Abstract
Background
Crohn’s disease (CD) is a form of inflammatory bowel disease (IBD) with different described behaviors, including stricture. At present, there are no laboratory studies that can differentiate stricturing CD from other phenotypes of IBD. We performed a pilot study to examine differences in the proteome among patients with stricturing Crohn’s disease, non-stricturing Crohn’s disease, and ulcerative colitis (UC).
Methods
Serum samples were selected from the Ocean State Crohn’s and Colitis Area Registry (OSCCAR), an established cohort of patients with IBD. Crohn’s disease patients with surgically-resected stricture were matched with similar patients with Crohn’s disease without known stricture, and with UC. Serum samples from each patient were digested and analyzed using liquid chromatography-mass spectrometry to characterize the proteome. Statistical analyses were performed to identify peptides and proteins that can differentiate CD with stricture.
Results
Samples from 9 patients in each group (27 total patients) were analyzed. Baseline demographic characteristics were similar among the three groups. We quantified 7668 peptides and 897 proteins for analysis. ROC analysis identified a subset of peptides with an area under the curve greater than 0.9, indicating greater separation potential. Partial least squares discriminant analysis was able to distinguish among the three groups with up to 70% accuracy by peptides, and up to 80% accuracy by proteins. We identified the significantly different proteins and peptides, and determined their function based on previously published literature.
Conclusions
The serum of patients with stricturing CD, non-stricturing CD, and UC are distinguishable via proteomic analysis. Some of the proteins that differentiate the stricturing phenotype have been implicated in complement activation, fibrinolytic pathways, and lymphocyte adhesion.
Keywords: Risk assessment, complication, scarring, fibrosis, intestinal obstruction, clinical proteomics
Introduction
Inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC), is a chronic condition of the gastrointestinal tract that has the potential to cause significant morbidity. The etiology of IBD is not known, but is thought to be the result of interaction between host genotype(1, 2) and external factors, acting as potential environmental triggers(3). Diagnosis of this condition at present relies on clinical, laboratory, endoscopic, histopathologic, and radiologic information. While these data are typically reliable for making the diagnosis of IBD, in some cases it may be difficult to differentiate between UC and CD(4). Furthermore, both of these disorders have a variety of phenotypes in terms of disease location, extent, and behavior(5), which can significantly affect treatment decisions and patient outcomes. The behaviors commonly used in CD classification are penetrating and stricturing disease.
Crohn’s disease strictures can be divided into inflammatory and fibrotic subtypes. It is presumed that inflammatory strictures progress to fibrotic strictures and become resistant to medical therapy(6). If the degree of resulting stenosis is severe enough, patients develop acute or chronic bowel obstruction requiring surgical intervention. At this time there is no way to predict whether a patient will develop this type of complication.
Biomarkers have the potential to yield valuable information about disease behavior and activity, either in conjunction with or in place of more invasive techniques. However, researchers have had limited success at uncovering biomarkers that can reliably differentiate IBD patients from healthy controls, CD from UC, or predict disease course or treatment response(7). In the past, biomarker discovery has been limited to proteins and metabolites that are already suspected to be involved in IBD pathogenesis. More recently, the use of mass spectroscopy and bioinformatic techniques has permitted the rapid analysis of thousands of peptides and proteins from the same sample. The results from these data-driven studies have the potential to identify candidate biomarkers deserving further evaluation and new hypotheses for mechanistic investigation(8).
The proteome is defined as the full complement of proteins encoded by a genome(9). Proteomics is the branch of systems biology examining the proteome, which seeks not only to catalog and quantify these proteins, but also to examine them in a holistic manner, capturing the full spectrum of protein forms and modifications, protein complexes, and interactions among them(10). Increasing knowledge of the proteome in health and in disease states has the potential to yield valuable insights into physiology and pathophysiology, as proteins are directly involved in nearly all physiologic processes. Their quantity and activity can vary markedly in different tissues, different stages of development, and illnesses(11, 12). The nature of the proteome makes proteomic studies particularly useful for discovering novel biomarkers or identifying potential drug targets (13).
In this pilot study, we attempted to determine the feasibility of using serum protein expression profiles to differentiate among patients with UC, inflammatory Crohn’s disease, and Crohn’s disease with fibrotic stricture. We hypothesized that individuals with a stricturing phenotype will express different proteins from the other two groups, and that some of these proteins may play a role in the development of fibrosis. Our aim is to use the information from this study to generate novel candidate serum markers for further validation and new hypotheses for future studies on stricturing Crohn’s disease.
Materials and Methods
Sample selection
The Ocean State Crohn’s and Colitis Area Registry (OSCCAR) is a community-based inception cohort that consists of 408 adult and pediatric patients diagnosed with inflammatory bowel disease between 2008 and 2012 in the state of Rhode Island(14). Patients were enrolled within one year of diagnosis of IBD, and had serum collected and stored at −80°C in BD P100 (BD Diagnostics) sample tubes at the time of enrollment, as well as yearly thereafter. The Montreal classification (MC) system was used to describe disease phenotype at baseline(5). Disease location in Crohn’s disease was designated as L1 (ileal), L2 (colonic), or L3 (ileocolonic) with L4 as a modifier designating concomitant upper tract disease. Upper tract disease (L4) was based on endoscopic findings of inflammation and ulceration proximal to the ligament of Treitz. Behavior was defined as B1 (inflammatory, non-stricturing and non-penetrating), B2 (stricturing), or B3 (fistulizing/penetrating) with a P modifier to describe concomitant perianal disease. Ulcerative colitis disease location was described as E1 (proctitis), E2 (left sided disease), or E3 (pancolitis). Classifications were based on all available endoscopic, radiographic and surgical data obtained within 3 months of IBD diagnosis. We selected patients in this cohort with Crohn’s disease who had an ileal resection for stricture, as described either on surgical report or pathology, as index patients. For each of these index patients, we matched two other patients from the cohort—one with Crohn’s disease but no stricture, and one with ulcerative colitis— based on age, gender, and medication exposure. For the index patients, we analyzed the serum sample drawn closest to the time of surgery. For the matched patients, we analyzed the sample drawn at the interval that best approximated the time from diagnosis to surgery for the index patient.
Proteomic Sample Preparation
Each serum sample (200 μL) was subjected to immunoaffinity depletion to remove the top 14 most abundant proteins using an IgY-14 LC5 column (Sigma-Aldrich) coupled with an Agilent 1100 series HPLC, under the conditions suggested by the manufacturer for mobile phases and flow rate. The flow-through fractions (low abundant proteins) were collected and concentrated in Amicon Ultra-15 concentrators (Millipore) with molecular weight cut off of 3 kDa, followed by a buffer exchange to 50 mM NH4HCO3 in the same unit according to the manufacturer’s instructions. The low abundant proteins were next sequentially denatured with 8 M urea, reduced with 5 mM dithiothreitol, alkylated with 20 mM iodoacetamide, and digested with trypsin (Promega) at a trypsin/protein ratio of 1:50; the peptide mixtures were then cleaned with C18 SPE cartridges (Sigma-Aldrich) and dried in vacuo before isobaric labeling of peptides. For preparation of whole serum protein digest, 20 μL of each serum sample was directly digested and cleaned up following the same procedures as described previously for the low abundant proteins.
Aliquots of the 27 individually digested samples were pooled to create a universal reference sample, which was labeled using iTRAQ 4-plex reagents (AB SciEx), together with equal amount of peptides derived from 3 clinical matched samples (Strictured Crohn’s, Crohn’s and Ulcerative Colitis), according to the manufacturer recommendations. These 4 individually iTRAQ labeled samples were then pooled to form one experiment; and in total 9 experiments each were created for low abundant protein and whole serum protein digests, respectively. To avoid systematic errors from the labeling reagent, labeling was randomized within experiments. The pooled sample of each experiment was subjected to automated offline high pH reversed phase fractionation as reported previously(15), and 12 fractions were generated in the final concatenation process. Each fraction was dried and reconstituted in 0.1% formic acid for a final concentration of 0.1 μg/μL. In total, 108 fractionated peptide samples were subjected to LC-MS/MS instrument analysis for peptide samples originated from the whole serum proteins and low abundant proteins, respectively.
Instrument Analysis
Five μL of each fractionated peptide sample was separated on a 2-column custom-built capillary LC system similar as reported previously(16). Reversed-phase separations were carried out on 35cm × 75 μm i.d. fused silica columns packed in house using 3μ Jupiter C18 particles (Phenomenex). Mobile phases consisted of 0.1% formic acid in water (A) and 0.1% formic acid in 100% acetonitrile (B) with a 100 min gradient. MS analyses were performed using an Orbitrap Velos Pro Hybrid mass spectrometer (ThermoScientific) outfitted with a custom electrospray ionization (ESI) interface, which was operated in data dependent MS/MS mode, with one high resolution (Resolution of 60,000 at 400 m/z) MS scan followed by HCD MS/MS scan events (resolution of 15,000 at 400 m/z) at normalized collision energy of 32.
Data Processing and Statistical Analysis
Protein identifications were performed using MSGF+ search engine against Uniprot database of human protein sequences (version of 2012-04-30) and the decoy database. Search parameters were set as follows: monoisotopic mass, peptide mass tolerance at ± 10 ppm and fragment mass tolerance at 0.1 Da, trypsin as the enzyme and allowing up to two missed cleavages. Lysine and N-term of peptides labeled by iTRAQ 4-plex and carbamidomethylation on cysteine were specified as fixed modifications, while oxidation of methionine was defined as variable modification. False discovery rate (FDR) of peptides identification was set to be < 1%. Protein identification was supported by at least one unique peptide identification. iTRAQ reporter ion intensities for identified peptides were extracted using MASIC (17).
Crosstab file that contains peptide ID and peptide abundance in each sample was imported to MatLab® for statistical analysis. The abundance ratio of each sample peptide to the reference pool peptide was log10 transformed and all samples were median centered for data normalization. Missing data were imputed using a Regularized Expectation Maximization (REM) algorithm. Peptides with a significant quantitative difference were calculated using repeated measures ANOVA with a Tukey post-hoc test. Partial least squares- discriminant analysis (PLS-DA) was employed as a supervised learning approach to evaluate the classification potential among the disease groups. PLS permits regression analysis of data sets that contain large numbers of variables compared to observations. Cross-validation was repeated 100 times to attain a robust estimate of the classification accuracy. The Area under a Receiver Operating Characteristic Curve (AUC) was also computed to look at the classification potential of each peptide for comparing between any two disease groups.
Significantly changed peptides in any of the analyses were correlated with the proteins from which they originate. The biological functions and the roles of these proteins in disease were derived from Uniprot.
Results
Patient characteristics
We analyzed three groups, each consisting of 9 patients. The three groups were similar in baseline characteristics (Table 1). Only 3 of the 9 CD patients with stricture resection were categorized as having the B2 (stricturing) phenotype by Montreal classification at enrollment. The one patient in the comparison CD group initially classified as B2 at diagnosis, has not had surgery 5 years after diagnosis, and has not been treated with a biologic agent.
Table 1.
Summary of patient characteristics. Age and disease duration data were recorded at the time of blood draw. Montreal classification obtained at enrollment.
| CD- confirmed fibrotic Stricture | CD-Control | UC | |
|---|---|---|---|
| # of patients | 9 | 9 | 9 |
| Characteristics | |||
| Age in years, mean (range) | 37 (8.8–78.1) | 34.5 (7.1–66.6) | 36.8 (10–74.9) |
| % Female | 44 | 44 | 44 |
| Duration of disease in months, mean (range) | 11.6(0.8–27.1) | 8.3(0.2–25.8) | 12(1.3–25.4) |
| Montreal Classification | |||
| L1 | 4 | 4 | N/A |
| L2 | 1 | 2 | N/A |
| L3 | 4 | 3 | N/A |
| L4* | 2 | 3 | N/A |
| B1 | 4 | 7 | N/A |
| B2 | 3 | 1 | N/A |
| B3 | 2 | 1 | N/A |
| P* | 0 | 1 | N/A |
| E1 | N/A | N/A | 1 |
| E2 | N/A | N/A | 4 |
| E3 | N/A | N/A | 4 |
Modifier designating concomitant upper tract and perianal disease
Group pair comparison
We quantified 7668 peptides and 897 proteins through the iTRAQ-labeling proteomics method. Table 2 shows a summary of the AUC values for peptides found to be significantly different between any two of the three groups. All of the comparisons revealed similar numbers of significantly different peptides as identified by ANOVA. Only 16 peptides had AUC values >0.9 for differentiating between CD with stricture and CD without stricture. The proteins containing these peptides, and a brief description of their function, are listed in Table 3. There were 143 and 135 peptides with AUC values >0.9 separating UC from stricturing CD and CD without stricture, respectively.
Table 2.
Results of the Area Under a Receiver Operating Curve analysis of peptides, separated by peptide analysis technique. This provides a summary of peptides that showed potential for differentiating between two groups. Significantly different peptides have ANOVA p-value of <0.05. See Table 3 for a description of the proteins differentiating stricturing CD from CD without stricture with AUC value >0.9. iTRAQ = Isobaric tags for relative and absolute quantitation (labeled peptides), WS = whole serum, DS = depleted serum
| Patient Group Comparison | Analysis method | Number of significantly different peptides between groups | Average AUC | Number of peptides with AUC >0.9 |
|---|---|---|---|---|
| Stricturing Crohn’s vs Crohn’s without stricture | iTRAQ DS | 192 | 0.68 | 8 |
| iTRAQ WS | 455 | 0.62 | 8 | |
| Stricturing Crohn’s vs ulcerative colitis | iTRAQ DS | 188 | 0.72 | 15 |
| iTRAQ WS | 456 | 0.83 | 128 | |
| Crohn’s without stricture vs ulcerative colitis | iTRAQ DS | 189 | 0.69 | 10 |
| iTRAQ WS | 456 | 0.83 | 125 |
Table 3.
List of proteins (based on peptide data in Table 2) that differentiate stricturing CD from CD without stricture with AUC value >0.9. Included are genes associated with the proteins, as well as brief description of protein function based on available literature.
| Protein name | Gene name | Reported Functions |
|---|---|---|
| Alpha-2-macroglobulin | A2M | Multiple: TNF, IL-1, and IL-8 binding; platelet activation; extracellular matrix organization; negatively regulates complement activation(31) |
| L-lactate dehydrogenase B chain | LDHB | Catalyzes fermentation of pyruvate to lactate(32) |
| Calpain small subunit 1 | CAPNS1 | Thrombosis, extracellular matrix organization and disassembly(33, 34) |
| Mannose-binding protein C | MBL2 | Enhances phagocystosis, activates complement pathway(35) |
| Apolipoprotein B-100 | APOB | Component of chylomicrons, LDL, and VLDL(36) |
| CD5 antigen-like | CD5L | Inhibits apoptosis, regulates lymphocyte binding(37) |
| Serum albumin | ALB | Ubiquitous |
| Filamin-A | FLNA | Promotes actin branching and linking(38) |
| Uteroglobin | SCGB1A1 | Inhibits phospholipase A2, anti-inflammatory effects(39) |
| Bromodomain testis-specific protein | BRDT | Spermatogenesis(40) |
| Cathepsin D | CTSD | Endopeptidase activity; mutations linked with several cancers(41) |
| Proteasome subunit alpha type-7 | PSMA7 | Part of proteinase complex with broad activity(42) |
| Ceruloplasmin | CP | Binds copper, involved in iron transport(43) |
| Oncoprotein-induced transcript 3 protein | OIT3 | Unknown; may be involved in hepatocellular function(44) |
| Prothrombin | F2 | Coagulation; converts fibrinogen to fibrin(45) |
| Putative tenascin-XA | TNXA | Unknown |
Partial least squares- discriminant analysis
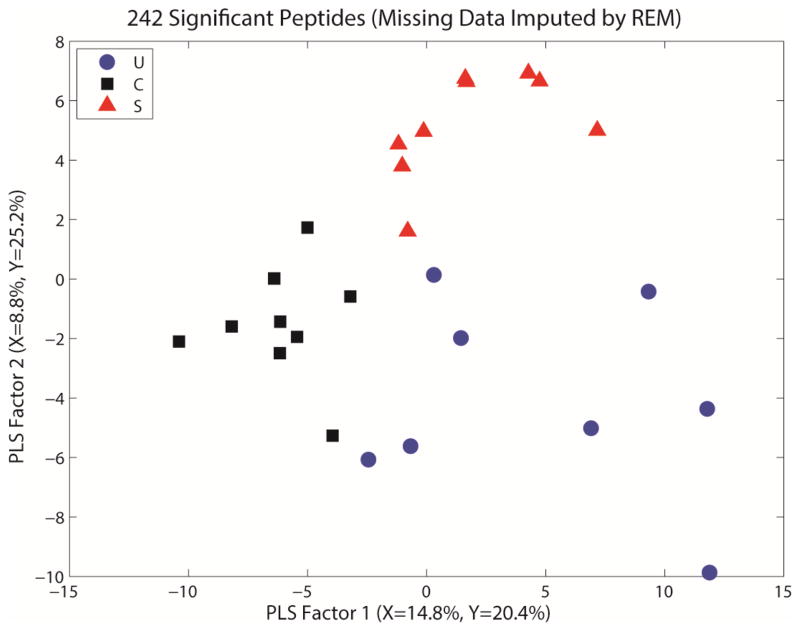
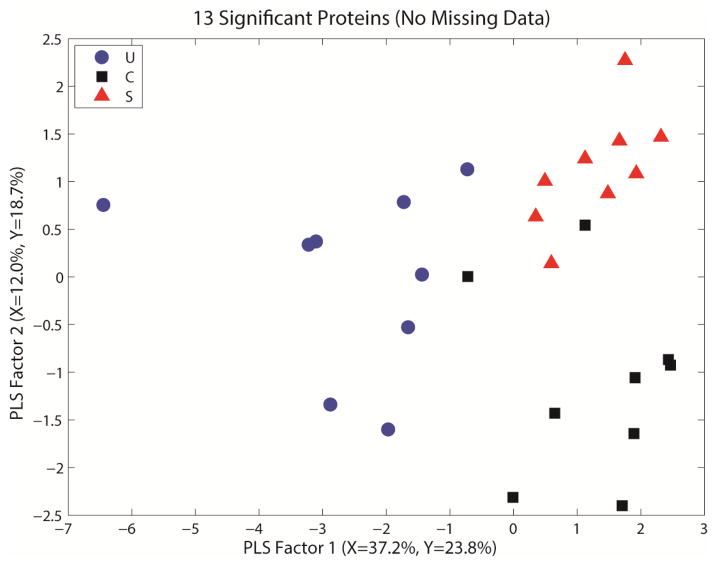
Figures 1 and 2 show representative PLS-DA regression plots for the peptide and protein data, respectively. These figures show each of the IBD phenotypes segregating in the regression graph, indicating different protein profiles. Based on these models, we determined the classification accuracy using different numbers of latent variables. For the peptide dataset, we observed that the best models use only 2 or 3 PLS factors, and yield 65–70% average accuracy. For the protein dataset, the best models also used 2 or 3 PLS factors, and yield 70–80% average accuracy.
Figure 1.
PLS plot for the whole serum iTRAQ dataset with missing data for peptides imputed via regularized expectation maximization for analysis. This provides a visual representation of the separation among the groups via regression modeling, showing each phenotype clustering in a different area of the regression graph. S= stricturing Crohn’s, C= Crohn’s without stricture, U= ulcerative colitis.
Figure 2.
PLS plot for the whole serum iTRAQ dataset using only proteins with complete data. This provides a visual representation of the separation among the groups via regression modeling, showing each phenotype clustering in a different area of the regression graph. S= stricturing Crohn’s, C= Crohn’s without stricture, U= ulcerative colitis.
Discussion
Our findings indicate that proteomic analysis by LC-MS is able to distinguish among patients with ulcerative colitis, non-stricturing Crohn’s disease, and stricturing Crohn’s disease. To our knowledge, correlating protein profile with IBD location, extent, or behavior represents novel findings.
As proteomics relates to IBD specifically, there are a number of studies that have attempted to gain insight into an aspect of IBD based on protein profiling(18–21). Nanni et al. found 20 peaks of protein expression in the serum that distinguished between subjects with IBD and healthy controls with a high degree of accuracy(22). Subramanian et al. examined the serum of CD and UC patients, and identified 12 peaks of protein expression that differentiated between the two(23). Hatsugai et al. studied serum mononuclear cells specifically, finding 58 proteins that helped discriminate between CD and UC(24). Other researchers, such as M’Koma et al, have evaluated colonic tissue directly, finding that protein expression differentiated between inflamed and non-inflamed mucosa, and between UC and CD patients(25). These studies commonly found peaks of protein expression that did not correlate with known proteins. For instance, in the Hatsugai study, only 11 of the 58 candidate proteins could be identified. Meuwis et al. performed a similar study to the above, using serum to distinguish among CD, UC, healthy controls, and other inflammatory conditions(26). Later, the Meuwis group also performed a pilot study to determine if proteome profiling could distinguish between those who did or did not respond to TNF-antagonist therapy, identifying platelet aggregation factor 4 as a potential biomarker for predicting non-response(27).
Our study found many proteins and peptides that are differentially expressed among the three groups. The PLS-DA analysis examines all of the proteins found to be significantly different by ANOVA together, and shows that patients with fibrotic stricture may have a protein profile that distinguishes them from other patients with IBD (Fig 1 and 2). Table 2 shows that many fewer peptides strongly differentiate between the two behaviors of CD than between UC and either phenotype of CD. The peptides with the greatest separation potential between stricturing and non-stricturing CD (see Table 3) are a much smaller set than those used in the PLS analysis, and may provide a more manageable list for targeting validation studies. Our study did not attempt to discern mechanisms by which these proteins may lead to fibrosis. Several of the proteins have multiple functions beyond those listed, and the role of these proteins in the pathogenesis of fibrotic strictures is not clear.
While the proteome contains a potential wealth of information at the basic functional level of an individual, there are still significant technical hurdles in proteomics techniques. Mass spectrometry is able to analyze and quantify thousands of proteins at a time, based on a unique mass-to-charge (m/z) ratio. However, not all proteins can be properly identified based on m/z ratio alone(28), or linked with the gene that gives rise to it. Analyzing unique peptide strings may capture the same protein in different conformations, but inference of protein quantity from peptide quantity is not always precise(29). The range of protein quantity is wide, varying up to 9 orders of magnitude from the most to least abundant proteins, and many important proteins such as interleukins are on the lower end of that range. Taken as a whole, other studies have estimated that 75% of the entire protein mass consists of a few thousand “housekeeping proteins”, found in all cells or tissues and carrying out basic roles in cell structure and function(30). Individual proteins often have multiple roles in a cell, and frequently interact with other proteins to achieve those roles, so a change in one protein may only be relevant if certain other proteins have changed as well. Finally, researchers have struggled with reproducibility issues, as there is no single broadly-accepted technique for analyzing a proteome sample.
There are a number of limitations to our study beyond those common to proteomic studies as a whole. As a pilot study, the sample size from each group is small, due to the size of the starting cohort and strict definition of stricture used for inclusion. We know that the cumulative incidence of stricture increases with time, and some of the patients considered to have non-stricturing Crohn’s disease may have been developing a fibrotic stricture at the time of the blood draw. Some patients had a stricture at initial presentation, but did not have their serum collected until after a resection was already performed, which may change their protein profile. Finally, many factors may influence protein expression, and disease behavior alone may not have caused the variability seen in this group.
This study highlights the need for further research into the proteome for patients with inflammatory bowel disease. Future studies may analyze larger groups of patients to confirm these results, and may be able to refine our findings or identify additional candidate proteins for evaluation as biomarkers. Finally, validation studies will help to determine whether candidate proteins are able to function as useful biomarkers.
Acknowledgments
This work was partially supported by the National Institutes of Health (DK095818); the Crohn’s and Colitis Foundation of America through a grant from the Centers for Disease Control and Prevention (1 UO1 DP000340-03). Portions of this work were performed at the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the Department of Energy’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory (PNNL) in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle for the DOE under Contract DE-AC05-76RLO 1830.
Footnotes
Author contributions: Development of question: B. Sands, N. LeLeiko, J. Shapiro, S. Shah, P. Townsend, Q. Zhang; Clinical design and sample collection: B. Sands, P. Townsend, J. Shapiro, N. Leleiko; R. Bright; Proteomics experiment design: Q. Zhang; Sample preparation and data acquisition: A. Schepmoes, K.K. Weitz; Statistical data analysis: L. Bramer, B.-J. Webb-Robertson, Q. Zhang; Drafting of manuscript: P. Townsend, Q. Zhang, N. LeLeiko, J. Shapiro, B. Sands. Critical review of manuscript: P. Townsend, Q. Zhang, J. Shapiro, B. Webb-Robertson, L. Bramer, A. Schepmoes, K. Weitz, M. Mallette, H. Moniz, R. Bright, A. Samad, T. Kamphaus, S.A. Shah, B.E. Sands, N. LeLeiko.
References
- 1.Anderson CA, Boucher G, Lees CW, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature genetics. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Toole A, Korzenik J. Environmental triggers for IBD. Current gastroenterology reports. 2014;16:396. doi: 10.1007/s11894-014-0396-y. [DOI] [PubMed] [Google Scholar]
- 4.Melmed GY, Elashoff R, Chen GC, et al. Predicting a change in diagnosis from ulcerative colitis to Crohn’s disease: a nested, case-control study. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2007;5:602–608. doi: 10.1016/j.cgh.2007.02.015. quiz 525. [DOI] [PubMed] [Google Scholar]
- 5.Silverberg MS, Satsangi J, Ahmad T, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 2005;19 (Suppl A):5a–36a. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 6.Li C, Kuemmerle JF. Mechanisms that mediate the development of fibrosis in patients with Crohn’s disease. Inflammatory bowel diseases. 2014;20:1250–1258. doi: 10.1097/MIB.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benor S, Russell GH, Silver M, et al. Shortcomings of the inflammatory bowel disease Serology 7 panel. Pediatrics. 2010;125:1230–1236. doi: 10.1542/peds.2009-1936. [DOI] [PubMed] [Google Scholar]
- 8.Han NY, Choi W, Park JM, et al. Label-free quantification for discovering novel biomarkers in the diagnosis and assessment of disease activity in inflammatory bowel disease. Journal of digestive diseases. 2013;14:166–174. doi: 10.1111/1751-2980.12035. [DOI] [PubMed] [Google Scholar]
- 9.Wilkins MR, Pasquali C, Appel RD, et al. From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and amino acid analysis. Bio/technology (Nature Publishing Company) 1996;14:61–65. doi: 10.1038/nbt0196-61. [DOI] [PubMed] [Google Scholar]
- 10.Tyers M, Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- 11.Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 12.Merlos Rodrigo MA, Zitka O, Krizkova S, et al. MALDI-TOF MS as evolving cancer diagnostic tool: A review. Journal of pharmaceutical and biomedical analysis. 2014;95C:245–255. doi: 10.1016/j.jpba.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Han NY, Kim EH, Choi J, et al. Quantitative proteomic approaches in biomarker discovery of inflammatory bowel disease. J Dig Dis. 2012;13:497–503. doi: 10.1111/j.1751-2980.2012.00625.x. [DOI] [PubMed] [Google Scholar]
- 14.Sands BE, LeLeiko N, Shah SA, et al. OSCCAR: Ocean State Crohn’s and Colitis Area Registry. Medicine and health, Rhode Island. 2009;92:82–85. 88. [PubMed] [Google Scholar]
- 15.Wang Y, Yang F, Gritsenko MA, et al. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics. 2011;11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigdel TK, Nicora CD, Hsieh SC, et al. Optimization for peptide sample preparation for urine peptidomics. Clin Proteomics. 2014;11:7. doi: 10.1186/1559-0275-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monroe ME, Shaw JL, Daly DS, et al. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput Biol Chem. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen CS, Sullivan S, Anderson T, et al. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Molecular & cellular proteomics : MCP. 2009;8:1765–1776. doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yau Y, Leong RW, Zeng M, et al. Proteomics and metabolomics in inflammatory bowel disease. Journal of gastroenterology and hepatology. 2013;28:1076–1086. doi: 10.1111/jgh.12193. [DOI] [PubMed] [Google Scholar]
- 20.Alex P, Gucek M, Li X. Applications of proteomics in the study of inflammatory bowel diseases: Current status and future directions with available technologies. Inflammatory bowel diseases. 2009;15:616–629. doi: 10.1002/ibd.20652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaiopoulou A, Gazouli M, Theodoropoulos G, et al. Current advantages in the application of proteomics in inflammatory bowel disease. Digestive diseases and sciences. 2012;57:2755–2764. doi: 10.1007/s10620-012-2291-4. [DOI] [PubMed] [Google Scholar]
- 22.Nanni P, Parisi D, Roda G, et al. Serum protein profiling in patients with inflammatory bowel diseases using selective solid-phase bulk extraction, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and chemometric data analysis. Rapid communications in mass spectrometry : RCM. 2007;21:4142–4148. doi: 10.1002/rcm.3323. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian V, Subramanian D, Pollok RC. S1182 Serum Protein Signatures Determined By Mass Spectrometry (SELDI-ToF) Accurately Distinguishes Crohn’s Disease (CD) from Ulcerative Colitis (UC) Gastroenterology. 2008;134:A-196. [Google Scholar]
- 24.Hatsugai M, Kurokawa MS, Kouro T, et al. Protein profiles of peripheral blood mononuclear cells are useful for differential diagnosis of ulcerative colitis and Crohn’s disease. Journal of gastroenterology. 2010;45:488–500. doi: 10.1007/s00535-009-0183-y. [DOI] [PubMed] [Google Scholar]
- 25.M’Koma AE, Seeley EH, Washington MK, et al. Proteomic profiling of mucosal and submucosal colonic tissues yields protein signatures that differentiate the inflammatory colitides. Inflammatory bowel diseases. 2011;17:875–883. doi: 10.1002/ibd.21442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meuwis MA, Fillet M, Geurts P, et al. Biomarker discovery for inflammatory bowel disease, using proteomic serum profiling. Biochem Pharmacol. 2007;73:1422–1433. doi: 10.1016/j.bcp.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Meuwis MA, Fillet M, Lutteri L, et al. Proteomics for prediction and characterization of response to infliximab in Crohn’s disease: a pilot study. Clinical biochemistry. 2008;41:960–967. doi: 10.1016/j.clinbiochem.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 29.Paulo JA. Practical and Efficient Searching in Proteomics: A Cross Engine Comparison. WebmedCentral. 2013:4. doi: 10.9754/journal.wplus.2013.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim MS, Pinto SM, Getnet D, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaMarre J, Wollenberg GK, Gonias SL, et al. Cytokine binding and clearance properties of proteinase-activated alpha 2-macroglobulins. Laboratory investigation; a journal of technical methods and pathology. 1991;65:3–14. [PubMed] [Google Scholar]
- 32.Adeva M, Gonzalez-Lucan M, Seco M, et al. Enzymes involved in l-lactate metabolism in humans. Mitochondrion. 2013;13:615–629. doi: 10.1016/j.mito.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 33.Tyagi T, Ahmad S, Gupta N, et al. Altered expression of platelet proteins and calpain activity mediate hypoxia-induced prothrombotic phenotype. Blood. 2014;123:1250–1260. doi: 10.1182/blood-2013-05-501924. [DOI] [PubMed] [Google Scholar]
- 34.Undyala VV, Dembo M, Cembrola K, et al. The calpain small subunit regulates cell-substrate mechanical interactions during fibroblast migration. Journal of cell science. 2008;121:3581–3588. doi: 10.1242/jcs.036152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nauta AJ, Raaschou-Jensen N, Roos A, et al. Mannose-binding lectin engagement with late apoptotic and necrotic cells. European journal of immunology. 2003;33:2853–2863. doi: 10.1002/eji.200323888. [DOI] [PubMed] [Google Scholar]
- 36.Olofsson SO, Wiklund O, Boren J. Apolipoproteins A-I and B: biosynthesis, role in the development of atherosclerosis and targets for intervention against cardiovascular disease. Vascular health and risk management. 2007;3:491–502. [PMC free article] [PubMed] [Google Scholar]
- 37.Tissot JD, Sanchez JC, Vuadens F, et al. IgM are associated to Sp alpha (CD5 antigen-like) Electrophoresis. 2002;23:1203–1206. doi: 10.1002/1522-2683(200204)23:7/8<1203::AID-ELPS1203>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 38.Ciobanasu C, Faivre B, Le Clainche C. Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. European journal of cell biology. 2013;92:339–348. doi: 10.1016/j.ejcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Antico G, Aloman M, Lakota K, et al. Uteroglobin, a possible ligand of the lipoxin receptor inhibits serum amyloid A-driven inflammation. Mediators of inflammation. 2014;2014:876395. doi: 10.1155/2014/876395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berkovits BD, Wolgemuth DJ. The role of the double bromodomain-containing BET genes during mammalian spermatogenesis. Current topics in developmental biology. 2013;102:293–326. doi: 10.1016/B978-0-12-416024-8.00011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Achour O, Bridiau N, Kacem M, et al. Cathepsin D activity and selectivity in the acidic conditions of a tumor microenvironment: Utilization in the development of a novel Cathepsin D substrate for simultaneous cancer diagnosis and therapy. Biochimie. 2013;95:2010–2017. doi: 10.1016/j.biochi.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Li N, Zhang Z, Zhang W, et al. Calcineurin B subunit interacts with proteasome subunit alpha type 7 and represses hypoxia-inducible factor-1alpha activity via the proteasome pathway. Biochemical and biophysical research communications. 2011;405:468–472. doi: 10.1016/j.bbrc.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 43.Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annual review of nutrition. 2002;22:439–458. doi: 10.1146/annurev.nutr.22.012502.114457. [DOI] [PubMed] [Google Scholar]
- 44.Xu ZG, Du JJ, Zhang X, et al. A novel liver-specific zona pellucida domain containing protein that is expressed rarely in hepatocellular carcinoma. Hepatology (Baltimore, Md) 2003;38:735–744. doi: 10.1053/jhep.2003.50340. [DOI] [PubMed] [Google Scholar]
- 45.Krishnaswamy S. The transition of prothrombin to thrombin. Journal of thrombosis and haemostasis : JTH. 2013;11 (Suppl 1):265–276. doi: 10.1111/jth.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]