Abstract
The anoxic saccharide-rich conditions of the earthworm gut provide an ideal transient habitat for ingested microbes capable of anaerobiosis. It was recently discovered that the earthworm Eudrilus eugeniae from Brazil can emit methane (CH4) and that ingested methanogens might be associated with this emission. The objective of this study was to resolve trophic interactions of bacteria and methanogens in the methanogenic food web in the gut contents of E. eugeniae. RNA-based stable isotope probing of bacterial 16S rRNA as well as mcrA and mrtA (the alpha subunit of methyl-CoM reductase and its isoenzyme, respectively) of methanogens was performed with [13C]-glucose as a model saccharide in the gut contents. Concomitant fermentations were augmented by the rapid consumption of glucose, yielding numerous products, including molecular hydrogen (H2), carbon dioxide (CO2), formate, acetate, ethanol, lactate, succinate and propionate. Aeromonadaceae-affiliated facultative aerobes, and obligate anaerobes affiliated to Lachnospiraceae, Veillonellaceae and Ruminococcaceae were associated with the diverse fermentations. Methanogenesis was ongoing during incubations, and 13C-labeling of CH4 verified that supplemental [13C]-glucose derived carbon was dissimilated to CH4. Hydrogenotrophic methanogens affiliated with Methanobacteriaceae and Methanoregulaceae were linked to methanogenesis, and acetogens related to Peptostreptoccocaceae were likewise found to be participants in the methanogenic food web. H2 rather than acetate stimulated methanogenesis in the methanogenic gut content enrichments, and acetogens appeared to dissimilate supplemental H2 to acetate in methanogenic enrichments. These findings provide insight on the processes and associated taxa potentially linked to methanogenesis and the turnover of organic carbon in the alimentary canal of methane-emitting E. eugeniae.
Introduction
Earthworms go mostly unnoticed because of their subsurface lifestyle. However, the propensity of earthworms to consume and transform their habitat has significant impact on terrestrial ecosystems and soil processes (Darwin, 1881; Lee, 1985; Edwards and Bohlen, 1996; Makeschin, 1997; Brown and James, 2006; Nechitaylo et al., 2010). For example, Lumbricus terrestris can consume the entire annual litter fall (300 g m−2) of forests it inhabits (Satchell, 1967). Earthworms can likewise be a dominant macrofauna of soil and up to 2000 individuals per square meter of soil have been recorded (Edwards and Bohlen, 1996). Assuming an average gut volume of approximately 250 mm3 (80 mm length × 1 mm2 radius × π), this number of earthworms would theoretically yield approximately 500 ml of gut contents per square meter of soil, illustrating that the alimentary canal of the earthworm can be an important component of soil.
The alimentary canal of the earthworm constitutes a mobile anoxic micro-compartment in aerated soils (Drake and Horn, 2007). The in situ conditions of the earthworm gut include anoxia, near neutral pH, relatively low redox potentials and millimolar concentrations of monosaccharide equivalents in the aqueous phase (Horn et al., 2003; Drake and Horn, 2007; Wüst et al., 2009a). The high concentration of saccharides in the gut appears to be derived from the saccharide-rich mucus that is secreted in the alimentary canal for facilitating gut passage and digestion of the ingested matter (Lavelle, 1986; Barois, 1987, Edwards and Bohlen, 1996; Trigo et al., 1999; Brown et al., 2000). The in situ conditions of the gut stimulate ingested obligate anaerobes and facultative aerobes that promote diverse fermentations along the alimentary canal, and fermentation-derived molecular hydrogen (H2) is emitted in vivo by earthworms (Karsten and Drake, 1995; Schmidt et al., 2001; Wüst et al., 2009a, 2011). Fermentation products in the gut may serve as substrates for soil-derived denitrifiers and be trophically linked to the in vivo emission of nitrous oxide (N2O) and dinitrogen (N2) (Karsten and Drake, 1997; Matthies et al., 1999; Depkat-Jakob et al., 2013). In this regard, earthworms can contribute to the capacity of soils to emit nitrogenous gases (Karsten and Drake, 1997; Matthies et al., 1999; Rizhiya et al., 2007; Lubbers et al., 2011), and it has been postulated that the emission of H2 by earthworms might drive energy-dependent processes in soil such as the fixation of N2 or carbon dioxide (CO2) (Wüst et al., 2009a). Soil iron is likewise subject to anaerobic redox transformations during gut passage (Wüst et al., 2009a). These findings illustrate that the alimentary canal of earthworms augments diverse anaerobic microbial activities that impact on the cycling of elements in soils.
Several studies have indicated that earthworms do not emit methane (CH4) (Hornor and Mitchell, 1981; Karsten and Drake, 1995; Drake and Horn, 2007; Šustr and Šimek, 2009). It was therefore surprising that certain earthworms in Brazil, in particular Eudrilus eugeniae, were recently discovered to emit CH4 in vivo (Depkat-Jakob et al., 2012). E. eugeniae is an epigeic species that is native to certain African soils but is commonly used in vermicomposting in other countries, including Brazil (Martinez, 1998; Domínguez, 2004; Oboh et al., 2007). The emission of CH4 by E. eugeniae appeared to be linked to the ingested methanogens of the families Methanosarcinaceae, Methanobacteriaceae and Methanomicrobiaceae (Depkat-Jakob et al., 2012). We hypothesized that the capacity of E. eugeniae to emit CH4 is linked to diverse fermentations in the alimentary canal, and the main objective of this study was to resolve anaerobic processes and associated taxa that can potentially drive methanogenesis in the gut of E. eugeniae.
Material and methods
Earthworms
Adult earthworms of the species E. eugeniae (Eudrilidae) were obtained in May 2012 from the distributer Minhobox (Juiz de Fora, Minas Gerais, Brazil). Earthworms were 2.3±0.2 g and 11–20 cm in length. The gut passage time of adult specimens of E. eugeniae is approximately 6 h (Mba, 1988).
In vivo emission of CH4
Individual specimens of E. eugeniae were washed with sterile deionized water, dried by blotting with tissue paper, weighed and placed into sterile 160-ml serum vials (one earthworm per replicate). Serum vials were sealed with butyl rubber septum stoppers and incubated for 5 h at room temperature in the dark. The gas phase was air.
Gut content microcosms
Earthworms were washed and exposed to ice-cold, sparkling mineral water. Gut content (approximately 25 g) was squeezed out from approximately 100 earthworms while gassing with 100% argon to minimize exposure of the gut content to air (Depkat-Jakob et al., 2012). Gut content was kept under an anoxic atmosphere of 100% argon in sterile serum bottles and mixed to a 1:10 (w/v) dilution with sterile anoxic sodium phosphate buffer (Wüst et al., 2011). The gut content homogenate was distributed into nine sterile anoxic 160-ml serum bottles that were crimp sealed with sterile butyl rubber stoppers and wrapped with tinfoil to minimize exposure to light; the gas phase was 100% argon. Each serum bottle contained 18-ml gut content homogenate (corresponding to approximately 1.8-g gut content) and was preincubated overnight in the dark at 25 °C to ensure that residual molecular oxygen (O2) was consumed. Replicates of three serum bottles were treated with either 2 ml of 100 mM [12C]-glucose (AppliChem GmbH, Darmstadt, Germany) or 2 ml of 100 mM [13C]-glucose (Sigma-Aldrich Chemie GmbH, Munich, Germany; 99 atom% 13C). Two milliliters of sterile anoxic water was added to three control bottles. Bottles were incubated in the dark at 25 °C. Samples were taken at different intervals after the addition of substrate with sterile syringes that had been flushed with sterile argon. Samples (1 ml) for molecular analyses were preserved in 9-ml RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) and stored at −20 °C. Gas samples (3 ml) were collected in Exetainers (Labco Limited, High Wycombe, UK). Aqueous samples (1 ml) for chemical analyses were filter sterilized through autoclaved nylon filters (0.2-μm pore size; Infochroma, Zug, Switzerland) and stored at −20 °C in sterile safe-lock tubes (Eppendorf, Hamburg, Germany).
Methanogenic-enrichment culture
The enrichment medium contained (in mg l−1) mineral salts (KH2PO4 10; NH4Cl 4.6; MgCl2•6H2O 10; CaCl2•2H2O 10, (modified from Wüst et al., 2009b)), trace metals (MnSO4•H2O 2.5; FeCl2•4H2O 0.7; CoCl2•2H2O 1; CaCl2•2H2O 1; ZnCl2 0.5; AlK(SO4)4•12H2O 0.2; H3BO3 0.1; Na2MoO4•2H2O 0.1; CuSO4•5H2O 0.1; Na2WO4•2H2O 0.05; NiCl2•2H2O 0.2; H2SeO3 0.5 (modified Balch et al., 1979)), 10 ml vitamin solution (Balch et al., 1979), 0.5 g yeast extract, 0.5 g tryptone, 15 g NaHCO3, 0.1 mg resazurin, 0.03 g cysteine and 0.03 g NaS. The pH was 7. Enrichments were incubated at 25 °C with a CO2 head space and supplemented with either 22 mM H2 or 5 mM acetate for 14 days. Unsupplemented microcosms served as controls. Enrichments were prepared in triplicate.
Analytical methods
The pressure in the incubation bottles was measured with a pressure transducer (DMG 2120, Ballmoos Elektronik AG, Horgen, Switzerland). The pH of liquid samples was measured with a pH-Meter (WTW pH 330, Wissenschaftliche Werkstätten, Weilheim, Germany) and was used for calculating CO2 production. The fresh weight (FW) of gut content at each sampling point was calculated by subtracting the amount of gut content removed at previous samplings. Gas concentrations of CO2, H2 and CH4 were determined with a SRI8610C gas chromatograph (SRI Instruments, Torrance, CA, USA). CH4 and H2 were separated with a Molecular Sieve Column (13 × , 2 m × 1/8 in; Restek, Bellefonte, PA, USA). CO2 was separated with a HayeSep-D column (2 m × 1/8 in; SRI instruments). The carrier gas was helium at a flow rate of 40 ml min−1 for CO2 and 20 ml min−1 for CH4 and H2. The oven temperature was 60 °C. The temperature of the detector (thermal conductivity detector) for CO2 was 175 °C. The temperature of the detector (helium ionization detector) for CH4 and H2 was 150 °C. Gas concentrations were calculated as previously described (Küsel and Drake, 1995). Organic compounds in the aqueous phase were analyzed by high performance liquid chromatography as previously described (Wüst et al., 2009a). The 13C content of CH4 and CO2 was determined by gas chromatograph combustion isotope-ratio mass spectrometry as previously described (Conrad et al., 2007). The CH4 emitted by living earthworms was measured with a Hewlett-Packard Co. 5980 series II gas chromatograph (Hewlett-Packard, Palo Alto, CA, USA) (Küsel and Drake, 1995).
Extraction of nucleic acids
Nucleic acids were extracted as previously described (Depkat-Jakob et al., 2012), and the extracts of each replicate were pooled per treatment. DNA was degraded by treatment with 1 U DNAse I per μl (Fermentas GmbH, St Leon-Rot, Germany), and RNA was purified by precipitation with 0.7 volume isopropyl alcohol and 0.1 volume of 5 mM sodium chloride (Merck KGaA, Darmstadt, Germany). The removal of DNA was confirmed by the inability to amplify 16S rRNA gene fragments by PCR without reverse transcription (see below).
RNA stable isotope probing (RNA SIP)
RNA SIP was performed per published protocol (Whiteley et al., 2007). A gradient solution (buoyant density of 1.793±0.002 g ml−1) was prepared by mixing 4.61 ml caesiumtrifluoroacetate solution (buoyant density: 2.0±0.05 g ml−1; GE Healthcare, Buckinghamshire, UK), 0.175 ml formamide and a variable amount of gradient buffer (100 mM KCl, 100 mM Tris, 1 mM EDTA, pH 8). The gradient solution was added to 68–506 ng RNA and placed in 4.9-ml OptiSeal Polyallomer Tubes (13 × 48 mm; Beckmann, Palo Alto, CA, USA). A centrifugation tube filled with gradient solution and 20 μl diethylpyrocarbonate (DEPC)–H2O served as blank for determining the densities of fractions after centrifugation. All gradients were set up with the same gradient solution. The separation of [12C]-RNA and [13C]-RNA was achieved by isopycnic centrifugation at 130 000 g (37 800 r.p.m.) at 20 °C for 67 h in a LE-70 ultracentrifuge (Beckman Coulter, Fullerton, CA, USA). The rotor stopped without braking. Ten fractions (0.45 ml) of each gradient were collected manually (Manefield et al., 2002) using a suction pump (Econo Pump1, Bio-Rad, Hercules, CA, USA). The densities of the fractions were determined by weighting at 25 °C. RNA in each fraction was precipitated according to Lottspeich and Engels (2006). In all, 200 μl of RNA was mixed with 130 μl of 3 M RNAse-free sodium acetate buffer (pH 5.2), 13.6 μl of a sterilized solution of 10 mg glycogen per ml and 1020 μl of ice-cold 96% ethanol. RNA was precipitated overnight at −20 °C and centrifuged for 20 min at 14 000 r.p.m. and 4 °C. The supernatant was removed, and the RNA-pellet was washed with 500 μl of ice-cold RNAse-free 70% ethanol. The purified RNA was eluted in DEPC–H2O. The concentration of RNA in each fraction was determined with a Quant-iT RiboGreen RNA Kit (Invitrogen, Eugene, OR, USA). RNA was stored at −80 °C.
Reverse transcription and PCR amplification
Extracted RNA was transformed into complementary DNA (cDNA) by reverse transcription with SuperScript III reverse transcriptase as previously described (Depkat-Jakob et al., 2012). PCR amplification of bacterial 16S rRNA genes from cDNA was performed with the primers 27F and 907RM (5′-AGAGTTTGATCMTGGCTC-3′ 5′-CCGTCAATTCMTTTGAGTTT-3′ Lane, 1991). The PCR conditions were as follows: initial denaturation at 95 °C for 5 min, 5 cycles at 95 °C for 60 s, at 40 °C for 60 s, and at 72 °C for 90 s, and 35 subsequent cycles at 95 °C for 60 s, at 50 °C for 30 s, and at 72 °C for 90 s. The final elongation was at 72 °C for 5 min. Final concentrations of PCR reagents were 1 × 5Prime Master Mix (5Prime, Hamburg, Germany), 1 mM magnesium chloride and 0.6 μM of each primer. cDNA of mcrA and mrtA transcripts of the RNA SIP analysis and mcrA and mrtA of the methanogenic H2–CO2 enrichment culture were amplified as previously described (Depkat-Jakob et al., 2012).
Sequence analyses
Cloning of PCR products for mcrA, mrtA and 16S rRNA cDNA retrieved from pooled fractions 2 and 3 (representing the ‘heavy' [labeled] RNA; buoyant density: 1.803±0.001–1.794±0.003 g ml−1; Supplementary Figure S1) and from pooled fractions 9 and 10 (representing the ‘light' [unlabeled] RNA; buoyant density: 1.743±0.003–1.735±0.004 g ml−1; Supplementary Figure S1), and also of PCR products obtained from the enrichment culture, was performed as previously described (Schmidt et al., 2014). PCR products of clones with correct inserts were selected for sequencing at Macrogen Europe (Amsterdam, the Netherlands). Sequences were analyzed with MEGA 5.1 (Tamura et al., 2011), ARB (Version 2005; Ludwig et al., 2004) and BLASTn (Altschul et al., 1990). McrA and mrtA clone sequences and reference sequences were translated in silico into amino-acid sequences and aligned with ARB, resulting in a final alignment of 130 aligned amino-acid positions. SINA Webaligner was applied to align 16S rRNA cDNA sequences, which were then merged with the 16S rRNA gene database from SILVA homepage (http://www.arb-silva.de/; last visit: 15/01/13; Pruesse et al., 2007). The resulting alignment contained 880 aligned nucleotide positions. Chimeric 16S rRNA cDNA gene sequences were identified as described (Schmidt et al., 2014). Potential chimeric sequences were blasted (BLASTn) and corrected by removing the shorter part of the sequence at the connection point of the different fragments. Retrieved amino-acid sequences were assigned to different Operational Taxonomic Units (OTUs) with DOTUR (Schloss and Handelsman, 2005). A conservative threshold value of 87.5% was used for determining family-level OTUs of 16S rRNA gene sequences (Yarza et al., 2008). A conservative threshold value of 85.7% was used for creating species-level OTUs of mcrA and mrtA sequences (Hunger et al., 2011); a maximal identity of ⩽85.7% is indicated by ‘spp.' after the genus name. The classification of 16S rRNA gene sequences was accomplished with the RDP classifier (Wang et al., 2007), by DOTUR analysis and via BLASTn. A sequence was assigned to a novel family when the maximum identity to a known sequence in the NCBI database was <87.5%. The coverage of the gene libraries was calculated according to Schloss et al. (2004), and rarefaction curves were constructed with aRarefact (http://www.uga.edu/~strata/software; last accessed: 8 January 2013) (Hurlbert, 1971; Heck et al., 1975).
Phylogenetic analyses
Phylogenetic trees of mcrA and mrtA were constructed with ARB by applying the algorithms neighbor-joining (Kimura correction (Saitou and Nei, 1987); 10 000 boostraps), maximum-likelihood (Dayhoff PAM modell, Phylip PROML; 100 bootstraps) and maximum-parsimony (Phylip PROTPARS; 500 bootstraps). Phylogenetic trees of mcrA and mrtA were based on a 100% similarity filter and 130 valid amino acids between positions 327 and 457 of mcrA of Methanocella paludicola SANAE. Phylogenetic trees of 16S rRNA cDNA sequences were constructed using neighbor-joining (Felsenstein correction (Felsenstein, 1985; Saitou and Nei, 1987); 10 000 bootstraps), AxML and maximum-parsimony methods and applying a 100% similarity filter of 880 valid nucleotides between positions 26 and 906 of the 16S rRNA gene sequence of Escherichia coli ATCC 11775.
Nucleotide sequence accession numbers
Sequences were submitted to the European Nucleotide Archive (accession numbers: HG964568–HG964633 (16S rRNA), HG964544–HG964567 (mcrA and mrtA of RNA SIP), and LK936462–LK936502 (mcrA and mrtA of methanogenic enrichment)).
Results
The experiments outlined below were designed to first demonstrate that E. eugeniae emitted CH4 in vivo and to subsequently examine the capacity of gut contents from E. eugeniae to dissimilate glucose, selected as a model saccharide found in the gut. [13C]-glucose was utilized as substrate (a) so that glucose-derived carbon could be traced to CH4 and (b) for the RNA SIP-based assessment of taxa involved in the methanogenic food web.
In vivo emission of CH4
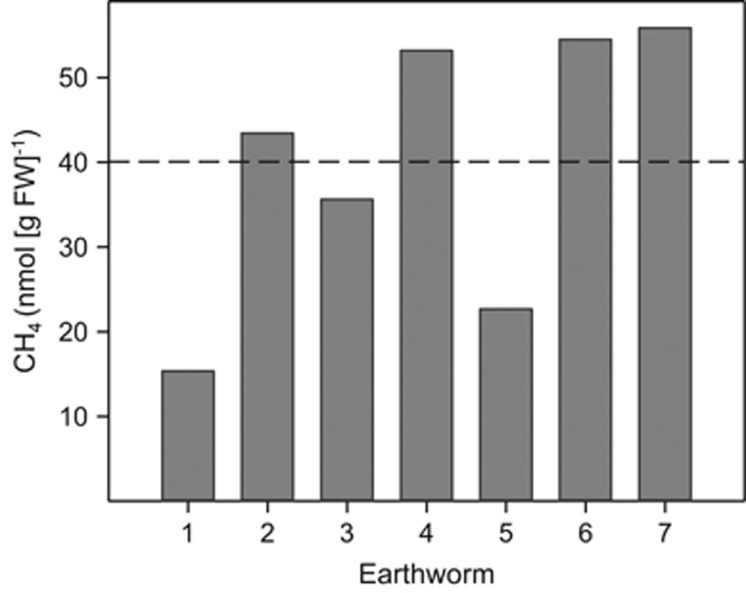
E. eugeniae emitted CH4, with the average in vivo emission approximating 40 nmol CH4 per gram FW in 5 h (Figure 1).
Figure 1.
In vivo emission of CH4 by seven individual specimens of E. eugeniae. Values represent the amount of CH4 emitted during a 5-h incubation period under an atmosphere of air. The dashed line indicates the mean value.
Dissimilation of glucose
Approximately 90 μmol supplemental glucose per gram FW gut content was consumed within 18 h in the [12C]- and [13C]-glucose treatments (Figure 2a). Glucose consumption occurred without apparent delay, indicating that microbes in the gut contents were poised to respond rapidly to nutrient input under anoxic conditions. Approximately 1 μmol CH4 per gram FW and 4 μmol CO2 per gram FW accumulated during the preincubation period.
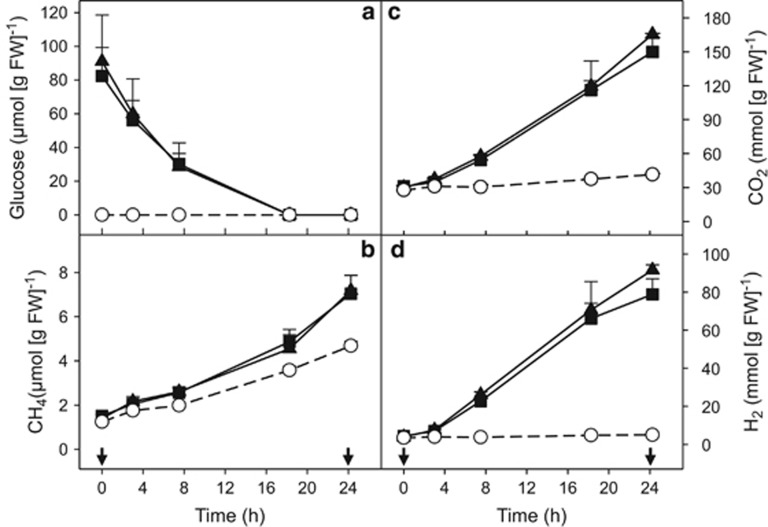
Figure 2.
Effect of supplemental glucose (a) on the production of CH4 (b), CO2 (c) and H2 (d) by the gut contents of E. eugeniae incubated under anoxic conditions. Symbols represent the mean of triplicate incubations, and error bars represent the s.d. Arrows indicate the time point at which the samples were taken for RNA-SIP analysis. Symbols: empty circles, control (no supplemental glucose); filled squares, [13C]-glucose treatments; filled triangles, [12C]-glucose treatments.
Gaseous (Figure 2) and soluble (Figure 3) products were similar for [12C]- and [13C]-glucose treatments. Up to approximately 88 and 74 μmol H2 per gram FW accumulated during the [12C]- and [13C]-glucose treatments, respectively, whereas only approximately 5 μmol H2 per gram FW was detected in unsupplemented controls (values are the difference between t0 and t24 time points) (Figure 2d). CO2 accumulation was rapid and relatively linear after the addition of glucose and yielded approximately 135 and 119 μmol CO2 per gram FW during the [12C]- and [13C]-glucose treatments, respectively (values are the difference between t0 and t24 time points) (Figure 2c). The accumulated CO2 in the [13C]-glucose treatment was strongly enriched in 13C, reaching 70 atom%. Approximately 14 μmol CO2 per gram FW accumulated in the unsupplemented controls (Figure 2c) (value is the difference between t0 and t24 time points).
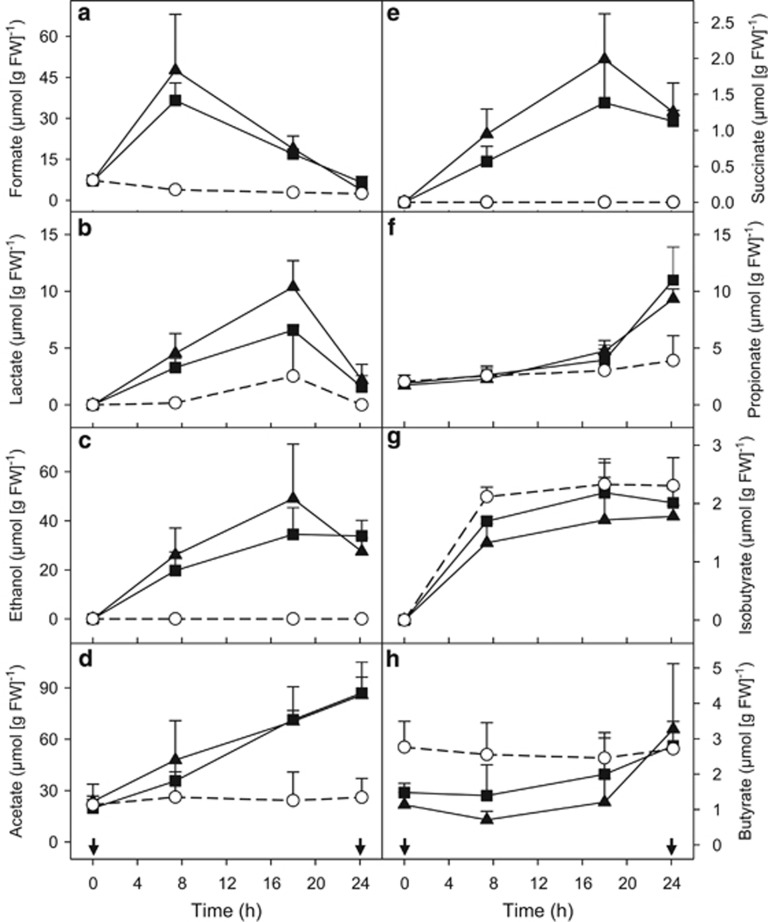
Figure 3.
Effect of supplemental glucose on the production of formate (a), lactate (b), ethanol (c), acetate (d), succinate (e), propionate (f), isobutyrate (g) and butyrate (h) by the gut contents of E. eugeniae incubated under anoxic conditions. Symbols represent the mean of triplicate incubations, and error bars indicate the s.d. Arrows indicate the time point at which the samples were taken for RNA-SIP analysis. Symbols: empty circles, control (no supplemental glucose); filled squares, [13C]-glucose treatments; filled triangles, [12C]-glucose treatments.
Approximately 6 μmol CH4 per gram FW accumulated in glucose treatments, whereas approximately half this much CH4 accumulated in unsupplemented controls (Figure 2b) (values are the difference between t0 and t24 time points). The accumulated CH4 in the [13C]-glucose treatment was strongly enriched in 13C, reaching 50 atom%. Based on a linear in vivo emission of CH4 (Depkat-Jakob et al., 2012) and an hourly in vivo emission rate of approximately 8 nmol CH4 per g FW earthworm (calculated from Figure 1), approximately 0.2 μmol CH4 per g FW earthworm would be emitted in vivo in 24 h. In contrast, approximately 3 μmol CH4 per g FW gut content was produced during the 24-h incubation without supplemental glucose (this value is the difference between the t0 and t24 time points). In addition, approximately 1 μmol CH4 per g FW gut content was produced during the overnight preincubation. Thus, on a FW basis, the gut content produced substantially more CH4 than did living earthworms, a result consistent with the fact that the gut content represents a very small amount of the total FW of the earthworm but is nonetheless the source of the CH4 that is emitted in vivo.
Diverse fermentations were substantially more stimulated by supplemental glucose than was methanogenesis (Figures 2 and 3). Formate, ethanol, lactate and succinate were produced and subject to subsequent consumption in glucose treatments; in contrast, acetate and propionate accumulated as end products in glucose treatments (Figure 3). Trace amounts of butyrate and isobutyrate were detected in controls and glucose treatments. At the end of the 24 h of incubation, approximately 69% and 61% of supplemented carbon and approximately 63% and 53% of supplemented electrons were recovered in the detected products of the [13C]- and [12C]-glucose treatments, respectively (Table 1). Unrecovered carbon and electrons may have been due in part to non-detected processes (for example, poor recovery of CO2 because of undetected carbonates, non-detected fermentation products or incomplete recovery of those detected, formation of storage polymers and assimilation of carbon).
Table 1. Recoveries of carbon (C-mol balance) and electrons (E-mol balance) in the detected products at the end of the 24-h incubation.
| Products |
Concentration of the detected products (μmol (g FW)−1) |
C-mol balance (%) |
E-mol balance (%) |
|||
|---|---|---|---|---|---|---|
| [12C]-Glc | [13C]-Glc | [12C]-Glc | [13C]-Glc | [12C]-Glc | [13C]-Glc | |
| Carbon dioxide | 121.4 | 105.1 | 22.2 | 21.3 | NA | NA |
| Methane | 2.3 | 2.1 | 0.4 | 0.4 | 0.9 | 0.8 |
| Hydrogen | 86.1 | 72.9 | NA | NA | 7.9 | 7.4 |
| Formate | 1.3 | 4.3 | 0.2 | 0.9 | 0.1 | 0.4 |
| Lactate | 2.2 | 1.6 | 1.2 | 1.0 | 1.2 | 1.0 |
| Ethanol | 27.5 | 33.9 | 10.1 | 13.7 | 15.1 | 20.6 |
| Acetate | 57.8 | 62.9 | 21.2 | 25.5 | 21.2 | 25.5 |
| Succinate | 1.3 | 1.1 | 0.9 | 0.9 | 0.8 | 0.8 |
| Propionate | 5.8 | 7.2 | 3.2 | 4.4 | 3.7 | 5.1 |
| Butyrate | 2.2 | 1.4 | 1.6 | 1.1 | 2.0 | 1.4 |
| Total | 307.9 | 292.1 | 61.0 | 69.1 | 52.8 | 62.9 |
Abbreviations: FW, fresh weight; NA, not applicable. The concentrations of the detected products represent means of triplicates and are based on the differences between the t0 and t24 time points, corrected by the concentration of the detected products of the unsupplemented controls. Balances are based on the amount of carbon or electrons recovered in the products versus the amount of carbon or electrons available in the glucose that was consumed.
Detected bacteria
A total of 528 bacterial 16S rRNA cDNA sequences were analyzed. The family-level coverage per clone library ranged between 85% and 94% (Supplementary Table S1), and rarefaction analyses confirmed that sequencing was sufficient for family-level coverage (Supplementary Figure S2A). Sequences from all clone libraries were affiliated with 46 different families and sub-orders; four families were defined as novel (Table 2). Families (and sub-orders) were assigned to the phyla Firmicutes (46.2%), Proteobacteria (19.5%, dominated by Gammaproteobacteria (11.4%)), Actinobacteria (16.5%), Fusobacteria (5.5%), Planctomycetes (5.5%), Tenericutes (4.5%), Spirochaetes (0.8%), Verrucomicrobia (0.8%), Bacterioidetes (0.6%) and Chloroflexi (0.2%) (Figure 4a and Table 2). Sequences obtained from cDNA at the start of incubation were related to 28 different families and sub-orders (Table 2). Those sequences were mostly related to the phyla Actinobacteria (21.7%), Fusobacteria (15.0%) and Firmicutes (28.3%), of which 39.2% were related to the family Peptostreptococcaceae (Figure 4a).
Table 2. Phylogenetic distribution of bacterial 16S rRNA cDNA sequences retrieved from RNA of [13C]- and [12C]-glucose treatments.
|
Phylogenetic affiliationa |
Relative abundance of sequences (%) |
||||||
|---|---|---|---|---|---|---|---|
| Phylum and classes | Order, sub-order (*) and family |
[13C]-glucose |
[12C]-glucose |
||||
| t0, L | t0, H | t24, L | t24, H | t24, L | t24, H | ||
| Actinobacteria (16.5) | Acidimicrobiales (2.2) | ||||||
| Acidimicrobiaceae | — | 3.3 | 1.2 | — | 1.1 | — | |
| Unclassified Acidimicrobineae | 3.4 | 1.1 | 1.2 | — | 2.2 | — | |
| Actinomycetales (13.3) | |||||||
| Corynebacterineae* (Mycobacteriaceae/Dietziaceae/Nocardiaceae) | 1.1 | 8.7 | 1.2 | 1.1 | 3.3 | 8.1 | |
| Micrococcineae* (Microbacteriaceae/Micrococcaceae) | 3.4 | 7.6 | 3.7 | — | 4.4 | 3.5 | |
| Propionibacterineae (Propionibacteriaceae/Nocardioidaceae) | 8.0 | 2.2 | 3.7 | 3.3 | 8.8 | 7.0 | |
| Coriobacteriales (0.2) | |||||||
| Coriobacteriaceae | — | 1.1 | — | — | — | — | |
| Rubrobacterales (0.8) | |||||||
| Rubrobacteraceae | 1.1 | 2.2 | — | — | 1.1 | — | |
| Bacterioidetes (0.6) | Bacteroidales (0.4) | ||||||
| Porphyromonadaceae | 1.1 | — | — | 1.1 | — | — | |
| Cytophagales (0.2) | |||||||
| Incertae sedis | — | — | — | — | 1.1 | — | |
| Chloroflexi (0.2) | Anaerolineales (0.2) | ||||||
| Anaerolineaceae | — | 1.1 | — | — | — | — | |
| Firmicutes (46.2) | Bacillales (2.1) | ||||||
| Bacillaceae | 1.1 | 1.1 | 4.9 | — | 1.1 | 3.5 | |
| Planococcaceae | 1.1 | — | — | — | — | — | |
| Clostridiales (42.0) | |||||||
| Clostridiaceae 1 | 5.7 | 5.4 | 3.7 | 1.1 | 6.6 | 8.1 | |
| Lachnospiraceae I(C. propionicum) | — | — | 3.7 | 27.8 | 16.5 | 23.3 | |
| Lachnospiraceae II | 4.6 | 4.4 | 8.6 | 3.3 | 4.4 | 2.3 | |
| Lachnospiraceae III (C. lentocellum) | 2.3 | 1.1 | 5.0 | 1.1 | 2.2 | 1.2 | |
| Peptostreptococcaceae | 10.2 | 12.0 | 12.4 | 18.9 | 14.3 | 10.5 | |
| Peptococcaceae | — | 1.1 | — | — | — | — | |
| Ruminococcaceae | 1.1 | — | 4.9 | 8.9 | 3.3 | 5.8 | |
| Veillonellaceae | — | — | — | 4.4 | 3.3 | — | |
| Erysipelotrichales (1.1) | |||||||
| Erysipelotrichaceae | 2.3 | 1.1 | 2.5 | — | — | 1.2 | |
| Lactobacillales (0.8) | |||||||
| Aerococcaceae | 2.3 | — | 1.2 | — | — | — | |
| Carnobacteriaceae | — | — | 1.2 | — | — | — | |
| Fusobacteria (5.5) | Fusobacteriales (5.5) | ||||||
| Fusobacteriaceae | 20.5 | 9.8 | — | — | 1.1 | 1.2 | |
| Planctomycetes (5.5) | Planctomycetales (5.5) | ||||||
| Planctomycetaceae I (Gemmata) | 2.3 | 8.7 | 1.2 | — | 1.1 | — | |
| Planctomycetaceae II (Rhodospirellula) | — | 1.1 | — | — | — | 2.3 | |
| Planctomycetaceae III (Pirellula) | 1.1 | 1.1 | 2.5 | — | — | — | |
| Planctomycetaceae IV (Isospaera/Sangulispaera) | — | 1.1 | 2.5 | — | — | 1.2 | |
| Planctomycetaceae V | 1.1 | 1.1 | 1.2 | — | — | 1.2 | |
| Novel familyb | 1.1 | — | — | — | 1.1 | — | |
| Proteobacteria (19.5) | |||||||
| Alphaproteobacteria (3.2) | Rhizobiales (1.7) | ||||||
| Hyphomicrobiaceae | — | 3.3 | 5.0 | — | — | — | |
| Phyllobacteriaceae | — | — | 2.5 | — | — | — | |
| Rhodobacterales (0.2) | |||||||
| Rhodobacteraceae | — | — | 1.2 | — | — | — | |
| Rhodospirillales (1.4) | |||||||
| Acetobacteraceae | — | 1.1 | 1.2 | — | 1.1 | 1.2 | |
| Rhodospirillaceae | — | 1.1 | 2.5 | — | — | — | |
| Betaproteobacteria (0.8) | Burkholderiales (0.6) | ||||||
| Oxalobacteraceae | — | 3.3 | — | — | — | — | |
| Neisseriales(0.2) | |||||||
| Neisseriaceae | — | — | 1.2 | — | — | — | |
| Deltaproteobacteria (4.2) | Myxococcales (4.2) | ||||||
| Novel familyb | — | — | 1.2 | — | — | — | |
| Novel familyb | — | — | — | — | — | 1.2 | |
| Polyangiaceae | 8.0 | 2.2 | 6.2 | — | 4.4 | 2.3 | |
| Gammaproteobacteria (11.4) | Aeromonadales (9.3) | ||||||
| Aeromonadaceae | 8.0 | 8.7 | — | 27.8 | 7.7 | 2.3 | |
| Enterobacteriales (1.3) | |||||||
| Enterobacteriaceae | — | — | — | 1.1 | 3.3 | 3.5 | |
| Methylococcales (0.8) | |||||||
| Methylococcaceae | — | — | 2.5 | — | 2.2 | — | |
| Spirochaetes (0.8) | Spirochaetales (0.8) | ||||||
| Leptospiraceae | — | — | — | — | 3.3 | 1.2 | |
| Tenericutes (4.5) | Mycoplasmatales (4.5) | ||||||
| Mycoplasmataceae | 8.0 | 4.4 | 9.9 | — | 1.1 | 4.7 | |
| Verrucomicrobia (0.8) | Novel familyb(0.8) | 1.1 | — | — | — | — | 3.5 |
Abbreviations: H, ‘heavy' fraction; L, ‘light' fractions; t0, sampling point at the start of incubation; t24 sampling point after 24 h of incubation.
Sequences were assigned to bacterial families by using the RDP Classifier, DOTUR analysis and BLASTn search. The number of sequences retrieved from cDNA of ‘heavy' and ‘light' fractions ranged from 81 to 92 (Supplementary Table S1). The percentage of relative abundances for phyla and orders are shown in parentheses.
Considered as a novel family based on gene sequence similarities <87.5%.
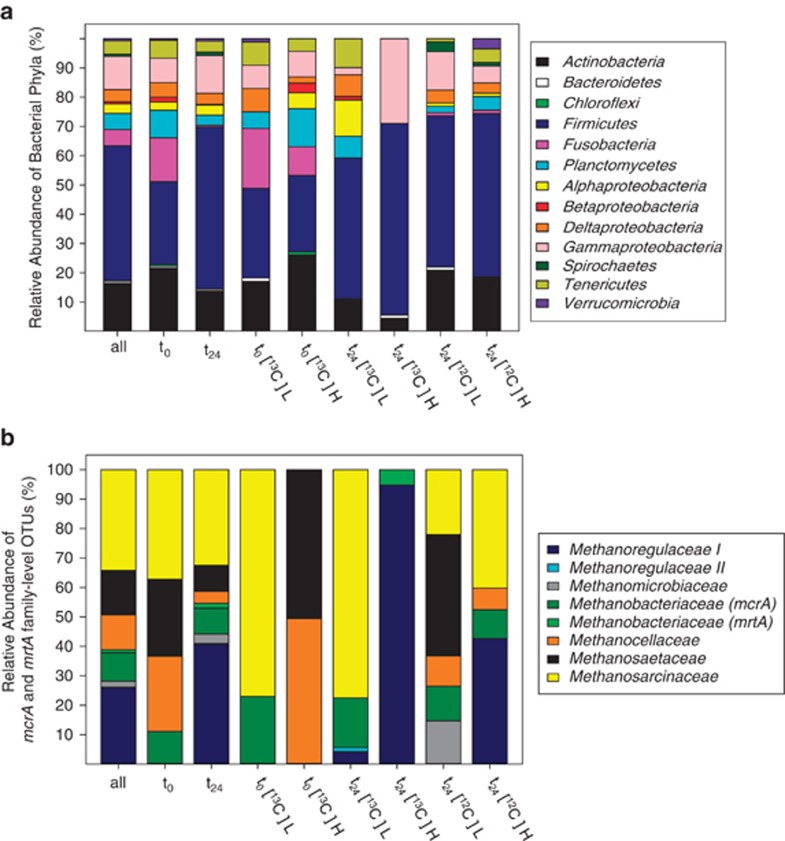
Figure 4.
Phylogenetic distribution of bacterial 16S rRNA cDNA (a) and mcrA and mrtA (b) sequences of [13C]- and [12C]-glucose treatments. Sequences were assigned to bacterial phyla with the RDP Classifier. Assignment of sequences to methanogenic families was based on ⩽75.4% gene sequence similarities (Hunger et al., 2011). As indicated in the lower axes, sequence analyses were performed for the total number of sequences (lane designated all), for all sequences at the start (t0) or at 24 h (t24) of incubation and for each fraction per time point and treatment. H, ‘heavy' fractions; L, ‘light' fraction; t0, sampling point at the start of incubation; t24, sampling point at 24 h of incubation; 13 and 12, [13C]- and [12C]-glucose treatments, respectively.
The bacterial 16S rRNA cDNA sequences obtained from the glucose treatments at the end of the 24-h incubation were affiliated with 41 different families and sub-orders (Table 2). Those sequences were mostly related to the phyla Firmicutes (55.2%), Proteobacteria (20.4%, dominated by Gammaproteobacteria (12.9%)) and Actinobacteria (13.8%) (Figure 4a). Lachnospiraceae I (18.1%), Peptostreptococcaceae (14.1%) and Aeromonadeaceae (9.8%) were the most abundant families.
Bacterial community composition
The shift of the buoyant densities of extracted RNA from [13C]-glucose treatments toward fractions with higher densities (Supplementary Figure S1) reinforces the likelihood that some of the glucose-derived carbon was assimilated by microorganisms in the E. eugeniae gut content. The bacterial community structure of ‘heavy' fractions of the [13C]-treatment was distinct from that of the ‘light' fractions of the [13C]-treatment and ‘heavy' fractions of the [12C]-glucose treatment (Figure 4a). 16S rRNA cDNA sequences obtained from ‘heavy' fractions of the [13C]-glucose treatment at the end of the 24-h incubation were affiliated with 12 different families and sub-orders, most of which were related to Aeromonadaceae (27.8%) and Lachnospiraceae I (27.8%) (Table 2). Sequences assigned to those families were closely related to Aeromonas hydrophila strain ANSE1 (GU296671, 99% similarity, Supplementary Table S3) and Clostridium propionicum strain JCM 1430 (AB649276, 97–99% similarity, Supplementary Table S3). Veillonellaceae- and Ruminococcaceae-affiliated sequences were also mainly abundant in the ‘heavy' fractions of the [13C]-glucose treatment (Table 2). Sequences assigned to Veillonellaceae and Ruminococcaceae were closely related to Succinispira mobilis strain 19gly1 (NR_028868, 99% similarity) and Clostridium viride strain DSM 6836 (NR_026204, 99% similarity, Supplementary Table S3), respectively. Approximately 19% of the sequences retrieved from ‘heavy' fractions of the [13C]-glucose treatment were affiliated to Peptostreptococcaceae (Table 2), with the closest known relatives (98–99% similarity, Supplementary Table S3) being Clostridium mayombei (FR733682) and Clostridium glycolicum strain CIN5 (AY007244).
Detected methanogens
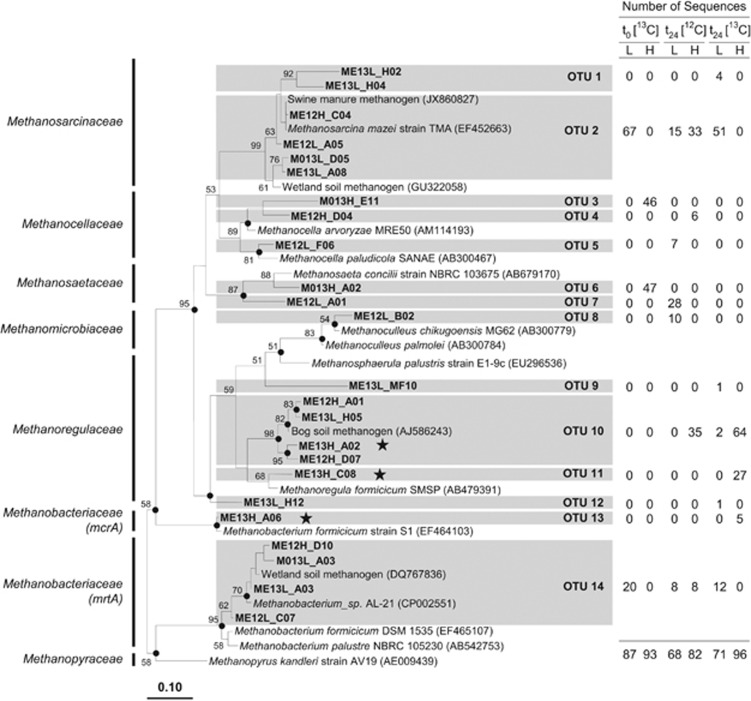
A total of 497 mcrA and mrtA sequences were assigned to 14 species-level OTUs. The coverage for each clone library was >97% (Supplementary Table S2), and rarefaction analyses confirmed that sequencing was sufficient for species-level coverage (Supplementary Figures S2B and C). Sequences retrieved at the start of incubation were affiliated with Methanosarcinaceae (37.2%), Methanocellaceae (25.6%), Methanosaetaceae (26.1%) and Methanobacteriaceae (11.1%) (Figure 4b). The active mcrA community structure changed after 24 h of incubation; sequences affiliated with Methanoregulaceae were detected, and the relative abundance of Methanosaetaceae and Methanocellaceae was decreased. McrA and mrtA sequences of the ‘heavy' fraction of [13C]-glucose treatments were assigned to the species-level OTUs 10, 11 and 13. Those OTUs were affiliated with Methanoregula spp. and Methanobacterium formicicum (Figure 5). Sequences related to those species were not detected or were not abundant in ‘light' fractions of [13C]-glucose treatments and ‘heavy' fractions of [12C]-glucose treatments.
Figure 5.
Phylogenetic neighbor-joining tree of representative mcrA and mrtA amino-acid sequences of [13C]- and [12C]-glucose treatments. Numbers next to the branches represent the percentages of replicate trees (>50%) in which the associated taxa clustered together in the bootstrap test (10 000 bootstraps). Dots at nodes indicate the confirmation of the tree topology by maximum-parsimony and maximum-likelihood calculations with the same data set. Bar represents a 0.1 estimated change per amino acid. Accession numbers of reference sequences are in parentheses. Stars identify labeled mcrA and mrtA phylotypes. The table outlines the origin of sequences per species-level OTU (based on 85.7% gene sequence similarity; Hunger et al., 2011). Abbreviations in sequence identifier code: H, ‘heavy' fractions; L, ‘light' fractions; M0, methanogen sequence from sampling point t0 (that is, at the start of incubation); ME, methanogen sequence from sampling point t24 (that is, at 24 h of incubation); 13 and 12, [13C]- and [12C]-glucose treatments, respectively.
Methanogenic-enrichment culture
The results above suggested that H2 rather than acetate was an important driver of methanogenesis. To evaluate this possibility in more detail, enrichment cultures prepared from the gut content of E. eugeniae were supplemented with H2–CO2 or acetate and incubated for 14 days. Methanogenesis was only stimulated by H2–CO2 (0.8 mM CH4 compared with 0.1 mM CH4 in controls lacking H2). Two species-level mrtA and four species-level mcrA phylotypes affiliated with Methanobacteriaceae were detected in the H2–CO2 enrichments. The relative abundance of phylotypes and affiliated species were: 39% Methanobacterium ivanovii (mrtA), 34% M. ivanovii (mcrA), 15% Methanobacterium sp. (mcrA), 5% Methanobacterium formicicum (mrtA), 5% M. formicicum (mcrA), and 2% Methanobacterium kanagiense (mcrA). It is noteworthy that acetate production was more significantly stimulated by H2–CO2 than was methanogenesis (13 mM acetate compared with 6 mM acetate in controls lacking H2), suggesting that H2 enriched not only methanogens from the gut contents of E. eugeniae but acetogens as well.
Discussion
The diverse anaerobic activities that occur in the alimentary canal of earthworms appear to be dominated by ingested microbes that become activated by the unique in situ conditions of the alimentary canal (Karsten and Drake, 1995; Matthies et al., 1999; Furlong et al., 2002; Horn et al., 2003; Drake and Horn, 2007; Knapp et al., 2009; Wüst et al., 2009a; Depkat-Jakob et al., 2012). This generalization for the earthworm gut is in contrast to other invertebrates whose guts are heavily colonized by novel endemic microbes (for example, termites; Brune, 2006; Dietrich et al., 2014). The methanogenic food webs of classic methane-emitting ecosystems such as ruminants and wetlands are primarily linked to the initial hydrolysis of autotrophically produced polymers (for example, the hydrolysis of plant-synthesized lignocelluloses; Morrison et al., 2009; Bridgham et al., 2013). In contrast, the methanogenic food web of E. eugeniae is likely linked to the hydrolysis of heterotrophically produced polymers, that is, the hydrolysis of earthworm-derived polysaccharide-rich mucus (Wüst et al., 2009a). Indeed, 100 mM monosaccharide equivalents, including >10 mM glucose can occur in the aqueous phase of gut contents (Horn et al., 2003; Wüst et al., 2009a). Glucose was therefore selected as a model saccharide to experimentally evaluate potential trophic interactions of prokaryotes that yield CH4 in the digestive system of E. eugeniae. The rapid and nearly linear production of H2 and CO2 by the gut contents of E. eugeniae in response to glucose (Figure 2) corroborate previous studies that demonstrated the capacity of microbes in the alimentary canal of various earthworms to augment anaerobic processes (Karsten and Drake, 1995; Matthies et al., 1999; Wüst et al., 2009a, 2011).
Fermentations and fermenters in E. eugeniae gut contents
Concomitant as well as successive fermentations were stimulated by supplemental glucose (Figure 3). The production of formate, acetate, succinate, lactate, ethanol, CO2 and H2 is indicative of mixed acid fermentation by facultative aerobes (Gottschalk, 1986). Most sequences retrieved from ‘heavy' fractions of [13C]-glucose treatments were affiliated with Aeromonadaceae, members of which are facultative aerobes (Martin-Carnahan and Joseph, 2005b). A. hydrophila was the closest cultured species to retrieved Aeromonadaceae-related sequences and is a facultative aerobe that can ferment sugars (including glucose) to ethanol, formate, acetate, succinate, CO2 and H2 (Martin-Carnahan and Joseph, 2005a). Certain sequences of ‘heavy fractions' from [13C]-glucose treatments were closely affiliated with C. viride (Ruminococcaceae), an obligate anaerobe that can ferment glucose to acetate, formate, succinate, lactate and ethanol (Ezaki, 1984; Buckel et al., 1994). C. viride is likewise able to ferment amino acids (Buckel et al., 1994), suggesting that bacteria related to C. viride might have additionally used endogenous amino acids in the earthworm gut contents (Drake and Horn, 2007). The formation of butyrate, isobutyrate and propionate (Figure 3) is indicative of the fermentation of amino acids (Nanninga and Gottschal, 1985; Gottschalk, 1986), and the high relative abundance of Peptostreptococcaceae-affiliated sequences suggests that members of this amino-acid-fermenting taxon (for example, Clostridium lituseburense) may have utilized amino acids (Hippe et al., 1992; Ezaki, 2009).
The production of propionate subsequent to the consumption of lactate and succinate (Figure 3) is indicative of Propionibacteria (Stackebrandt et al., 2006). However, propionate production could have also been linked to members of the detected family Lachnospiraceae I that form propionate when utilizing lactate (for example, C. propionicum) (Leaver et al., 1955). Members of Veillonellaceae can also produce propionate as well as various volatile fatty acids, CO2 and H2 (Morrison, 1984). Certain sequences were closely related to the Veillonellaceae-affiliated species S. mobilis, a strict anaerobe that can ferment organic and amino acids (but not carbohydrates such as glucose) and produce formate, acetate, propionate, CO2 and H2 (Janssen, 1984), compounds detected in the incubations (Figures 2 and 3).
Trophic links of methanogens in the gut contents of E. eugeniae
CH4 production was not as significantly stimulated by supplemental glucose as was fermentation (Figures 2 and 3). However, 13C-labeling of CH4 verified that supplemental [13C]-glucose-derived carbon was dissimilated to CH4, reinforcing the likelihood that glucose-derived carbon was also assimilated by methanogens. High amounts of the methanogenic substrates H2 and acetate (Zinder, 1993) accumulated in glucose treatments (Figures 2 and 3). These combined findings indicate that methanogens in the alimentary canal of E. eugeniae are closer to substrate saturation than are fermentative taxa.
Methanogens of the families Methanosarcinaceae, Methanobacteriaceae and Methanomicrobiaceae were previously detected in the E. eugeniae gut contents (Depkat-Jakob et al., 2012), and the detection of mcrA sequences related to these taxa (Figure 4b) suggest that these taxa are linked to methanogenesis in the alimentary canal. Members of Methanosarcinaceae are acetoclastic, that is, can convert acetate to CO2 and CH4 (Boone et al., 1993; Hedderich and Whitman, 2006). However, mcrA sequences related to the genus Methanosarcina were not detected in ‘heavy' fractions of [13C]-glucose treatments in which acetate accumulated (Figure 3). Thus acetate derived from supplemental glucose did not appear to be a dominant substrate for methanogenesis.
Analysis of mcrA sequences from ‘heavy' fractions of [13C]-glucose treatments indicated that methanogens closely related to M. formicicum and Methanoregula species were likely involved in the consumption of glucose-derived fermentation products. M. formicicum and species of Methanoregula utilize H2–CO2 and formate (Schauer and Ferry, 1980; Bräuer et al., 2011; Yashiro et al., 2011). However, which source of reductant (H2 or formate) was used for methanogenesis remains unclear. Four molecules of H2 or four molecules of formate are needed to produce one molecule of CH4 (Hedderich and Whitman, 2006). Approximately 80 μmol H2 per gram FW accumulated, and approximately 34 μmol formate per gram FW transiently accumulated and were subsequently consumed in the [13C]-glucose treatments (Figures 2 and 3), values that could theoretically yield approximately 20 μmol CH4 per gram FW and 8 μmol CH4 per gram FW, respectively. That only an additional 2 μmol CH4 per gram FW accumulated in glucose treatments compared with the amount of CH4 that accumulated in unsupplemented controls indicates that relatively little of the glucose-derived H2 and formate was linked to methanogenesis.
Four molecules of formate are required to synthesize one molecule of acetate via acetogenesis (Drake et al., 2008). Thus approximately 9 μmol acetate per gram FW could have been formed from the approximate 34 μmol formate per gram FW transiently formed from glucose in [13C]-glucose treatments. However, because acetate synthesis was continuous during the entire incubation period and far exceeded this value (Figure 3d), it is not possible to accurately correlate the synthesis of acetate via acetogenesis with the consumption of glucose-derived formate. Nonetheless, the occurrence of acetogens in the gut contents was indicated by (a) the detection of sequences in ‘heavy' fractions of [13C]-glucose treatments that were related to the acetogens C. glycolicum (Küsel et al., 2001) and C. mayombei (Kane et al., 1991) and (b) the H2-dependent stimulation of acetate synthesis in methanogenic enrichments. Thus acetate production may have been linked to both fermentation and acetogenesis. C. glycolicum and C. mayombei can utilize diverse substrates for acetogenesis, including H2–CO2, carbohydrates (for example, glucose), alcohols and organic acids (for example, formate) (Kane et al., 1991; Küsel et al., 2001), illustrating the potential of acetogens to facilitate multiple trophic links during the flow of carbon in the gut contents. C. mayombei can also convert succinate to propionate and CO2 (Kane et al., 1991). The capacity of acetogens to utilize a variety of electron donors and electron acceptors (Drake et al., 2008) reinforce the likelihood that acetogens could be metabolically active in the complex milieu of the gut contents of E. eugeniae.
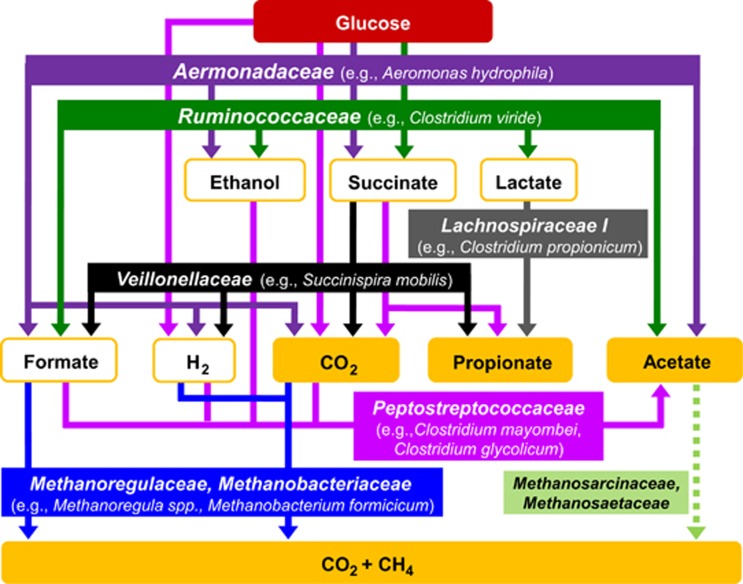
Conclusions
The collective findings of this study provide the basis for a hypothetical model of the processes and microbial taxa linked to the production of CH4 (Figure 6). Facultative aerobes of the family Aeromonadaceae and obligate anaerobes of the families Lachnospiraceae I, Veillonellaceae and Ruminococcaceae were stimulated by supplemental glucose and are conceived to be representative of taxa involved in the consumption of saccharides in the gut of E. eugeniae. Fermentation-derived fatty acids may serve as nutrients for the earthworm under in situ conditions (Wüst et al., 2009a).
Figure 6.
Hypothetical model of the methanogenic food web of the glucose-supplemented gut contents of the CH4-emitting earthworm E. eugeniae. The model is based on detected processes and known functions of the detected taxa. Compounds that accumulated as end products are shown in orange boxes. Species names in brackets represent the closest cultured relatives to retrieved sequences. The dashed light green arrow identifies a potential reaction that was not clearly resolved.
Different H2- and fatty acid-producing fermentations can be spatially distributed along the alimentary canal (Wüst et al., 2009a). Thus methanogenesis might be favored in parts of the alimentary canal where specific physico-chemical conditions favor methanogenesis. Fermentation-derived intermediates such as formate and H2 were likely utilized for methanogenesis by members of the families Methanobacteriaceae and Methanoregulaceae (Figure 6). Although acetoclastic methanogens (for example, Methanosarcinaceae and Methanosaetaceae) were detected, assimilation of glucose-derived acetate by such methanogens was not apparent, and acetate did not appear to stimulate acetoclastic methanogens in the SIP experiment or enrichments. However, it cannot be excluded that acetoclastic methanogens are active in the alimentary canal under in situ conditions.
Glucose was utilized in the present study as a representative saccharide found in the gut contents. However, the occurrence of other diverse saccharides such as maltose, mannose, galactose, arabinose and rhamnose in the alimentary canal of earthworms (Wüst et al., 2009a) suggests that the in situ methanogenic food web in the alimentary canal is more complex than that resolved in the present study. In addition, a more in-depth sequencing will be required to gain a more complete understanding of the prokaryotic species-level diversities of the microbial community associated with the methanogenic food web in the alimentary canal. Current studies are focused on the potential effects that different saccharides might have on trophic interactions that drive methanogenesis and on the in situ spatial orientation and regulation of methanogenesis in the alimentary canal of E. eugeniae.
Acknowledgments
We thank Anita S Gößner, Peter S Depkat-Jakob, Oliver Schmidt, Marcus A Horn (University of Bayreuth) and Peter Claus (Max-Planck-Institute for Terrestrial Microbiology) for technical assistance and helpful discussions. Support for this study was provided by the Deutsche Forschungsgemeinschaft (DR310/4-1, DR310/4-2), the University of Bayreuth, Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Embrapa Florestas, FAPESP (2008/58114-3, 2011/50914-3) and the University of São Paulo.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens—re-evaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barois I. Interactions entre les Vers de Terre (Oligochaeta) tropicaux geophages et la microflore pour l'exploitation de la matiere organique du sol. Travaux des Chercheurs de la Station de Lamto (Côte d'Ivoire) 1987;7:152–155. [Google Scholar]
- Boone DR, Whitman WB, Rouvière P.1993Diversity and taxonomy of methanogensIn: Ferry JG (ed).Methanogenesis Chapman and Hall: New York, NY, USA; 35–80. [Google Scholar]
- Bridgham SD, Cadillo-Quiroz H, Keller JK, Zhuang Q. Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob Chang Biol. 2013;19:1325–1346. doi: 10.1111/gcb.12131. [DOI] [PubMed] [Google Scholar]
- Brown GG, Barois I, Lavelle P. Regulation of soil organic matter dynamics and microbial activity in the drilosphere and the role of interactions with other edaphic functional domains. Eur J Soil Biol. 2000;36:177–198. [Google Scholar]
- Brown GG, James SW. Earthworm biodiversity in Sao Paulo state, Brazil. Eur J Soil Biol. 2006;42:145–149. [Google Scholar]
- Brune A.2006Symbiotic associations between termites and prokaryotesIn: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds).The Prokaryotes3rd edn.Springer-Verlag: New York, NY, USA; 439–474. [Google Scholar]
- Bräuer SL, Cadillo-Quiroz H, Ward RJ, Yavitt JB, Zinder SH. Methanoregula boonei gen. nov., sp. nov., an acidiphilic methanogen isolated from an acidic peat bog. Int J Syst Evol Microbiol. 2011;61:45–52. doi: 10.1099/ijs.0.021782-0. [DOI] [PubMed] [Google Scholar]
- Buckel W, Janssen PH, Schuhmann A, Eikmanns U, Messner P, Sleytr U, et al. Clostridium viride sp. nov., a strictly anaerobic bacterium using 5-aminovalerate as growth substrate, previously assigned to Clostridium aminovalericum. Arch Microbiol. 1994;162:387–394. [Google Scholar]
- Conrad R, Chan OC, Claus P, Casper P. Characterization of methanogenic Archaea and stable isotope fractionation during methane production in the profundal sediment of an oligotrophic lake (Lake Stechlin, Germany) Limnol Oceanogr. 2007;52:1393–1406. [Google Scholar]
- Darwin C. The Formation of Vegetable Mould Through the Action of Worms with Observations on Their Habits. John Murray: London, UK; 1881. [Google Scholar]
- Depkat-Jakob PS, Brown GG, Tsai SM, Horn MA, Drake HL. Emission of nitrous oxide and dinitrogen by diverse earthworm families from Brazil and resolution of associated denitrifying and nitrate-dissimilating taxa. FEMS Microbiol Ecol. 2013;83:375–391. doi: 10.1111/j.1574-6941.2012.01476.x. [DOI] [PubMed] [Google Scholar]
- Depkat-Jakob PS, Hunger S, Schulz K, Brown GG, Tsai SM, Drake HL. Emission of methane by Eudrilus eugeniae and other earthworms from Brazil. Appl Environ Microbiol. 2012;78:3014–3019. doi: 10.1128/AEM.07949-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich C, Köhler T, Brune A. The cockroach origin of the termite gut microbiota: patterns in bacterial community structure reflect major evolutionary events. Appl Environ Microbiol. 2014;80:2261–2269. doi: 10.1128/AEM.04206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez J.2004State of the art and new perspectives on vermicomposting researchIn: Edwards CA (ed).Earthworm Ecology2nd ednCRC Press: Boca Raton, FL, USA; 401–424. [Google Scholar]
- Drake HL, Gossner AS, Daniel SL. Old acetogens, new light. Ann NY Acad Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- Drake HL, Horn MA. As the worm turns: the earthworm gut as a transient habitat for soil microbial biomes. Annu Rev Microbiol. 2007;61:169–189. doi: 10.1146/annurev.micro.61.080706.093139. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Bohlen PJ. Biology and Ecology of Earthworms. vol. 3. Chapman and Hall: London, UK; 1996. [Google Scholar]
- Ezaki T.1984Genus I. RuminococcusIn: Krieg NR, Holt JG (eds).Bergy's Manual of Determinative Bacteriology The Williams and Wilkins Co.: Baltimore, MD, USA; 1015–1017. [Google Scholar]
- Ezaki T.2009Genus I. PeptostreptococcusIn: De Vos P, Garrity GM, Jones D, Krieg NR, Ludwig W, Rainey FA (eds).Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York, NY, USA; 1008–1009. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Furlong MA, Singleton DR, Coleman DC, Whitman WB. Molecular and culture-based analyses of prokaryotic communities from an agricultural soil and the burrows and casts of the earthworm Lumbricus rubellus. Appl Environ Microbiol. 2002;68:1265–1279. doi: 10.1128/AEM.68.3.1265-1279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk G.1986Bacterial Metabolism2nd edn.Springer: New York, NY, USA [Google Scholar]
- Heck KL, Vanbelle G, Simberloff D. Explicit calculation of rarefaction diversity measurement and determination of sufficient sample size. Ecology. 1975;56:1459–1461. [Google Scholar]
- Hedderich R, Whitman WB.2006Physiology and biochemistry of the methane-producing ArchaeaIn: Falkow S, Rosenberg E, Dworkin M (eds).The Prokaryotes3rd edn.Springer: New York, NY, USA; 1050–1079. [Google Scholar]
- Hippe H, Andreesen JR, Gottschalk aG.1992The genus Clostridium—nonmedicalIn: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer K-H (eds).The Prokaryotes2nd edn.Springer: New York, NY, USA; 1800–1825. [Google Scholar]
- Horn MA, Schramm A, Drake HL. The earthworm gut: an ideal habitat for ingested N2O-producing microorganisms. Appl Environ Microbiol. 2003;69:1662–1669. doi: 10.1128/AEM.69.3.1662-1669.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornor SG, Mitchell MJ. Effect of the earthworm Eisenia foetida (Oligochaeta) on fluxes of volatile carbon and sulfur compounds from sewage sludge. Soil Biol Biochem. 1981;13:367–372. [Google Scholar]
- Hunger S, Schmidt O, Hilgarth M, Horn MA, Kolb S, Conrad R, et al. Competing formate- and carbon dioxide-utilizing prokaryotes in an anoxic methane-emitting fen soil. Appl Environ Microbiol. 2011;77:3773–3785. doi: 10.1128/AEM.00282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlbert SH. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- Janssen PH.1984Genus XXXIV: SuccinispiraIn: Krieg NR, Holt JG (eds).Bergy's Manual of Determinative Bacteriology The Williams and Wilkins Co.: Baltimore, MD, USA; 1117–1118. [Google Scholar]
- Kane MD, Brauman A, Breznak JA. Clostridium mayombei sp. nov., an H2/CO2 acetogenic bacterium from the gut of the African soil-feeding termite Cubitermes speciosus. Arch Microbiol. 1991;156:99–104. [Google Scholar]
- Karsten GR, Drake HL. Comparative assessment of the aerobic and anaerobic microfloras of earthworm guts and forest soils. Appl Environ Microbiol. 1995;61:1039–1044. doi: 10.1128/aem.61.3.1039-1044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten GR, Drake HL. Denitrifying bacteria in the earthworm gastrointestinal tract and in vivo emission of nitrous oxide (N2O) by earthworms. Appl Environ Microbiol. 1997;63:1878–1882. doi: 10.1128/aem.63.5.1878-1882.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp B, Podmirseg S, Seeber J, Meyer E, Insam H. Diet-related composition of the gut microbiota of Lumbricus rubellus as revealed by a molecular fingerprinting technique and cloning. Soil Biol Biochem. 2009;41:2299–2307. [Google Scholar]
- Küsel K, Drake HL. Effects of environmental parameters on the formation and turnover of acetate by forest soils. Appl Environ Microbiol. 1995;61:3667–3675. doi: 10.1128/aem.61.10.3667-3675.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küsel K, Karnholz A, Trinkwalter T, Devereux R, Acker G, Drake HL. Physiological ecology of Clostridium glycolicum RD-1, an aerotolerant acetogen isolated from sea grass roots. Appl Environ Microbiol. 2001;67:4734–4741. doi: 10.1128/AEM.67.10.4734-4741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds).Nucleic Acid Techniques in Bacterial SystematicsJohn Wiley and Sons: Chichester, UK, pp115–175.
- Lavelle P. Associations mutualistes avec la microflore du sol et richess espécifique SOLIS les tropiques: l'hypothèse du premier maillon. Comptes Rendues de la Acadamie des Sciences de Paris. 1986;302:11–14. [Google Scholar]
- Leaver FW, Wood HG, Stjernholm R. The fermentation of three carbon substrates by Clostridium propionicum and Propionibacterium. J Bacteriol. 1955;70:521–530. doi: 10.1128/jb.70.5.521-530.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KE. Earthworms: Their Ecology and Relationships with Soils and Land Use. Academic Press: Sydney, Australia; 1985. [Google Scholar]
- Lottspeich F, Engels J. Bioanalytik. Spektrum Akademischer Verlag: Heidelberg, Germany; 2006. [Google Scholar]
- Lubbers I, Brussaard L, Otten W, Van Groenigen J. Earthworm-induced N mineralization in fertilized grassland increases both N2O emission and crop-N uptake. Eur J Soil Sci. 2011;62:152–161. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar A, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeschin F.1997Earthworms (Lumbricidae: Oligochaeta): important promoters of soil development and soil fertilityIn: Benckiser G (ed.)Fauna in Soil EcosystemsMarcel Dekker, Inc.: New York, NY, USA, pp173–223.
- Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Carnahan A, Joseph SW.2005Genus I. AeromonasIn: Brenner DJ, Krieg NR, Staley JT (eds)Bergey's Manual of Systematic Bacteriology2nd edn.Springer: New York, NY, USA; 557–578. [Google Scholar]
- Martin-Carnahan A, Joseph SW. Aeromonadaceae. Bergey's Manual of Systematic Bacteriology. 2005. pp. 556–578.
- Martinez AA.1998A Grande e Poderosa Minhoca: Manual Prático do Minhocultor4th edn.FUNEP: Jaboticabal, Brazil [Google Scholar]
- Matthies C, Grießhammer A, Schmittroth M, Drake HL. Evidence for involvement of gut-associated denitrifying bacteria in emission of nitrous oxide (N2O) by earthworms obtained from garden and forest soils. Appl Environ Microbiol. 1999;65:3599–3604. doi: 10.1128/aem.65.8.3599-3604.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mba CC. Biomass and vermicompost production by the earthworm Eudrilus eugeniae (Kinberg) Rev Biol Trop. 1988;37:11–13. [Google Scholar]
- Morrison M, Pope PB, Denman SE, McSweeney CS. Plant biomass degradation by gut microbiomes: more of the same or something new. Curr Opin Biotechnol. 2009;20:358–363. doi: 10.1016/j.copbio.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Morrison R.1984Family I. VeillonellaceaeIn: Krieg NR, Holt JG (eds).Bergy's Manual of Determinative Bacteriology The Williams and Wilkins Co.: Baltimore, MD, USA [Google Scholar]
- Nanninga HJ, Gottschal JC. Amino acid fermentation and hydrogen transfer in mixed cultures. FEMS Microbiol Ecol. 1985;31:261–269. [Google Scholar]
- Nechitaylo TY, Yakimov MM, Godinho M, Timmis KN, Belogolova E, Byzov BA, et al. Effect of the earthworms Lumbricus terrestris and Aporrectodea caliginosa on bacterial diversity in soil. Microb Ecol. 2010;59:574–587. doi: 10.1007/s00248-009-9604-y. [DOI] [PubMed] [Google Scholar]
- Oboh BO, Akintobi DO, Ejidereonwu C. Morphometric studies in Eudrilus eugeniae populations from different locations in Lagos, Nigeria. Nat Sci. 2007;5:16–21. [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acid Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhiya E, Bertora C, van Vliet PC, Kuikman PJ, Faber JH, van Groenigen JW. Earthworm activity as a determinant for N2O emission from crop residue. Soil Biol Biochem. 2007;39:2058–2069. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Satchell JE.1967LumbricidaeIn: Burges A, Raw F (eds).Soil Biology Academic Press: London, UK; 259–322. [Google Scholar]
- Schauer NL, Ferry JG. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980;142:800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Larget BR, Handelsman J. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol. 2004;70:5485–5492. doi: 10.1128/AEM.70.9.5485-5492.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt O, Horn MA, Kolb S, Drake HL.2014Temperature impacts differentially on the methanogenic food web of cellulose-supplemented peatland soil Environ Microbioldoi: 10.1111/1462-2920.12507 [DOI] [PubMed]
- Schmidt O, Wüst PK, Hellmuth S, Borst K, Horn MA, Drake HL. Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl Environ Microbiol. 2001;77:5842–5850. doi: 10.1128/AEM.05432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Cummins CS, Johnson JL.2006Familiy Propionibacteriaceae: the genus PropionibacteriumIn: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds).The Prokaryotes3rd edn.Springer: New York, NY, USA; 400–418. [Google Scholar]
- Šustr V, Šimek M. Methane release from millipedes and other soil invertebrates in Central Europe. Soil Biol Biochem. 2009;41:1684–1688. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigo D, Barois I, Garvin MH, Huertal E, Irisson S. Mutualism between earthworms and soil microflora. Pedobiologia. 1999;43:866–873. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley AS, Thomson B, Lueders T, Manefield M. RNA stable-isotope probing. Nat Protoc. 2007;2:838–844. doi: 10.1038/nprot.2007.115. [DOI] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Drake HL. In situ hydrogen and nitrous oxide as indicators of concomitant fermentation and denitrification in the alimentary canal of the earthworm Lumbricus terrestris. Appl Environ Microbiol. 2009;75:1852–1859. doi: 10.1128/AEM.02745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Drake HL. Clostridiaceae and Enterobacteriaceae as active fermenters in earthworm gut content. ISME J. 2011;5:92–106. doi: 10.1038/ismej.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Henderson G, Janssen PH, Rehm BH, Drake HL. Gut-associated denitrification and in vivo emission of nitrous oxide by the earthworm families Megascolecidae and Lumbricidae in New Zealand. Appl Environ Microbiol. 2009;75:3430–3436. doi: 10.1128/AEM.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüst PK, Horn MA, Henderson G, Janssen PH, Rehm BH, Drake HL. Gut-associated denitrification and in vivo emission of nitrous oxide by the earthworm families Megascolecidae and Lumbricidae in New Zealand. Appl Environ Microbiol. 2009;75:3430–3436. doi: 10.1128/AEM.00304-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza P, Richter M, Peplies J, Euzeby J, Amann R, Schleifer KH, et al. The all-species living tree project: A 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst Appl Microbiol. 2008;31:241–250. doi: 10.1016/j.syapm.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Yashiro Y, Sakai S, Ehara M, Miyazaki M, Yamaguchi T, Imachi H. Methanoregula formicica sp. nov., a methane-producing archaeon isolated from methanogenic sludge. Int J Syst Evol Microbiol. 2011;61:53–59. doi: 10.1099/ijs.0.014811-0. [DOI] [PubMed] [Google Scholar]
- Zinder SH.1993Physiological ecology of methanogensIn: Ferry JG (ed).Methanogenesis Chapman and Hall: New York, NY, USA; 128–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.