous presence in the upper marine water column (Figure 3, Supplementary Table S2). At sites near the coast, the abundance of free-living MGII 16 S rRNA genes was orders of magnitude higher than in POM-attached fractions, reaching a maximum abundance of 3.2 (± 0.04) × 104 genes ml−1 at 20-m depth at station H3 (Figure 3a and b). The abundance of free-living MGII 16 S rRNA genes across all depths decreased exponentially with distance from shore (R2=0.45, P=0.002, Figure 3a and b), and correlates significantly with Chl a (Figure 3a) (least squares regression: P<0.01, R2=0.49). Offshore, however, MGII 16 S rRNA gene abundance was highest in the 0.8–3 μm fractions (5.9 (± 0.22) × 103 ml−1; t-test: P<0.05; Figure 3c and f) at most depths at stations 67–70, 67–80 and 67–90. This suggests that most MGII were physically associated with particles in the 0.8–3-μm size range in offshore waters. Furthermore, at 5 m at station 67–90 and 20 m at 67–90 there were more MGII detected in the >3-μm fraction compared with the free-living fraction (Figure 3b and f), suggesting that compared with free-living MGII, there were more MGII cells attached to particles>3 μm at these locations.
Figure 3.
Abundance of MGII 16 S rRNA genes in three size-fractions along a 300 km transect extending into and beyond the California Current System (CCS) measured using qPCR. (a, b) 0.2–0.8 μm, (c, d) 0.8–3 μm, (e, f)>3 μm. (a, c, e) Background contour shows the concentration of chlorophyll a (μg l−1), interpolated using the weighted-average gridding algorithm in Ocean Data View (http://odv.awi.de; version 4.5.6). The diameter of each black circle is in proportional to the abundance of MGII 16 S rRNA genes ml−1. (b, d, f) Points (n=19) represent mean values across three technical replicates and dashed lines represent 95% prediction interval. Note the differences in y axis scales. See Supplementary Table S2 for raw qPCR data.
There was an exponential (P<0.007, R2=0.48) decrease in MGII abundance in the >3-μm size fraction with increasing distance from shore (Figure 3f), which is consistent with the exponential decrease in concentration and size of large particles (4–100 μm) with distance from shore (Hood et al., 2007). However, MGII attached to smaller particles in the 0.8–3 μm size range did not decrease with increased distance from shore (Figure 3c and f). Similarly, MGII at shallower depths (5 and 20 m) in the >3-μm fractions also did not decline markedly with increased distance from shore (Figure 3f).
Influence of depth, geography and POM on MGII distributions
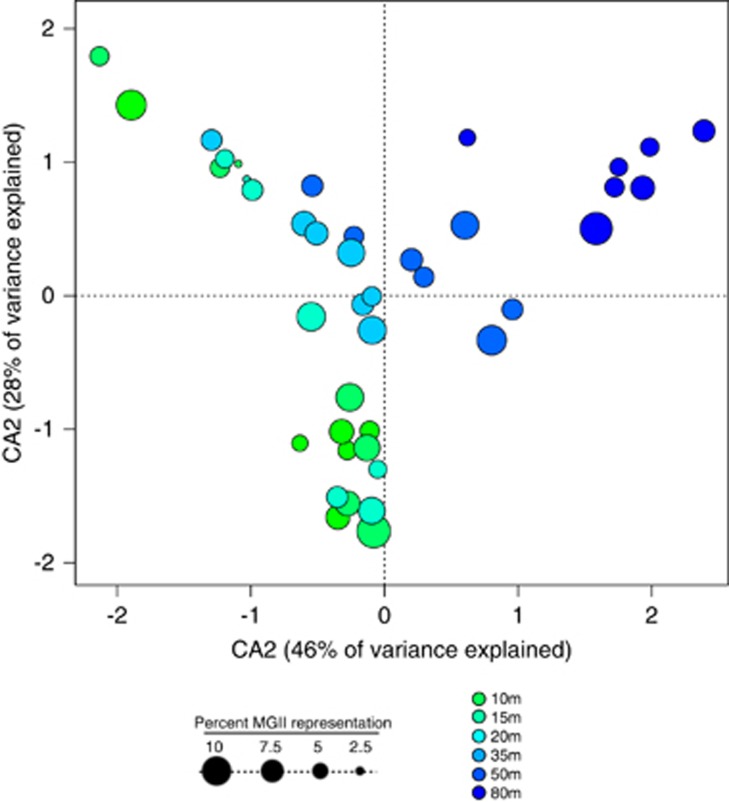
To investigate the spatial ecology of MGII across the CCS, we analyzed distributions of both Illumina-sequenced barcoded MGII 16 S rRNA gene amplicons and Sanger-sequenced MGII 16 S rRNA genes. Barcoded Illumina sequencing results indicate that MGII are a significant relative fraction of the total community, representing on average 7% (± 5%) of total sequences per sample (Figure 4). The relative abundance of MGII in the amplicon libraries should be interpreted with some caution, as a recent analysis suggests that SAR11 genotypes, which should make up a large fraction of the surface microbial community, are underrepresented by the 515F–806R primer set (A. Apprill, personal communication), potentially leading to a large overestimation of the relative abundance of MGII in our samples. The relative abundance of MGII in the amplicon libraries, however, is within the range of relative MGII abundance reported from in situ metatranscriptomes from central California coast waters (Ottesen et al., 2013). Multivariate statistical analysis demonstrates that different depths contain discrete MGII phylotypes (ANOSIM: P=0.001) with adjacent water depths containing more similar MGII populations (Figure 4). This gradient in MGII community structure through the euphotic zone correlates significantly with environmental parameters that covary with depth, such as salinity, temperature and macronutrient concentrations including particles (using beam attenuation as a proxy) (Supplementary Table S3). These data suggest niche specificity within the pelagic MGII archaea, and the partitioning of MGII ecotypes across relatively short vertical distances (∼80 m) in the water column. Shallow MGII ecotypes (Figure 4) may be able to maintain population size on particles under oligotrophic conditions (Figure 3e and f).
Figure 4.
Canonical correspondence analysis of MGII 16 S rRNA genes from barcoded amplicon libraries (Illumina MiSeq) across depth gradients in the CCS. Samples are represented in the biplot by dots, the size and color of which indicate MGII relative abundance and water depth, respectively. Correlations of MGII OTUs with geochemical measurements are presented in Supplementary Table S3.
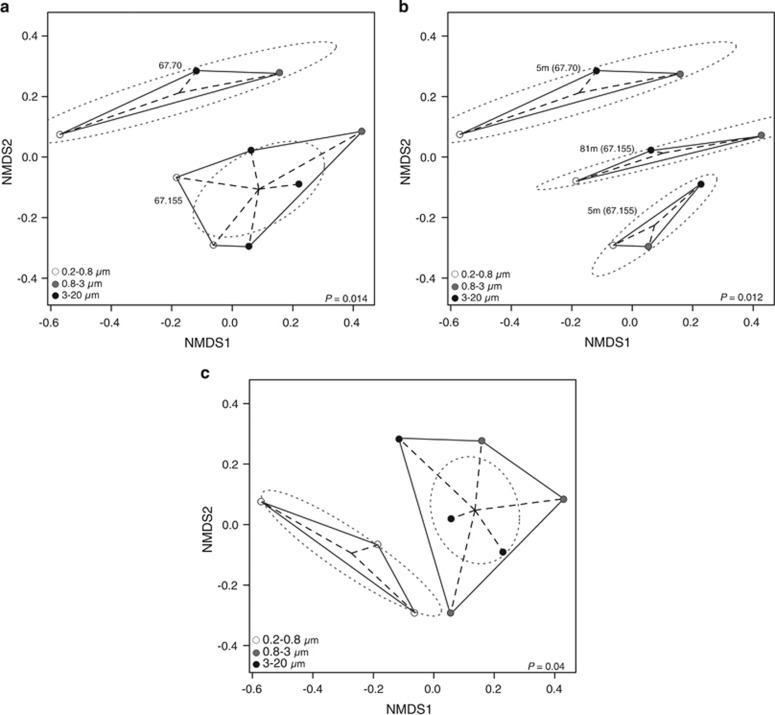
Based on the Sanger 16 S rRNA gene data from only sites 67–70 and 67–155, MGII community composition differs significantly by site (Figure 5a; ANOSIM: P=0.014) and water depth (Figure 5b; ANOSIM: P=0.012) suggesting a partitioning of MGII communities between mesotrophic and oligotrophic offshore waters. Similar observations of non-random MGII distributions have been reported from the Arctic and Columbia River (Crump and Baross, 2000; Wells et al., 2006) and through seasonal cycles at coastal time series sites (Galand et al., 2010; Hugoni et al., 2013). No significant differences were observed when samples from the near-shore station H3 were included in the analysis because non-metric multidimensional scaling could only explain <5% of the variation. The inability of our statistical methods to resolve differences between near-shore (H3) and off-shore (67–70 and 67–155) MGII communities may stem from increased community complexity when considering both communities simultaneously.
Figure 5.
Multivariate statistical analysis and visualization of size-fractionated, Sanger sequenced MGII 16 S rRNA genes. Plots show significantly different MGII 16 S rRNA gene OTUs found at different (a) geographic locations (ANOSIM: P=0.014), (b) water depths (ANOSIM: P=0.012), and (c) size fractions (ANOSIM: P=0.04). Dotted lines represent 95% confidence intervals based on OTU point scores.
MGII are represented by four previously defined subclades: MGII.A, MGII.B, MGII.C and MGII.D (Massana et al., 2000; Hugoni et al., 2013). The MGII.A clade dominated the 16 S rRNA clone libraries in the CCS and was most abundant at station 67–70 and in fractions >0.8 μm (Supplementary Figure S2). This finding is consistent with an archaeal 16 S rRNA survey of Arctic waters that also detected MGII.A in particle associated fractions (Galand et al., 2008) and strong seasonal patterns showing a high abundance of MGII.A during summer (Hugoni et al., 2013). Most sub-clades of MGII.A and MGII.C (6/9 and 6/10, respectively) were relatively more abundant in 16 S libraries from >0.8 μm fractions. No sequences affiliated with the MGII.B clade were detected in our Sanger data set. Sequences affiliated with the MGII.D clade were detected exclusively in the 3–20 μm fractions (Supplementary Figure S2), whereas no MGII clades were detected exclusively in the free-living fractions. Based on the clone library data, free-living and POM-attached MGII populations are significantly different (ANOSIM: P=0.04, Figure 5c). Thus, just as particle-attached bacterial populations are genetically distinct (DeLong et al., 1993; Hunt et al., 2008; Shapiro et al., 2012), POM-attached MGII populations are evolutionarily divergent from free-living MGII.
Richness of POM-attached MGII in the CCS is an order of magnitude higher (Supplementary Figure S3) than marine Arctic samples (Galand et al., 2008), and the mean alpha diversity (based on the Shannon Index) of POM-attached MGII is greater, relative to free-living MGII (Supplementary Figure S3). This suggests that POM can support MGII communities that are as, if not more, diverse than those found in free-living fractions. A more diverse community of MGII on POM is consistent with a size-fractionated metagenomic study of an oxygen minimum zone that found an overall increase in microbial diversity on particles (Ganesh et al., 2014). Diverse POM-attached MGII ecotypes could partly explain the large cell-to-cell variability in MGII transcriptional responses to environmental cues (Ottesen et al., 2013).
The metabolic profile of POM-attached MGII is distinct from free-living MGII
Metabolic adaptations to particle attachment at the community level are only beginning to be understood (Hunt et al., 2008; Shapiro et al., 2012; Yawata et al., 2014), and taxon-specific comparisons of POM-attached versus free-living ecotypes are lacking. Given the distinct quantitative and spatial ecology of free-living and POM-attached MGII discussed above, they are logical targets for such analyses. To better understand the difference in metabolic potential between free-living and POM-attached MGII, we mapped metagenomic data obtained from size-fractionated samples (Monier et al., 2012) to a closed assembly representing a genome of an uncultivated MGII taxon from Puget Sound (Washington, USA; Iverson et al., 2012). Six percent of total metagenomic reads (n=298 001) were recruited to the Puget Sound MGII genome (>60% amino-acid identity) with hits to 77% of genes (n=1694) in the genome. The mean (±s.d.) number of reads mapping to the MGII genome in metagenomes from the free-living, 0.8–3 μm and 3–20 μm fractions was 9118 (±3051), 3220 (±1252) and 1801 (±678). The lower raw read coverage in the POM fractions likely reflects both the lower absolute abundance of MGII and higher relative abundance of large eukaryotic genomes in the larger fractions. Consistent with this interpretation, the mean (±s.d.) SCG abundance in metagenomes from the free-living, 0.8–3 μm and 3–20 μm fractions was 18 (±5), 7.8 (±4.5), 2.1 (±0.7), respectively. MGII homologs to 85% of the 35 SCG were detected in the free-living and 0.8–3 μm fractions. Seventy-three percent of the SCG was detected in the metagenomes from 3–20 μm fractions, indicating lower MGII genome coverage in the largest size fraction.
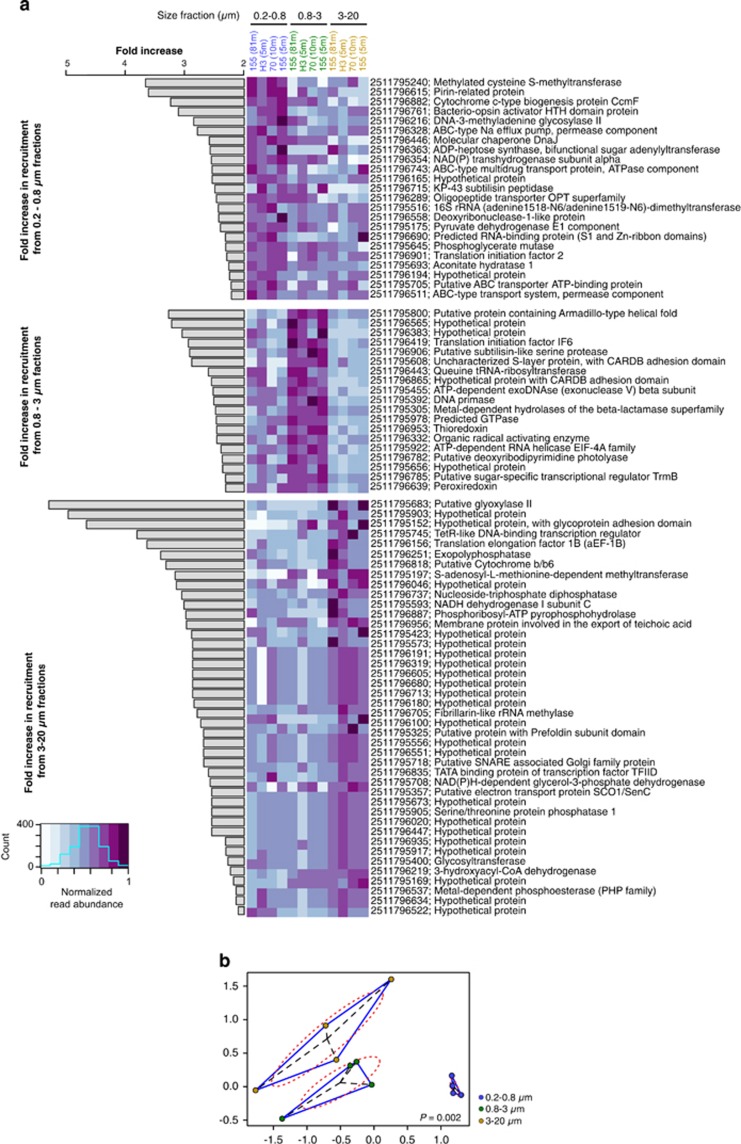
After normalizing metagenomic read recruitment to SCG abundance, 23 homologs were significantly overrepresented exclusively in the free-living fractions, 19 in the 0.8–3-μm fractions, and 42 in the 3–20-μm fractions (Figure 6a). Because the data are normalized to SCG abundances, these significant differences are likely a function of variation in copy number of MGII gene homologs in the different size fractions (as opposed to variation in cell abundances). Thaumarchaeota are unlikely to recruit to the MGII genome in our analysis as even the most highly conserved archaeal core genes between Nitrosopumilus maritimus and the MGII genome share <50% amino-acid identity (Iverson et al., 2012). A high number of significantly overrepresented MGII homologs (25%) in free-living metagenomes encode components of ABC transmembrane transporters (Figure 6a). In contrast, the majority (59%) of MGII homologs significantly overrepresented in POM fractions encode hypothetical proteins of unknown function (Figure 6a), suggesting that many novel proteins are important for MGII metabolism of, and attachment to, POM. Indeed, three of these hypothetical proteins (IMG# 2511795152, 2511795608 and 2511796865) contain conserved domains involved in cell adhesion to surfaces (Figure 6a), supporting a previous study that demonstrated genes encoding adhesion proteins are selected for in some particle-attached populations of Vibrio cyclitrophicus (Shapiro et al., 2012). Homologs to many other MGII hypothetical proteins containing calcium-dependent adhesion (adhesin) domains were also detected in POM fractions (Supplementary Table S4). MGII homologs encoding transcriptional regulators and proteins involved in lipid catabolism were only significantly (Kruskal–Wallis test: P<0.05) overrepresented in>0.8-μm fractions (Figure 6), further highlighting genomic and potential metabolic differences between free-living and POM-attached MGII. We acknowledge that the limited genomic reference data for MGII and lower MGII abundance in>0.8 μm fractions have likely caused some MGII genes to be missed in the metagenomic data, leading to potential errors in calculations of overrepresented genes. Despite these caveats, the genomic differences we report among the different size fractions appear to represent adaptations to these contrasting chemical environments.
Figure 6.
(a) Fold increase in recruitment (see Materials and Methods) of significantly different homologs (fold increase>2, P<0.05) to the Puget Sound MGII genome (Iverson et al., 2012) across size-fractionated metagenomes (blue=0.2–0.8 μm, green=0.8–3 μm, yellow=3–20 μm). Sample IDs are present at the top of the heatmap and are grouped and color-coded by size fraction. IMG database identifiers (integer strings) for gene products are specified in front of the gene product descriptions. Relative to free-living fractions, particle-associated fractions contain more overrepresented homologs coding for enzymes involved in hydrolysis of high molecular weight substrates (exopolyphosphatase, metal-dependent phosphoesterase and 3-hydroxyacyl-CoA dehydrogenase), transcription factors (TetR, TFIID and TrmB) and hypothetical proteins containing surface adhesion domains. (b) Non-metric multidimentional scaling (NMDS) of homologs to previously sequenced MGII fosmids in the size-fractionated metagenomes. Blue lines connect size fraction samples, dashed red lines represent 95% confidence intervals based on OTU point scores, and dashed black lines connect sample points to their centroid (mean of point scores) value.
Contrasting ecophysiologies between free-living and particle-attached MGII may underlie the genomic differences observed here. POM-attached MGII appear to have an increased capacity for transcriptional regulation as evidenced by higher copy number of transcription factors TetR, TFIID and TrmB (Figure 6a). This may reflect a need to respond to the variable composition of POM. The significant increase in MGII homologs encoding proteins responsible for catabolism of fatty acids and lipids (metal-dependent phosphoesterase and 3-hydroxyacyl-CoA dehydrogenase) and polyphosphate (exopolyphosphatase) in metagenomes from >0.8-μm fractions (Figure 6a) points toward an increased MGII metabolic capacity for degradation of high molecular weight substrates on POM. Homologs to many additional genes involved in fatty-acid degradation pathways were detected in the 3–20-μm fractions including two acyl-CoA dehydrogenases, three acetyl-CoA acetyltransferases, an enoyl-CoA hydratase and a glutaryl-CoA dehydrogenase (Supplementary Table S4). These findings are consistent with an in situ gene expression study from the central California coast, where MGII exhibited a metabolic profile consistent with binding and hydrolysis of high molecular weight substrates, such as cell membrane components, which in turn may feed other members of the microbial community specialized on the uptake of smaller carbon compounds (Ottesen et al., 2013). Because POM is enriched with lipids relative to seawater (Wakeham and Canuel, 1988) it may represent a niche for POM-attached MGII ecotypes with an increased genomic capacity for fatty-acid oxidation.
Previous metagenomic studies have suggested that many MGII are photoheterotrophic, using proteorhodopsin as a proton pump to supplement their capacity to generate adenosine triphosphate with sunlight under oligotrophic conditions (Frigaard et al., 2006; Martin-Cuadrado et al., 2008). Proteorhodopsin may be used to supplement heterotrophic growth when recovering from carbon starvation (Steindler et al., 2011), thus conferring a fitness advantage for oligotrophs. Contigs encoding MGII proteorhodopsin from 0.8 to 3-μm fractions are phylogenetically distinct from their closest homologs in GenBank (MGII proteorhodopsin from Puget Sound (Iverson et al., 2012) and the Mediterranean Sea (Ghai et al., 2010) and those recovered from free-living fractions (Supplementary Figure S4). Furthermore, MGII homologs encoding a bacterio-opsin activator protein are significantly enriched in metagenomes from free-living fractions (Figure 6a). Such differences suggest that free-living MGII genomes may be relatively enriched with genes involved in photoheterotrophy, and further support the conclusion that the life history of POM-attached MGII is distinct from free-living MGII.
To investigate the metabolic capacity of POM-attached MGII that may not be represented by the Puget Sound MGII genome, we also compared the size-fractionated metagenomes to previously sequenced metagenomic MGII fosmid sequences. Size-fractionated metagenomic sequences homologous to open-reading frames on previously sequenced MGII fosmids are significantly different across the three fractions (ANOSIM: P=0.002) (Figure 6b). Moreover, the variation in MGII functional potential is two orders of magnitude higher in the POM fractions (σ0.8–3 μm=0.026 and σ3–20 μm=0.075) relative to the free-living fractions (σ0.2–0.8 μm=0.00012). This implies that POM-attached MGII have more diverse forms of metabolism compared with free-living MGII and supports the conclusion that the metabolic potential of free-living and POM-attached MGII is significantly different.
Interactions between MGII and living POM
The metagenomic evidence presented above suggests a linkage between MGII ecophysiology and POM, but does not definitively indicate whether MGII detected in >0.8 μm fractions are attached to living picoeukaryotes, in addition to detrital particles. To investigate this possibility, populations of chlorophyll-containing (n=3 populations, 240–900 cells per population) and heterotrophic picoeukaryotes (those staining with Lysotracker Green dye, n=900 cells) were flow sorted at sea and subjected to whole-genome amplification. No PCR amplification of archaeal 16 S rRNA genes was obtained from picoeukaryote whole-genome amplification templates and no MGII cells were seen physically attached to picoeukaryotes with CARD-FISH. Although these negative data are difficult to draw conclusions from, we suggest that most POM-attached MGII in the CCS are attached to detrital POM (for example, Figure 2e–g) and not living eukaryotes. Although MGII may not be physically attached to phytoplankton, the abundance of free-living MGII do correlate with chlorophyll (P<0.05, Figure 3a), suggesting a connection between free-living MGII growth and phytoplankton abundance or activity.
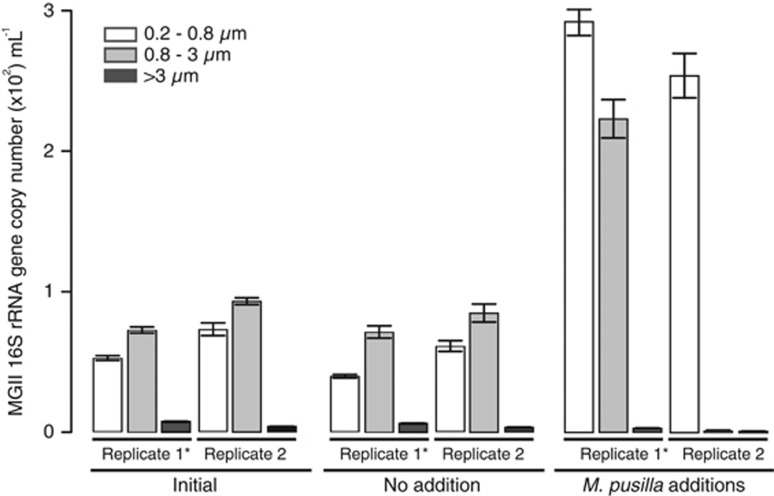
To further investigate the possibility that free-living MGII abundance is stimulated by living phytoplankton, we monitored the net growth (growth plus loss) of free-living and POM-attached MGII (via qPCR) in bottle experiments where the cultured photosynthetic picoeukaryote M. pusilla was added. After 72 h, MGII 16 S rRNA gene copies in free-living fractions increased by fivefold (relative to control bottles) in replicate bottles (Figure 7). This corresponds to a free-living MGII net growth rate of 0.44–0.67 d−1. This is slightly lower than steady state generation times reported for free-living marine bacterioplankton (Ducklow, 2000). However, they are an order of magnitude faster than generation times reported for total Euryarchaeota in unamended seawater batch cultures (Herndl et al., 2005). The mechanism behind the MGII growth enhancement is unknown, but may be the result of utilization of (i) specific compounds contained in or released by eukaryotic phytoplankton, such as reduced nitrogen compounds or vitamins or (ii) labile organic material in general. We do not have evidence that supports one of these possibilities over the other. The stimulation of free-living MGII growth in the presence of phytoplankton could explain the positive correlations of free-living MGII with phytoplankton biomass reported above and in a coastal observatory (Galand et al., 2010), and support the bloom-type MGII dynamics in surface waters observed previously (Mincer et al. 2007). Variability among the treatment bottles, especially in the larger size fractions, may stem from the presence of gelatinous zooplankton identified as a Salpa sp. in replicate 1. In this bottle, MGII had a net growth rate of 0.47 d−1 in the 0.8–3 μm fraction (Figure 7). As these are net growth rates (includes losses), it is likely that the actual growth rates for free-living and POM-attached MGII are higher. Suspended POM resulting from zooplankton grazing is readily colonized and remineralized by marine microbes (Karl et al., 1988), and could facilitate the growth of POM-attached MGII ecotypes in the presence of Salpa (Figure 7). The significantly higher number of ABC transporters possessed by free-living MGII (Figure 6) could be a partial explanation for the relatively faster net growth rate of free-living MGII in the presence of M. pusilla (Figure 7). Higher numbers of ABC transporters may enable free-living MGII to take up certain phytoplankton-derived labile organic compounds faster, and thereby outcompete particle-attached MGII. Specific microbial interactions that induce bloom-type dynamics of MGII are poorly understood but warrant further investigation.
Figure 7.
Quantification of MGII 16 S rRNA genes (qPCR) in bottle experiments in which the eukaryotic phytoplankter M. pusilla was added to seawater from the CCS (72 h incubation). Error bars represent s.d. across three technical replicates (three qPCR reactions). The ‘*' indicates the bottle that contained Salpa sp.
Conclusions
Our analyses provide metagenomic and physiological evidence for the widely observed correlations between Chl a and MGII Euryarchaeota in the ocean, and suggest that particle attachment is an important factor in the ecology and evolution of some members of the MGII. We speculate that by associating with particles, some POM-attached MGII ecotypes can maintain population size under oligotrophic conditions and outnumber free-living MGII. POM-attached MGII populations are phylogenetically distinct from free-living MGII and have a relatively increased genomic capacity for transcriptional regulation and degradation of high molecular weight substrates. We demonstrate that growth of free-living MGII is stimulated in the presence of phytoplankton, whereas growth of particle-attached MGII may be stimulated by zooplankton (salps). These analyses demonstrate that the ecophysiology of MGII is linked to interactions with both phytoplankton and POM, and imply that marine Euryarchaeota have a role in elemental cycling through POM transformations in the ocean.
Acknowledgments
We thank the captain and crew of the Monterey Bay Aquarium Research Institute (MBARI) vessel R/V Western Flyer during cruises CN207, C0912 and CN13ID. We thank Valeria Jimenez, Noriko Okamoto, Tim Pennington and Marguerite Blum for assistance with sample collection at sea, Sebastian Sudek for preparing the M. pusilla culture, and Francisco Chavez, chief scientist on cruise C0912. We thank the MBARI Biological Oceanography Group for Chl a and dissolved nutrient data, Christopher Francis for use of the barcoded amplicon data, and Nicholas Nidzieko for assistance with ocean color data. We thank Laurence P Madin for discussions about tunicate and Salpa physiology, Amy Apprill for advice and information about amplicon libraries, and Patrick Keeling and Thomas Richards for helpful discussions throughout the project. This work was funded by the Gordon and Betty Moore Foundation through grants GBMF3307 to AES and AZW, GBMF1668 to AZW, National Science Foundation Grants OCE-1260006 and DBI-1318455 to AES, and the David and Lucile Packard Foundation (in support of cruises as well as JMS, HMW, JES and AZW contributions). This is University of Maryland Center for Environmental Science contribution 4919.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP, et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol. 2000;2:516–529. doi: 10.1046/j.1462-2920.2000.00133.x. [DOI] [PubMed] [Google Scholar]
- Buesseler KO, Trull TW, Steinberg DK, Silver MW, Siegel DA, Saitoh SI, et al. VERTIGO (VERtical Transport in the Global Ocean): a study of particle sources and flux attenuation in the North Pacific. Deep Sea Res. 2008;55:1522–1539. [Google Scholar]
- Button DK. Kinetics of nutrient-limited transport and microbial growth. Microbiol Rev. 1985;49:270–297. doi: 10.1128/mr.49.3.270-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charette MA, Moran SB, Bishop JKB. 234Th as a tracer of particulate organic carbon export in the subarctic northeast Pacific Ocean. Deep Sea Res II. 1999;46:2833–2861. [Google Scholar]
- Cho BC, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature. 1988;332:441–443. [Google Scholar]
- Collins CA, Pennington JT, Castro CG, Rago TA, Chavez FP. The California Current system off Monterey, California: physical and biological coupling. Deep Sea Res II. 2003;50:2389–2404. [Google Scholar]
- Crump BC, Baross JA. Archaeaplankton in the Columbia River, its estuary and the adjacent coastal ocean, USA. FEMS Microbio Ecol. 2000;31:231–239. doi: 10.1111/j.1574-6941.2000.tb00688.x. [DOI] [PubMed] [Google Scholar]
- Delong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong EF, Franks DG, Alldredge AL. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol Oceanogr. 1993;38:924–934. [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Ducklow H.2000Bacterial production and biomass in the oceansIn: Kirchman DL (ed)Microbial Ecology of the Oceans1st edn,Wiley-Liss: New York, NY, USA; 85–120. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N, Martinez A, Mincer T, DeLong E. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Davis AA. Widespread archaea and novel bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar Ecol Prog Ser. 1997;150:275–285. [Google Scholar]
- Fuhrman JA, McCallum K, Davis AA. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Galand PE, Gutièrrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory) Limnol Oceanogr. 2010;55:2117–2125. [Google Scholar]
- Galand PE, Lovejoy C, Pouliot J, Vincent WF. Heterogeneous archaeal communities in the particle-rich environment of an arctic shelf ecosystem. J Mar Syst. 2008;74:774–782. [Google Scholar]
- Ganesh S, Parris DJ, DeLong EF, Stewart FJ. Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J. 2014;8:187–211. doi: 10.1038/ismej.2013.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghai R, Martin-Cuadrado A-B, Molto AG, Heredia IG, Cabrera R, Martin J, et al. Metagenome of the Mediterranean deep chlorophyll maximum studied by direct and fosmid library 454 pyrosequencing. ISME J. 2010;4:1154–1166. doi: 10.1038/ismej.2010.44. [DOI] [PubMed] [Google Scholar]
- Giering SL, Sanders R, Lampitt RS, Anderson TR, Tamburini C, Boutrif M, et al. Reconciliation of the carbon budget in the ocean's twilight zone. Nature. 2014;507:480–483. doi: 10.1038/nature13123. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Herndl GJ, Reinthaler T. Microbial control of the dark end of the biological pump. Nat Geosci. 2013;6:718–724. doi: 10.1038/ngeo1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, et al. Contribution of archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes RM, Aminot A, Kèrouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- Hood RR, Abbott MR, Huyer A, Korso PM. Surface patterns in termperature, flow, phytoplankton biomass, and species composition in the coastal transition zone off Northern California. J Geophys Res. 2007;95:18,081–18,094. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hugoni M, Taib N, Debroas D, Domaizon I, Dufournel IJ, Bronner G, et al. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc Natl Acad Sci USA. 2013;110:6004–6009. doi: 10.1073/pnas.1216863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- Ingalls AE, Shah SR, Hansman RL, Aluwihare LI, Santos GM, Druffel ERM, et al. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc Natl Acad Sci USA. 2006;103:6442–6447. doi: 10.1073/pnas.0510157103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–590. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- Karl DM, Knauer GA, Martin JH. Downward flux of particulate organic matter in the Ocean—a particle decomposition paradox. Nature. 1988;332:438–441. [Google Scholar]
- Kawai M, Futagami T, Toyoda A, Takaki Y, Nishi S, Hori S, et al. High frequency of phylogenetically diverse reductive dehalogenase-homologous genes in deep subseafloor sedimentary metagenomes. Front Microbiol. 2014;5:80. doi: 10.3389/fmicb.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lindemann SR, Moran JJ, Stegen JC, Renslow RS, Hutchison JR, Cole JK, et al. The epsomitic phototrophic microbial mat of Hot Lake, Washington: community structural responses to seasonal cycling. Front Microbiol. 2013;4:323. doi: 10.3389/fmicb.2013.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T, Salzberg S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado A, Rodriguez-Valera F, Moreira D, Alba J, Ivars-Martìnez E, Henn M, et al. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- Massana R, DeLong E, Pedros-Alio C. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl Environ Microbiol. 2000;66:1777. doi: 10.1128/aem.66.5.1777-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Murray AE, Preston CM, Delong EF. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mincer TJ, Church MJ, Taylor LT, Preston C, Karl DM, DeLong EF. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ Microbiol. 2007;9:1162–1175. doi: 10.1111/j.1462-2920.2007.01239.x. [DOI] [PubMed] [Google Scholar]
- Monier A, Welsh RM, Gentemann C, Weinstock G, Sodergren E, Armbrust E, et al. Phosphate transporters in marine phytoplankton and their viruses: cross domain commonalities in viral host gene exchanges. Environ Microbiol. 2012;14:162–176. doi: 10.1111/j.1462-2920.2011.02576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira D, Rodríguez-Valera F, López-García P. Analysis of a genome fragment of a deep-sea uncultivated Group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ Microbiol. 2004;6:959–969. doi: 10.1111/j.1462-2920.2004.00644.x. [DOI] [PubMed] [Google Scholar]
- Olson RJ, Chisholm SW, Zettler ER, Armbrust EV. Pigments, size, and distribution of Synechococcus in the North-Atlantic and Pacific Oceans. Limnol Oceanogr. 1990;35:45–58. [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA, et al. Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA. 2013;110:E488–E497. doi: 10.1073/pnas.1222099110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-J, Ghai R, Martín-Cuadrado A-B, Rodríguez-Valera F, Chung W-H, Kwon K, et al. Genomes of two new ammonia-oxidizing archaea enriched from deep marine sediments. PLoS One. 2014;9:e96449. doi: 10.1371/journal.pone.0096449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A, McNichol A, Benitez-Nelson B, Hayes J, Eglinton T. Origins of lipid biomarkers in Santa Monica Basin surface sediment: a case study using compound-specific [Δ] 14C analysis. Geochim Cosmochim Acta. 2001;65:3123–3137. [Google Scholar]
- Pennington J, Chavez FP. Seasonal fluctuations of temperature, salinity, nitrate, chlorophyll and primary production at station H3/M1 over 1989–1996 in Monterey Bay, California. Deep Sea Res Pt II. 2000;47:947–973. [Google Scholar]
- Preston CM, Marin R, Jensen SD, Feldman J, Birch JM, Massion EI, et al. Near real-time, autonomous detection of marine bacterioplankton on a coastal mooring in Monterey Bay, California, using rRNA targeted DNA probes. Environ Microbiol. 2009;11:1168–1180. doi: 10.1111/j.1462-2920.2009.01848.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Allan DH, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA. 2014;111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reysenbach AL, Flores G. Electron microscopy encounters with unusual thermophiles helps direct genomic analysis of Aciduliprofundum boonei. Geobiology. 2008;6:331–336. doi: 10.1111/j.1472-4669.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- Reysenbach AL, Pace NR.1995Reliable amplification of hyperthermophilic archaeal 16S rRNA genes by the polymerase chain reactionIn: Robb FT, Place AR (eds)Archaea: A Laboratory Manual-Thermophiles Cold Spring Harbour Laboratory Press: New York, NY, USA; 101–107. [Google Scholar]
- Rich VI, Konstantinidis K, DeLong EF. Design and testing of ‘genome-proxy'microarrays to profile marine microbial communities. Environ Microbiol. 2008;10:506–521. doi: 10.1111/j.1462-2920.2007.01471.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto CM, Friederich GE, Codispoti LA. MBARI Technical Report, 90-2. Monterey Bay Aquarium Research Institute: Moss Landing, CA, USA; 1990. [Google Scholar]
- Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. Isotopic composition of N2O produced by marine ammonia-oxidizing archaea. Science. 2011;333:1282. doi: 10.1126/science.1208239. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL. Enrichment and characterization of ammonia-oxidizing archaea from the open ocean: phylogeny, physiology, and stable isotope fractionation. ISME J. 2011;5:1796–1808. doi: 10.1038/ismej.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol. 2010;12:1989–2006. doi: 10.1111/j.1462-2920.2010.02205.x. [DOI] [PubMed] [Google Scholar]
- Schlitzer R.2013. Ocean Data View. Available at http://odv.awi.de .
- Shapiro JB, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabo G, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Casciotti KL, Chavez FP, Francis CA. Differential contributions of archaeal ammonia oxidizer ecotypes to nitrification in coastal surface waters. ISME J. 2014;8:1704–1714. doi: 10.1038/ismej.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MW, Allen LZ, Allen AE, Herfort L, Simon HM. Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol. 2013;4:120. doi: 10.3389/fmicb.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, Amann R.1991Development and application of nucleic acid probesIn Stackebrandt E., Goodfellow M. (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons, Ltd.: Chichester, England; 205–248. [Google Scholar]
- Steinberg DK, Van Mooy BAS, Buesseler KO, Boyd PW, Kobari T, Karl DM. Bacterial vs. zooplankton control of sinking particle flux in the ocean's twilight zone. Limnol Oceanogr. 2008;53:1327–1338. [Google Scholar]
- Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One. 2011;6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudek S, Everroad RC, Gehman AM, Smith JM, Poirier CL, Chavez FP, Worden AZ.2015Cyanobacterial distributions along a physico-chemical gradient in the Northeastern Pacific Ocean Environ Microdoi: 10.1111/1462-2920.12742 [DOI] [PubMed]
- Suzuki MT, Taylor LT, DeLong EF. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5'-nuclease assays. Appl Environ Microbiol. 2000;66:4605–4614. doi: 10.1128/aem.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiera E, Reinthaler T, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeham SG, Canuel EA. Organic geochemistry of particulate matter in the eastern tropical North Pacific Ocean: Implications for particle dynamics. J Mar Res. 1988;46:183–213. [Google Scholar]
- Wells LE, Cordray M, Bowerman S, Miller LA, Vincent WF, Deming JW. Archaea in particle-rich waters of the Beaufort Shelf and Franklin Bay, Canadian Arctic: Clues to an allochthonous origin. Limnol Oceanogr. 2006;51:47–59. [Google Scholar]
- Wilke A, Harrison T, Wilkening J, Field D, Glass EM, Kyrpides NC, et al. The M5nr: a novel non-redundant database containing protein sequences and annotations from multiples sources and associated tools. BMC Bioinformatics. 2012;13:141. doi: 10.1186/1471-2105-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata T, Corder OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA. 2014;111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.