Abstract
TMPRSS2 gene rearrangements occur at DNA breaks formed during androgen receptor-mediated transcription and activate expression of ETS transcription factors at the early stages of more than half of prostate cancers. NKX3.1, a prostate tumor suppressor that accelerates the DNA repair response, binds to androgen receptor at the ERG gene breakpoint and inhibits both the juxtaposition of the TMPRSS2 and ERG gene loci and also their recombination. NKX3.1 acts by accelerating DNA repair after androgen-induced transcriptional activation. NKX3.1 influences the recruitment of proteins that promote homology-directed DNA repair. Loss of NKX3.1 favors recruitment to the ERG gene breakpoint of proteins that promote error-prone nonhomologous end-joining. Analysis of prostate cancer tissues showed that the presence of a TMPRSS2-ERG rearrangement was highly correlated with lower levels of NKX3.1 expression consistent with the role of NKX3.1 as a suppressor of the pathogenic gene rearrangement.
Keywords: NKX3.1, TMRPSS2-ERG, androgen receptor, prostate cancer, gene rearrangement
Introduction
Prostate cancer occurrence increases with age to a greater degree than any other solid tumor. Early molecular events in prostate cancer often include both loss of NKX3.1 expression and gene rearrangement involving members of the ETS family (1, 2). Here we show that NKX3.1 protein loss that occurs in regions of prostate inflammation and intraepithelial neoplasia predisposes to the predominant ETS gene rearrangement involving TMPRSS2 and ERG. The TMRPSS2 and ERG rearrangement on chromosome 21 that results in the placement of the ERG gene downstream from the androgen-responsive TMPRSS2 promoter is the most frequent chromosomal rearrangement in prostate cancer (3). NKX3.1 protein loss occurs as a result of genetic deletion or gene methylation. Moreover, inflammation, a common finding in the aging prostate, can cause NKX3.1 loss by triggering protein ubiquitination and proteasomal degradation (4, 5).
TMPRSS2-ERG rearrangement is mediated by the action of androgen receptor that binds to both genes and brings their DNA in juxtaposition at the rearrangement breakpoints (6, 7). After binding to androgen response elements, androgen receptor initiates transcription via a process that is mediated by DNA breakage (8). In the course of transcriptional activation by steroid hormone receptors a ligand-bound steroid hormone receptor will complex with chromatin and generate hydrogen peroxide during demethylation of histone H3 dimethylysine (8). Hydrogen peroxide reacts with DNA to form 8-oxoguanine. Repair of the 8-oxo-G modification follows initiation of steroid hormone-induced transcription as indicated by involvement of 8-oxo-G glycolase-1. Thus the finding that base excision repair is a functional component of steroid-receptor mediated transcription is consistent with the observation that AR mediates rearrangement of the TMPRSS2 and ERG genes (6).
NKX3.1 exerts tumor suppressive effects in part by activating cellular response to oxidation and by enhancing repair of DNA damage. Nkx3.1 gene targeted mice have attenuated response to cellular oxidative stress (9). NKX3.1 activates expression of genes associated with DNA repair (10). Moreover, NKX3.1 itself is involved in DNA repair. NKX3.1 enhances cell survival after DNA damage and migrates to sites of DNA damage to recruit DNA repair proteins and augment their activation (11, 12). NKX3.1 also binds to and activates topoisomerase I (13). It was recently shown that NKX3.1 and topoisomerase I colocalize to AR enhancer binding sites to mediate the AR transcriptional program (14). Thus in the prostate NKX3.1 is a ubiquitous component of the AR transcriptional machinery. The experiments described here show that NKX3.1 colocalizes with androgen receptor at sites of TMPRSS2 and ERG juxtaposition and rearrangement. Moreover, we now show that NKX3.1 loss increases the frequency of TMPRSS2 and ERG juxtaposition induced by androgen receptor and NKX3.1 loss predisposes to gene rearrangement by affecting the assembly of the DNA repair protein complex at the break site. These data establish a sequence of pathogenic events in early prostate carcinogenesis, place NKX3.1 loss upstream from TMPRSS2-ERG rearrangement, and emphasize that NKX3.1 protein levels are a target for prevention and treatment strategies in early prostate cancer.
Materials and Methods
Cell culture, transfection and reagents
LNCaP, cell culture and transfection have been previously described as have the LNCaP derivative lines with NKX3.1 knock down, LNCaP(siLuc), LNCaP(si471), and LNCaP(si3098) (11). PC-3 and PC-3(NKX3.1) stable cell lines were cultured in IMEM (Invitrogen) supplemented with 10% FBS (15). LAPC4 cells were provided by Dr. Robert Reiter, UCLA. Antibodies used were as follows: anti-AR, ATM, ATR, DNA-PK, Ku80 and Ku70, NBS1, OGG1, BRCA2, Histone H1 were from Santa Cruz, anti-β-actin from Sigma, anti-BrdU from BD Biosciences, anti-γH2AX, LSD1, 8-oxo-G from Millipore, anti-MRE1 and RAD51 from GeneTex, RPA from Calbiochem, APE1, XRCC1 and XRCC4 were from Abcam, and anti-ToPro3 from Invitrogen.
Constructs
hNKX3.1-LZRSΔ-IRES-GFP plasmid was generated by removing Flag-mNkx3.1 fragment from Flag-Nkx3.1- LZRSΔ-IRES-GFP plasmid (kindly provided by Dr. Cory Abate-Shen, Columbia University) by restriction enzyme digestion at BamH1 and XhoI sites. Full length hNKX3.1fragment produced by PCR reaction was inserted into the plasmid at BamH1 and XhoI sites (Plasmid backbone was LZRSpBMN-Z-β).
Retrovirus infection
Phoenix E packaging cells were transfected with NKX3.1-LZRSΔ-IRES-GFP or LZRSΔ-IRES-GFP vectors with Fugene HD. Virus containing supernatant was mixed with 10 µg/ml Polybrene and applied to LNCaP(si471) cells. Cells were infected twice at 24-hr intervals and lysed for immunoblotting.
Fluorescence in situ hybridization (FISH)
LNCaP(siLuc) and LNCaP(si471) cells were cultured in 35mm dishes in phenol red-free IMEM containing 10% FBS. To detect TMPRSS2 and ERG gene juxtaposition induced by androgen, cells were cultured in IMEM with 10% CCS for 3 days, followed by synchronization with 5 µM α-amanitin for 2 hrs. Cells were then washed with SFM containing 10% CCS for 60 min and subsequently exposed to vehicle (ethanol) or 100nM DHT for 60 min. To detect chromosomal rearrangements, cells were treated with 10Gy, 100 µM etoposide, or 1 µM doxorubicin after DHT exposure. Interphase cells were prepared for FISH analysis using 0.075 M KCl hypotonic treatment at 37°C for 30 min followed by fixation with cold 3:1 CH3OH:glacial acetic acid. Slides were left at room temperature for 24 hrs prior to application of probes.
FISH hybridization was performed using biotin (Biotin-Nick Translation Mix, Roche) and digoxigenin (Dig-Nick Translation Mix, Roche) labeled BAC DNA probes. BAC DNA plasmids (BACPAC Resources Center, Children’s Hospital Oakland Research Institute) were amplified and purified using QIAGEN Plasmid Midi Kit. To assay androgen-induced locus juxtaposition we used a TMPRSS2-ERG probe that contained a 5’ digoxigenin-tagged TMPRSS2 fragment (RP11-35C4) and a 3’ biotin-tagged ERG fragment (RP11-476D17). To assay gene rearrangement we used an ERG split probe that contained a digoxigenin-tagged 5’ ERG fragment (RP11-95I21) and a 3’ biotin-tagged ERG fragment (RP11-476D17) (6). Approximately 100 ng of each probe was used for hybridization. Prior to hybridization, the slides were pretreated by incubating with 2×SSC at 37°C for 15 min, then with 2×SSC at 75°C, 5 min, followed by successive rinses with 70%, 80%, 95% and 100% ethanol. Probes and slides were denatured at 75°C for 5 min separately. Slide denaturation was performed in prewarmed 70% formamide/2×SSC. Hybridization was at 37°C overnight. Three post hybridization washes were performed in prewarmed 50% formamide/2×SSC at 37°C, followed by two washes in prewarmed 0.5× SSC/0.3% NP-40 at 60°C for 2 min, one wash in 2×SSC/0.1% NP-40 at 25°C, one wash in 1× PBS containing 0.1% Tween 20. After washing slides were blocked 30 min at 25°C with 100 µl CAS block (ZYMED) with 1% goat serum. FISH signals were detected by 30-minute exposure to streptavidin-Alexa fluor 594 conjugate (Invitrogen) and fluoresceinated anti-digoxigenin antibodies (Roche) diluted in blocking reagent at 25°C. Slides were counterstained with DAPI in PBS and mounted in Antifade Mounting Medium (Molecular Clones). Fluorescence images were captured with 100× objective with a PALM wide-field fluorescence microscope. Image analysis was done using Image J or Fiji processing software.
Paraffin-embedded tissues were deparaffinized and then immersed in citrate buffer (10 mM sodium citrate, pH6.0, 0.05% Tween-20,) at 94°C for 40 min, followed by incubation with 0.5% pepsin in 10 mM HCl at 37°C for 10 min. Slides were fixed with 4% formaldehyde for 10 min, rinsed successively in 70%, 80%, 90%, and 100% ethanol washes and air-dried. Denatured slides were treated with preheated 10 mM Tris-HCl, pH8.9, 0.1 M KCl, 50 µg/ml BSA, 0.05% Tween 20 at 94°C for 5 min and then rinsed successively with 70%, 80%, 90%, and 100% ethanol. Denatured probes preheated at 80°C for 5 min were applied on slides preheated at 60°C for 5 min. and incubation proceeded at 37°C overnight. For cultured cells treatment was identical following probe hybridization.
Reverse transcription PCR (RT-PCR) and real time PCR (QPCR)
Total RNA was isolated from cells using the RNAeasy kit (QIAGEN). For RT-PCR cDNA was generated from 1 µg RNA in a 25 µl reaction with SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen) in the presence of oligodT primers. Subsequently 1 µl cDNA reaction was used for PCR with Platinum Blue PCR Supermix. Conventional PCR was performed at an annealing temperature of 60°C for 35–40 cycles. PCR products were resolved on 2% agarose. Primers for conventional PCR reactions are shown in Table S1. For QPCR, 1 µl cDNA reaction was amplified with Fast SYBR Green Master Mix (Applied Biosystems) and detected with 7500 fast Real-Time PCR System (Applied Biosystems). Primers for real time PCR are shown in Table S2.
Immunoblotting was performed as previously described (11).
TMPRSS2-ERG ChIP
Cultured cells were grown to 70% confluence in phenol red-free modified IMEM supplemented with 10% FBS, and then changed to IMEM with 10% charcoal-stripped fetal bovine serum (CCS) for at least 3 days. Following the addition of 100 nM DHT for 60 min, cells were treated with 100 µM etoposide and then pulsed with 10 µM BrdU for 1 hour. Cells were cross-linked with 0.75% formaldehyde at 25°C for 8 min while rocking. Quenching was done with 125 mM glycine for 5 min. followed by two rinses with ice-cold PBS. To prepare crude nuclei, cells were resuspended in hypotonic lysis buffer with protease inhibitor cocktail (Roche) (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2) on ice for 10 min. Cells were ruptured with 20 strokes in a Dounce homogenizer and the nuclei were centrifuged and suspended in RIPA buffer. Nuclei were ruptured by sonication at output level 4, 15 times for 10 sec in an ice water bath with 1 min breaks between sonications with a Micromix sonicator 3000. Each ChIP assay was performed with 50 µg of chromatin DNA. Supernatants were collected and diluted 1:5 in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1). One percent chromatin was used as the input sample. One ml of lysate was precleared with 10 µg sheared salmon sperm DNA (Invitrogen), 10 µl serum, and protein A-sepharose (50 µl of 50% slurry in 10 mM Tris-HCl, pH 8.1, 1 mM EDTA) for 1–2 hr at 4°C. Immunoprecipitation was performed overnight at 4°C with 2 µg of antibody. After immunoprecipitation 30 µl protein A-Sepharose and 10 µg of salmon sperm DNA and 10 µl of serum were added and the incubation was continued for 1–2 hr at 4°C. Precipitates were washed sequentially for 5 min each in TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), and buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Precipitates were then washed twice with TE buffer. The chromatin was eluted twice with 1% SDS, 0.1 M NaHCO3 for 15 min was then disassociated in 225 µM NaCl overnight at 65 °C. The DNA was sequentially treated with 50 µg/ml RNase at 37°C for 30 min and then 100 µg/ml protease K at 37°C for 30 min and then purified over a QIAquick spin column, per manufacturer’s instruction. For QPCR, we used 1 µl from a 40 µl DNA extract and 40–50 cycles of PCR amplification. Primers are shown in Table S3.
ChIP Re-IP and real-time PCR
Complexes were eluted from the primary immunoprecipitation by incubation with 10 mM DTT at 37°C for 30 min and diluted 1:50 in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1) followed by reimmunoprecipitation with secondary antibodies. ChIP Re-IP of supernatants was done essentially as were the primary IPs. For PCR, 60 cycles of amplification were used. For ChIP-re-ChIP assay, cells were pretreated with 100 nM bortezomib for 1 hour to prevent degradation of proteins during etoposide treatment (16, 17).
Chromosome conformation capture (3C) assay
The chromosome conformation capture assay was performed according to published procedures with minor modifications (18, 19). Cells in 100 mm dishes were treated with 2% formaldehyde for 10 minutes at 25°C. Glycine was added to a final concentration of 0.125 M for 5 min. Nuclei were extracted with lysis buffer (10 mM Tris HCl pH8.0, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EGTA) containing protease inhibitors on ice for 20 min, then ruptured with 20 strokes in a Dounce homogenizer. The nuclei were resuspended in restriction buffer (NEB), then add to 0.3% SDS and incubated at 37°C for 1 h. Triton X-100 was added to 1.8% and the reaction was incubated an additional hour at 37°C. Intact nuclei were digested for 12 h while shaking at 37°C by addition of 200 U of restriction enzyme EcoRI (NEB). The digestion was stopped at 65°C for 20 min after addition of SDS to a concentration of 1.6%. The reaction was then diluted 12-fold with T4 DNA ligase buffer to an approximate DNA concentration of 3.7 ng/µl. Triton X-100 was added to 1%, then the reaction was incubated at 37°C for 1 hr. DNA fragments were ligated with 100 U of T4 ligase for 2 hrs at room temperature and then overnight at 16°C. The cross-links were reversed by overnight incubation at 65°C in the presence of 5 µg/ml proteinase K, and the DNA was pretreated with RNase (10 µg/ml) at 37°C for 30 min. DNA was purified by phenol-chloroform extraction and ethanol precipitation. The DNA concentration was determined using a NanoDrop. The control template was generated as follows. Purified genomic DNA was digested with the same restriction enzyme that used for generation of the cross-linked template, and restriction fragments were randomly ligated without dilution using a DNA concentration of 300 ng/µl. Ligated fragments were identified by PCR using primers listed in Table S4.
Immunofluorescence of prostate cancer tissues
Primary prostate cancer tissues were obtained anonymously as described previously (1) and from the Molecular Pathology Shared Resource of the Herbert Irving Comprehensive Cancer Center. Tissues were used by IRB approval. Tissues were not selected in any way other than the diagnosis of prostate cancer that had been treated by prostatectomy. Deparaffinized slides were pretreated in 10 mM citrate buffer pH 6.0 in a steamer for 40 min. The cells were permeabilized with 0.5% Triton X-100 for 15 min and then 0.3% H2O2 in PBS for 60 min to quench endogenous peroxidase activity. The slides were incubated with rabbit NKX3.1 (1:2000) antibody and murine histone H1 (1:1000) overnight, rinsed with PBS+ 5% Tween 20, then incubated with biotinylated-anti-mouse IgG antibody (1:200) for 30 min. After three rinses with PBS + 5% Tween 20 + 1% goat serum for 5 min each, the slides were incubated with Texas-read-avidin (1:200) and secondary anti-rabbit IgG-horseradish peroxidase (1:200) antibody for 30 min. Fluorescence-plus-Tyramide (PerkinElmer) was applied on slides for 6 min, and ToPro3 (1:1000) for 5 min. Cover slips were mounted with Vectasheld (Vector).
Statistical Analyses
Comparisons shown in figures by brackets were analyzed by two-side t-test and p-values are indicated where differences are significant. For ChIP assays in Figures 6 and 8 we also used analysis of variance (ANOVA) to test the overall statistical significance of the observed differences between LNCaP(siLuc) and LNCaP(si471) cell levels at different time points. The null hypothesis assumed that the expression level of a gene may vary over time but there would be no differences between the LNCaP(siLuc) groups and the LNCaP(si471) groups. The alternative assumed that the two groups may exhibit different trends over time.
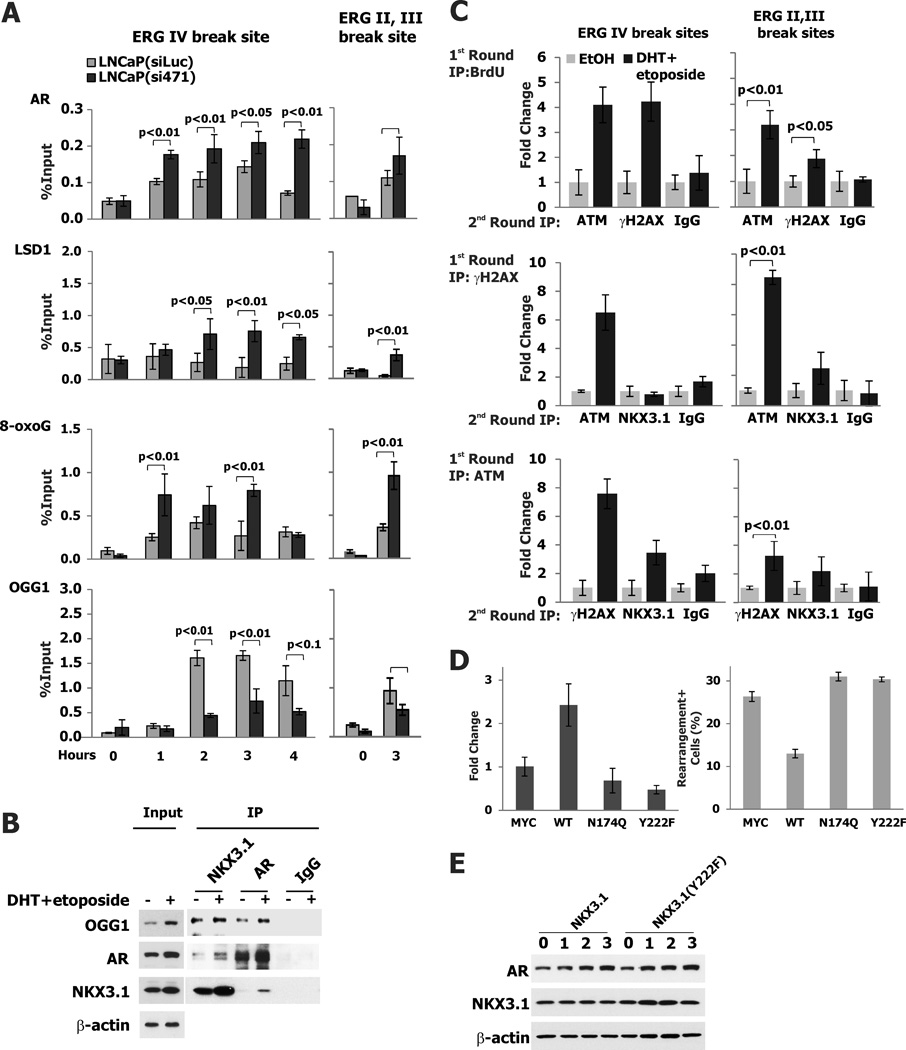
Figure 6. Oxidative DNA damage and ATM Activation at the ERG gene breakpoint.
A. Assembly of proteins involved in initiation of AR-mediated transcription was analyzed by ChIP assay in LNCaP(siLuc) and LNCaP(si471) cells at the time points indicated. Cells were exposed to etoposide for up to 4 hours (ERG IV) and for 3 hours (ERG II,III) and then were exposed to 100 nM DHT. Cells were analyzed at the time of DHT treatment and hourly thereafter in the case of ERG IV. B. Immunoprecipitation and immunoblotting demonstrating association of NKX3.1, AR, and OGG1. LNCaP cells were exposed either to ethanol or 100 nM DHT plus 10 µM etoposide for 2 hr. Immunoprecipitation followed by immunoblotting was performed as shown. See Table S5 for p-values of ANOVA comparing data for LNCaP(siLuc) and LNCaP(si471) for each antibody. C. ChIP-re-ChIP with BrdU antibody in first round IP with LNCaP cells exposed to 100 nM DHT and 100 µM etoposide followed by extraction and re-ChIP with either ATM or γH2AX antibodies (upper panel). The middle and lower panels show reciprocal ChIP-re-ChIP assays demonstrating ATM complexes with γH2AX and NKX3.1, but NKX3.1 complexes only with ATM, consistent with the expectation that NKX3.1 activation of ATM is upstream from γH2AX. D. LNCaP(si471) cells were cotransfected with GFP and wild type or mutant NKX3.1 expression plasmids as indicated. GFP positive cells were sorted isolated and treated as above prior to processing for ChIP analysis with ATM antibody. Thus only native NKX3.1 was active in ATM recruitment to the ERG breakpoint (upper panel). The cells were also analyzed for TMPRSS2-ERG rearrangement and only native NKX3.1 was shown to affect the gene rearrangement likelihood in GFP+ cells (lower panel). E. LNCaP(si471) cells were transfected with either wild type or NKX3.1(Y222F) expression vectors and, 24 hours later, exposed to 100 nM DHT. Cells were sampled for immunoblotting as shown.
Results
NKX3.1 affects AR-induced juxtaposition and rearrangement of TMPRSS2 and ERG
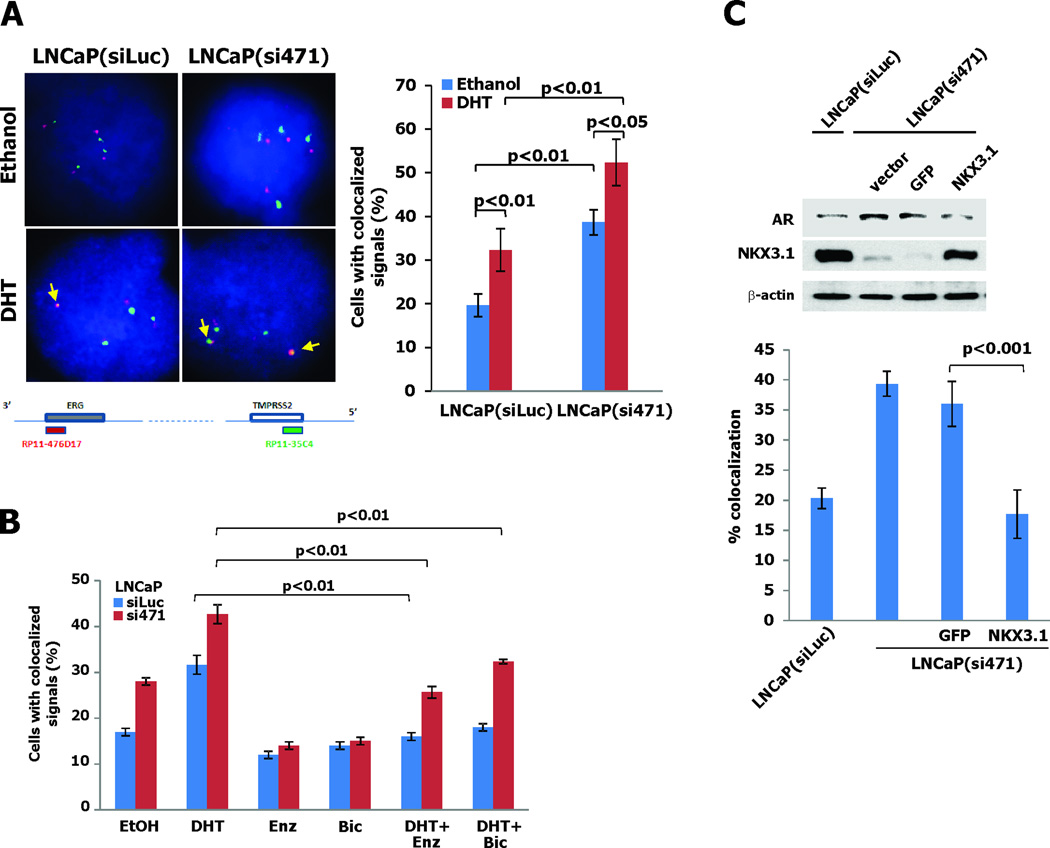
Androgen receptor can mediate ligand-dependent juxtaposition of ERG and TMPRSS2 in cultured LNCaP cells, suggesting that the highly prevalent TMPRSS2-ERG rearrangement in prostate cancer is facilitated or perhaps induced by AR action (6). To determine whether NKX3.1 can affect the gene rearrangement we examined the TMPRSS2 and ERG juxtaposition in LNCaP cells with stable NKX3.1 knockdown (11). Increased juxtaposition of TMPRSS2 and ERG loci was seen in cells with NKX3.1 knockdown (Figure 1A). The androgen-dependence of this effect was confirmed since the antiandrogens bicalutamide and enzalutamide both decreased the frequency of TMPRSS2 and ERG juxtaposition in both NKX3.1(siLuc) and NKX3.1(si471) cells extending the observations that TMPRSS2-ERG fusions are AR-dependent in cell lines (20). No basal suppression was seen with antiandrogens alone (Figure 1B). Moreover, restoration of NKX3.1 expression in the LNCaP(si471) knockdown cells restored the background level of TMPRSS2-ERG colocalization (Figure 1C).
Figure 1. NKX3.1 decreases AR-induced juxtaposition of TMPRSS2 and ERG.
A. Juxtaposition of TMPRSS2 (green dot) and ERG (red dot) in LNCaP cells treated for one hour with 100 nM DHT as detected by FISH assay. The map below the micrographs shows the probes applied to assay TMPRSS2 and ERG proximity. The brackets indicate significant differences with corresponding p-values. B. Effect of antiandrogens on DHT-dependent TMPRSS2-ERG juxtaposition in LNCaP(siLuc) and LNCaP(si471) cells. For the experiments with combined treatment cells were exposed to 25 µM enzalutamide or 10 µM bicalutamide for 1 hour prior to DHT exposure. C. LNCaP(si471) cells were infected with retroviruses derived from NKX3.1-LZRSΔ-IRES-GFP or LZRSΔ-IRES-GFP vectors. Cells were infected twice and lysed for immunoblotting as shown in the top panel. LNCaP(siLuc) and infected LNCaP(si471) cells were exposed to DHT FISH analysis for TMPRSS2-ERG gene proximity was performed.
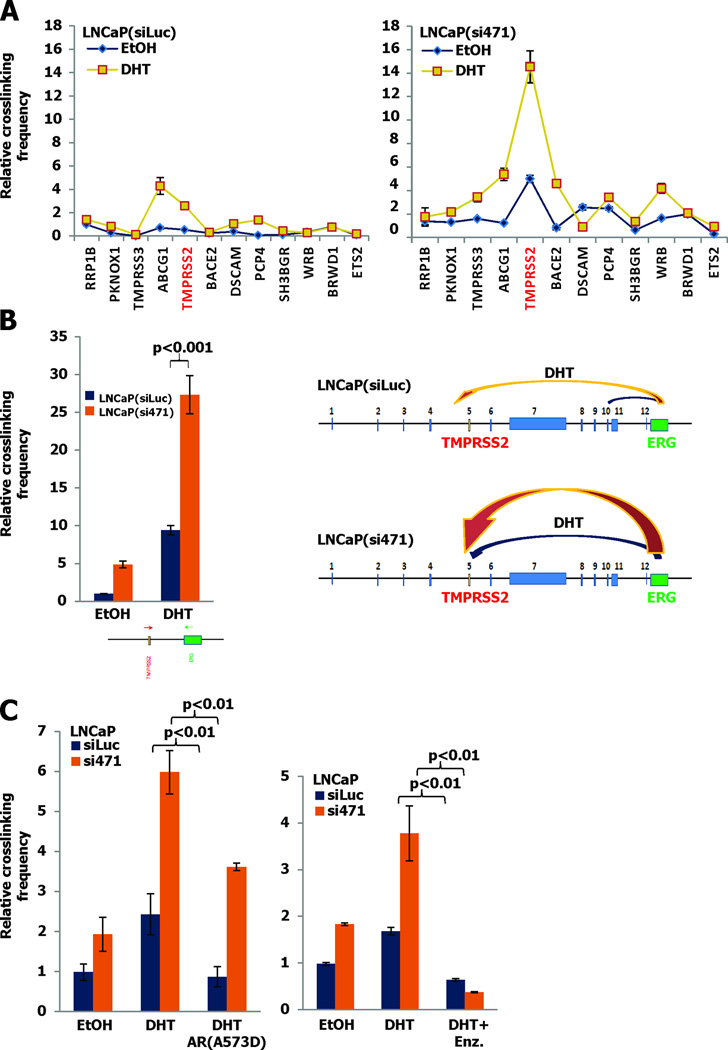
To define more precisely the region of chromosome 21 juxtaposition mediated by AR we used a chromosome conformational change (3C) assay to detect fusion between the first exon of the ERG gene and twelve candidate loci upstream. The 3C assay clearly showed that AR favored juxtaposition of ERG and TMPRSS2 in preference to eleven other loci (Figure S1). We then found that the degree of TMPRSS2-ERG crosslinking by 3C was much greater in LNCaP(si471) than in control cells confirming that loss of NKX3.1 favored the gene rearrangement (Figures 2A and B). We also noted that in LNCaP(si471) cells the favored breakpoint site was in the TMRPSS2 gene compared to ABCG1 in LNCaP(siLuc) cells (Figure 2A). The TMPRSS2-ERG juxtaposition detected by 3C was dependent on the action of AR as demonstrated by inhibition of crosslinking by a dominant-negative AR lacking DNA binding capacity (18). Also, TMPRSS2-ERG juxtaposition was inhibited by treatment with enzalutamide, further confirming the role of AR in directing the chromosomal conformation that preceded TMPRSS2-ERG recombination (Figure 2C).
Figure 2. Specific association of TMPRSS2 and ERG is attenuated by NKX3.1.
A. LNCaP(siLuc) and LNCaP(si471) cells were exposed to 100 nM DHT for 1 hour and analyzed by 3C assay for chromosome 21q22.3. The graphs show crosslinking frequency between ERG and TMPRSS2. B. Quantitation of the crosslinking interaction at the TMPRSS2 site demonstrating a 3-fold difference in crosslinking between LNCaP(si471) and LNCaP(siLuc) cells. This is further demonstrated by the cartoon on the right of 2B. C. Cells were transfected with the AR DNA-binding mutant (A573D) (left panel) or exposed to with 25 µM enzalutamide (right panel) and subjected to 3C assay.
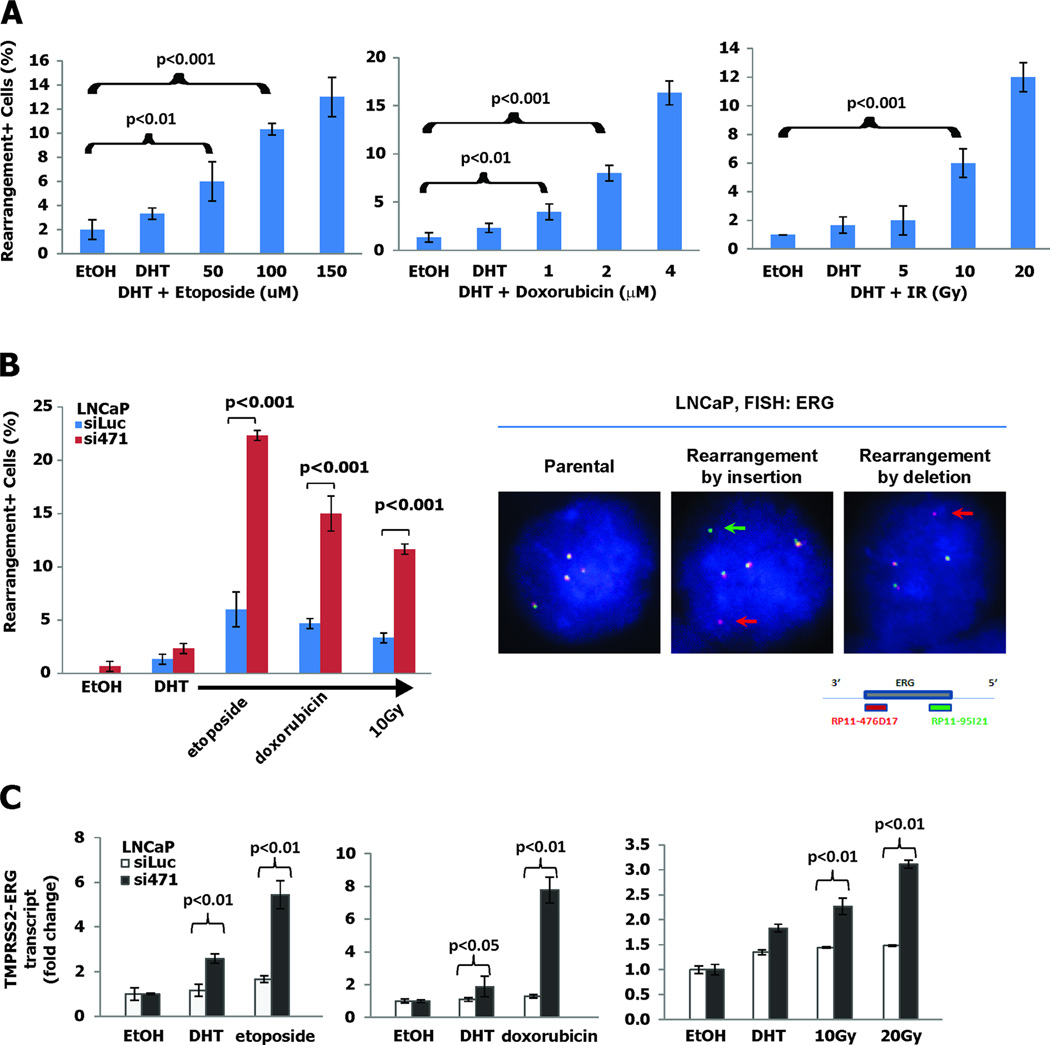
Having demonstrated the effect of NKX3.1 on AR-induced chromosomal conformation, we next examined TMPRSS2-ERG rearrangement in vitro. As was described by Mani et al, AR-induced TMPRSS2-ERG rearrangements were more easily detected when the cultured cells were exposed to DNA damaging agents (Figure 3A). Knockdown of NKX3.1 in LNCaP cells markedly increased the frequency of TMPRSS2-ERG rearrangement induced by DHT and DNA damaging agents (Figure 3B). When compared with the degree of TMPRSS2-ERG juxtaposition seen in Figure 2B, actual gene rearrangement is less frequent unless augmented by prolonged DHT exposure or the induction of DNA damage (20). This result was not due to off-target effects of the knockdown construct since the same effect of NKX3.1 knockdown was seen with a second knockdown construct (Figure S2A) (11). The fused TMPRSS2 and ERG genes generated chimeric transcripts in vitro as shown by RT-PCR analysis of fusion transcripts in derivative LNCaP cells treated with DHT and DNA damaging agents (Figure 3C). The specificity of NKX3.1 in suppressing gene rearrangement was demonstrated further by restoration of NKX3.1 expression to LNCaP(si471) cells (Figure S2B). Also, we could demonstrate a similar effect of NKX3.1 knockdown in an independent NKX3.1-expression cell line, LAPC4 which are known to be susceptible to AR-dependent TMPRSS2-ERG rearrangement (Figure S2C) (18). The fusion of TMPRSS2 and ERG was completely dependent on the presence of AR. Using PC-3 cells and derivative PC-3 cells that were engineered to express NKX3.1 we saw no induction of TMPRSS2-ERG fusions or expression of chimeric RNA (Figure S3A–D). However, the presence of androgen receptor, but not AR(A573D), a DNA binding mutant, induced TMPRSS2-ERG rearrangement in PC-3 cells that was also attenuated by NKX3.1 expression (Figure S3E). Thus the specificity of the fusion phenomenon was determined by the presence of AR and its ability to bind DNA.
Figure 3. NKX3.1 prevents TMPRSS2-ERG gene rearrangement.
A. LNCaP cells were exposed to etoposide, doxorubicin or γ-irradiation 24 hr prior to fixation and were treated with 100 nM DHT for 1 hour prior to fixation. TMPRSS2-ERG rearrangements were assayed by in situ hybridization. B. Experiment as in panel A but LNCaP(siLuc) and LNCaP(si471) cells to 100 nM DHT plus 100 µM etoposide, 1 µM doxorubicin, or 10 Gy. FISH assay to detect gene rearrangements was performed with 3’ probe (red dot) and 5’ probe (green dot) of the ERG gene detecting four paired dots in parental LNCaP cells (left), split green and red dots in a rearrangement by insertion (center), and loss of a green dot in rearrangement by deletion (right). C. TMPRSS2-ERG gene rearrangement was interrogated by assay for fusion transcripts in LNCaP(siLuc) and LNCaP(si471) cells. Fusion transcripts were measured by quantitative RT-PCR with primers overlapping both TMPRSS2 and ERG. The brackets indicate differences that were statistically significant where p-values are shown.
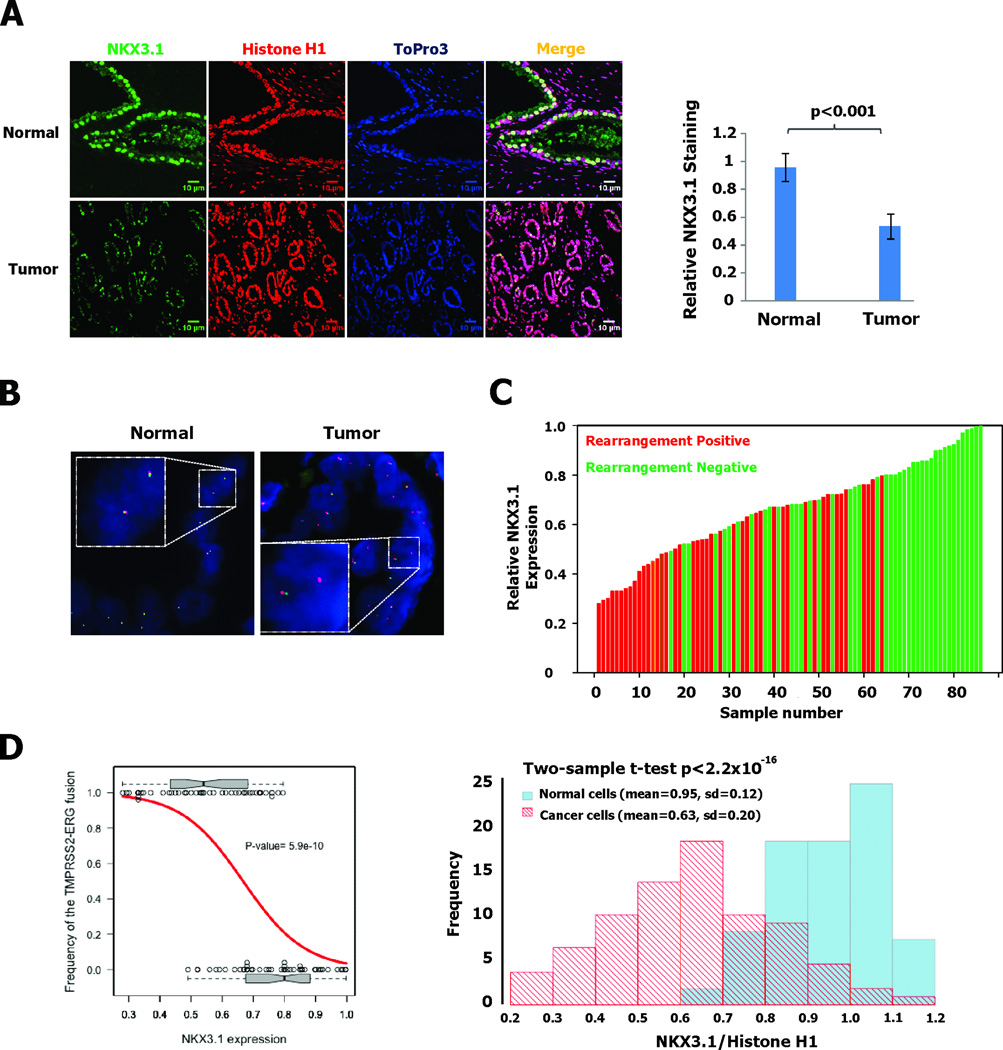
NKX3.1 expression and TMPRSS2-ERG rearrangement in tumor specimens
To determine if the effect of NKX3.1 on TMPRSS2-ERG rearrangement in vitro was relevant for the pathogenesis of prostate cancer, we investigated the relationship between NKX3.1 expression and the presence of TMPRSS2-ERG rearrangement in cancer tissues. We previously published measurements of NKX3.1 expression levels relative to histone H1 in primary prostate cancer tissues and compared to adjacent normal prostate epithelial cells (1). We assayed NKX3.1 expression in 86 primary prostate cancer samples in which we also determined TMPRSS2-ERG rearrangement by FISH. An example of staining of prostate cancer and adjacent normal tissue is shown in Figure 4A. The average intensities of NKX3.1 expression in the specimen shown are displayed quantitatively in the histogram on the left of Figure 4A. Using ERG split probes that contain 5’ERG (digoxigenin-labeled RP11-95I21) and 3’ ERG (biotin-labeled RP11-476D17), we identified the tissues that had TMPRSS2-ERG rearrangements. An example of chromosomal rearrangement from a single specimen and hybridization in adjacent normal cells is shown in Figure 4B. TMPRSS2-ERG rearrangement was infrequently seen in the samples with the highest levels of NKX3.1 staining, but we detected gene rearrangements more frequently in the majority of samples with lower NKX3.1 staining (Figure 4C). We computed Spearman's ρ estimated to be −0.6 to assess the correlation between NKX3.1 expression and TMPRSS2-ERG fusion. A logistic regression model was used to study the dependence of TMPRSS2-ERG fusion on NKX3.1 expression levels and we found that the odds of a TMPRSS2-ERG fusion increased by a factor of 2.7 (95% confidence interval is [1.9, 4.5]) as the relative expression level of NKX3.1 dropped by 0.1. The likelihood ratio test was carried out between the fitted model and the null model of no association between TMPRSS2-ERG fusion and NKX3.1 expression, which reported a p=5.9 × 10−10. When the NKX3.1:Histone H1 ratios are plotted by histogram the frequency distributions for normal and cancer cells are different to a high degree of statistical significance (p<2.2×10−14) (Figure 4D).
Figure 4. Reduced NKX3.1 expression in prostate cancer specimens correlates with TMPRSS2-ERG rearrangement.
A. Immunofluorescence staining with NKX3.1 antiserum and with monoclonal histone H1 antibody shows that NKX3.1 expression was down regulated in prostate cancer region compared to adjacent normal prostate epithelial tissue. On the right is shown an example of the quantitative comparison between normal and cancer regions on the same slide. B. FISH rearrangement assay with ERG split probes shows normally paired signals in normal prostate epithelial tissue and gene rearrangement with deletion in regions of malignant cells. C. 86 prostate cancer specimens were analyzed for both relative NKX3.1 expression and TMPRSS2-ERG rearrangement. D. Logistic regression was fitted between the frequency of the rearrangement-positive samples and NKX3.1 expression. The observed incidences of TMPRSS2-ERG fusion were plotted against their corresponding NKX3.1 expression level (black circles). Two boxplots were used to summarize the distributional difference of NKX3.1 expression level between the two rearrangement outcomes. The estimated frequency by the logistic regression model is shown (red curve), with the p=5.9×10−10 from the likelihood ratio test on the logistic regression model. On the right a histogram is shown of the frequency of NKX3.1:Histone H1 intensity ratios for normal and cancer cells. The difference by 2-sample t-test has a p<2.2×10−16.
The data from this analysis allowed confirmation of the correlation between NKX3.1 expression and the cancer grades estimated by Spearman's ρ=-0.42. We observed that higher Gleason grades contained lower levels of NKX3.1 expression (Figure S4A). To test the statistical significance of this negative association, we applied the F test of one-way analysis of variance. Our results suggested that the mean expression level of NKX3.1 is statistically significant different between different cancer grades (p = 1.1 × 10−5). The side-by-side box plots in Figure S4B display the decreasing trend of NKX3.1 expression as the cancer grade increases. We were also able to examine the correlation between TMPRSS2-ERG fusion and cancer grade by Spearman's ρ= 0.44. (Figure S4C). To test the statistical significance of this positive association, we applied the chi-squared test of independence (p=0.00026). Our results suggest that the proportion of TMPRSS2-ERG fusion is statistically significantly different between different cancer grades. The incidence of TMPRSS2-ERG fusion increase as the cancer grade increases (Figure S4C).
Assembly of AR transcription elements at sites of a TMPRSS2-ERG breakpoint
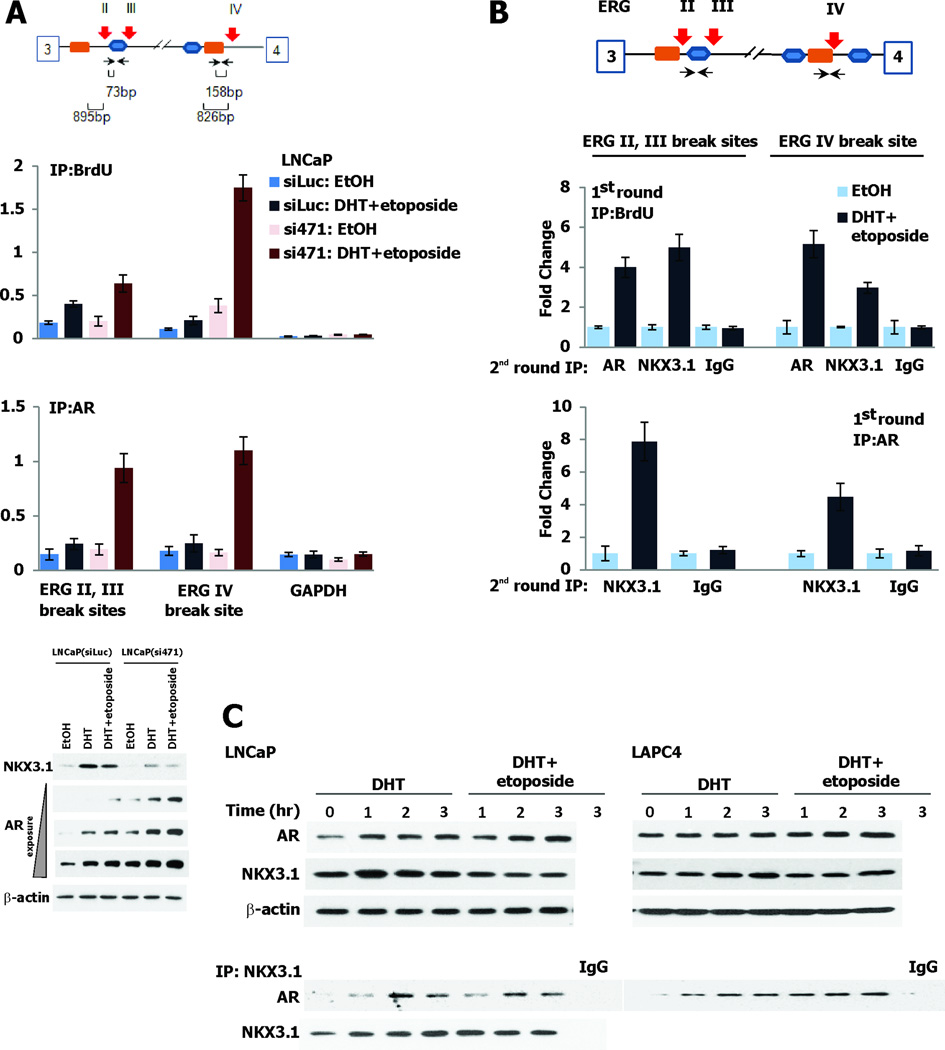
The fusion of TMPRSS2 and ERG genes that is common in prostate cancer is mediated by binding of AR at sites of genetic recombination (7). We sought to examine the involvement of NKX3.1 at these recombination sites so we first showed by ChIP assay that we could identify the same sites on TMPRSS2 and ERG as Lin et al originally found were bound by AR (Figure S5A). We were also able to demonstrate that NKX3.1 did not localize to a site in ERG intron 1 that was not a breakpoint for genetic recombination thus providing a negative control. We then demonstrated in LNCaP cells that NKX3.1 expression affected the incorporation of BrdU and the binding of AR at recombination sites in the ERG gene (Figure 5A). By coimmunoprecipitation of chromatin (re-ChIP) we showed that NKX3.1 and AR formed a complex at the same ERG gene breakage sites where BrdU was incorporated after cells had been exposed to DHT and etoposide (Figure 5B). A search for candidate ARE and NKE (NKX3.1 binding sites) identified several adjacent sites that had markedly lower frequency of recombination underscoring the specificity of the recombination for the ERG intron II,III site (Figure S5B). Complex formation between AR and NKX3.1 was also demonstrated by coimmunoprecipitation in both LNCaP cells and in a second AR-expressing cell line, LAPC4 treated with DHT or with DHT+etoposide (Figure 5C).
Figure 5. AR and NKX3.1 form a complex at ERG break sites.
A. The schematic shows the break sites in intron 3 of ERG and the adjacent NKX3.1 DNA binding sequence (TAAGTA, orange box) and the androgen response element (ARE) (TGTCCT, blue box). BrdU incorporation is increased at breakpoints and further augmented by knockdown of NKX3.1. AR is also found at the breakpoint in the presence of DHT. The immunoblots at the bottom of panel A show expression of NKX3.1 and AR under the experimental conditions in the two derivative cell lines. B. ChIP-re-ChIP using BrdU antibody in the first round IP (top panel) followed by AR or NKX3.1 antibodies in the second round IP indicates that both AR and NKX3.1 are recruited to the break sites. ChIP-re-ChIP with anti-AR antibody at the first round IP (lower panel), and NKX3.1 antibody at the second round indicates that AR and NKX3.1 form a complex at the break site. C. The upper section of the panel shows immunoblots with monoclonal AR antibody and affinity-purified NKX3.1 antiserum probing extracts from LNCaP and LAPC4 cells. The lower section shows immunoprecipitation/immunoblot analysis of the AR and NKX3.1 interaction.
Nuclear receptors have the capacity to cause mutations by errors in the process of transcriptional initiation. Initiation of transcription by nuclear receptors is tightly linked to activation of the lysine-specific demethylase LSD1 that generates H2O2 locally in the course of histone demethylation (21). The newly-generated peroxide oxidizes DNA leading to formation of 8-oxoguanine adducts at the hormone-response element that, after modification by OGG1, are targets for base excision repair (8). Transcription is initiated at the site of base excision in a process that is completed within one hour of nuclear hormone exposure. We demonstrated that NKX3.1 affected the association of OGG1 and LSD1 with DNA-bound AR. Using ChIP assays in NKX3.1-knockdown cells we showed that NKX3.1 loss resulted in increased AR recruitment to both the ERG IV and ERG II,III break sites and caused increased association with LSD1 resulting in greater 8-oxoG accumulation. Concomitantly there was reduced recruitment of OGG1 to the ERG breakpoint sites in the absence of NKX3.1 further favoring DNA breakage and genetic recombination (Figure 6A). As suggested by the ChIP assays, OGG1 could be shown directly to associate with both AR and NKX3.1 (Figure 6B). Furthermore, OGG1 localization to the breakpoints was enhanced by other methods of DNA damage and decreased by enzalutamide, further emphasizing the roles of NKX3.1 and AR on the process of TMPRSS2-ERG rearrangement (Figure S6). Thus, when NKX3.1 levels were reduced AR-mediated DNA breakage increased.
Moreover, although AR was essential for TMPRSS2-ERG rearrangement, transcription was not required. We treated LNCaP(siLuc) and LNCaP(si471) cells with α-amanitin to block engagement of RNA polymerase II. This resulted in complete shutdown of TMPRSS2 transcription, but had essentially no effect on either TMPRSS2-ERG juxtaposition or gene rearrangement (Figure S7). Therefore AR acts in a ligand-dependent way to enable gene rearrangement that is moderated and inhibited by the presence of NKX3.1 but is independent of gene transcription.
NKX3.1 affects DNA repair processes at the ERG breakpoint
NKX3.1 itself affects the DNA repair response by activating ATM kinase and ATM recruitment to sites of DNA damage (12). We hypothesized that NKX3.1 could affect TMPRSS2-ERG recombination not only by interacting with AR, but also by influencing the response to DNA damage that ensued from AR binding. ATM accumulated at both the ERG IV and the ERG II,III breakpoints as did γH2AX. Moreover, with re-ChIP assays ATM and NKX3.1 were shown to associate at the breakpoint as did ATM and γH2AX. However, NKX3.1 did not associate with γH2AX since phosphorylation of H2AX was downstream of NKX3.1 (Figure 6C). NKX3.1 augmentation of ATM recruitment to the ERG IV breakpoint was abrogated by mutation of tyrosine 222 that is phosphorylated seconds after DNA damage and is required for ATM activation (12). Moreover, DNA binding by NKX3.1 was also required for ATM activation at the ERG IV breakpoint since a missense NKX3.1 mutation abrogating DNA binding also abrogated the effect of NKX3.1 on ATM recruitment to the breakpoint (Figure 6D). Importantly, although mutation of tyrosine 222 abrogated the effect of NKX3.1 on TMPRSS2-ERG rearrangement, there was no difference between wild type and NKX3.1(Y222F) on androgen receptor expression levels thus showing that differences in TMPRSS2-ERG rearrangement were not due to differences in AR levels but were related directly to the role of NKX3.1 in DNA repair (Figure 6E).
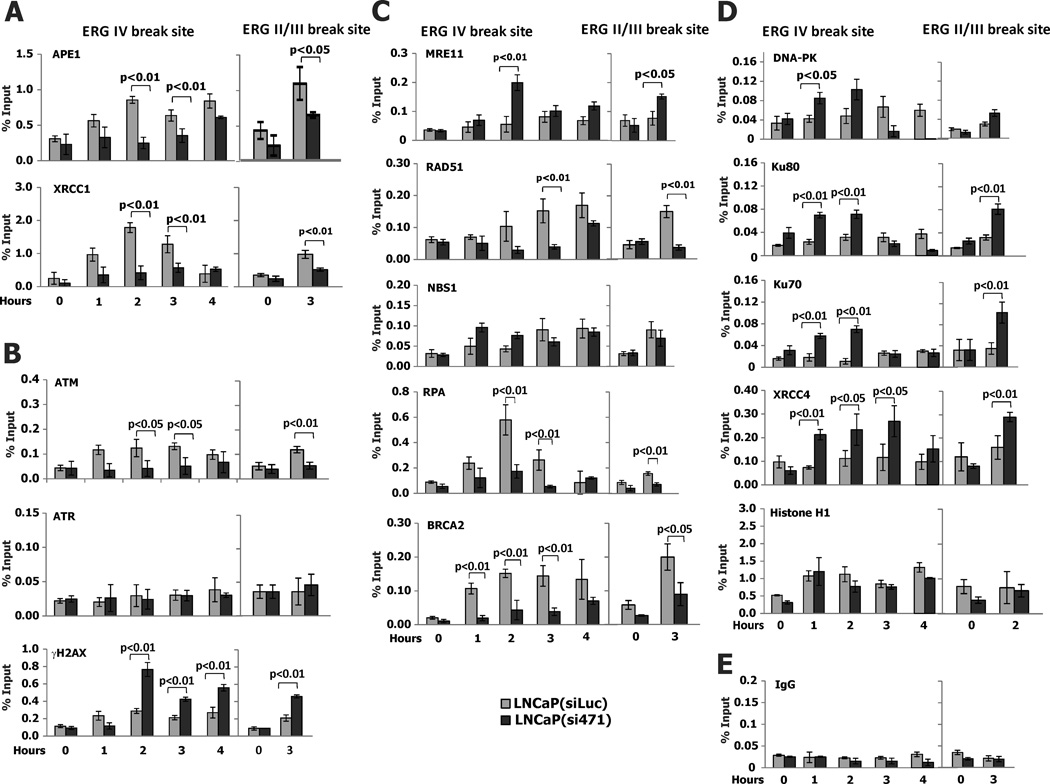
NKX3.1 also influenced the recruitment of DNA repair proteins to the ERG IV and ERG II,III breakpoint sites. In NKX3.1 knockdown cells there were reduced levels of APE1 and XRCC1 recruited to the breakpoint sites (Figure 7A). Both of these proteins are involved in base excision repair (22, 23). Consistent with our prior demonstration that NKX3.1 activated ATM, but had minimal effect on ATR, we found that NKX3.1 knockdown reduced ATM accumulation at the breakpoint sites. The formation of γH2AX was initially reduced, but subsequently increased, consistent with the delayed processing of DNA repair affected by NKX3.1 loss. The increase of γH2AX accumulation at the break site in the presence of NKX3.1 knockdown was also consistent with the residence of DNA-PK at that site as will be seen below. DNA-PK is a kinase for histone H2AX (24, 25) and its presence at the break site was increased by NKX3.1 (Figures 7B and 7D). Proteins generally thought to be associated with homology-directed DNA repair had reduced association with the ERG IV and II,III breakpoints in the presence of NKX3.1 knockdown (Figure 7C). Although there were small effects of NKX3.1 on MRE11 and NBS1, there was substantial reduction in the association of RAD51 with the breakpoints in the presence of NKX3.1 knockdown. RPA and BRCA2 recruitment was also affected by NKX3.1.
Figure 7. NKX3.1 affects the mechanism of DNA repair at the ERG break site.
ChIP assays were all performed with LNCaP(siLuc) and LNCaP(si471) cells after exposure to 100 nM of DHT and 100 µM etoposide. A. ChIP assay with APE1 and XRCC1 antibodies after treatment shows the slower and less robust assembly of APE1 and XRCC1 in LNCaP(si471) cells, indicating that loss of NKX3.1 resulted in down regulation of base excision repair at the break site. B. Faster initiation of ATM and γH2AX recruitment at the break site in LNCaP(siLuc) cells reflected the effect of NKX3.1 on ATM. Increased γH2AX recruitment at later time points in LNCaP(si471) cell may suggest activation of alternative DNA repair pathways in the face of reduced ATM signaling. C. Three proteins associated with homology-directed DNA repair are favored in LNCaP(siLuc) cells after DNA breakage. MRE11 and NBS1 are involved in both HR and NHEJ thus the higher level of recruitment in LNCaP(si471) cells may indicate relative degrees of activation in the two derivative cells. D. Chip assay indicates the assembly of protein involved in nonhomologous end-joining. ChIP assay with anti-DNA-PK, Ku70 and 80, and XRCC4, demonstrates greater extent of assembly of the proteins responsible for conventional NHEJ pathway. No difference in Histone H1 recruitment after DHT plus etoposide stimulus infers lack of involvement of NKX3.1 in the alternative NHEJ pathway. E. A control experiment done with irrelevant IgG for ChIP assay. See Table S5 for p-values of ANOVA comparing data for LNCaP(siLuc) and LNCaP(si471) for each antibody.
In contrast to the effects of NKX3.1 knockdown on proteins involved in homology-directed DNA repair, there was increased recruitment of proteins involved in nonhomologous end-joining (Figure 7D). DNA-dependent protein kinase, Ku80, Ku70, and XRCC4 were all increased at the ERG IV breakpoint site (26, 27). Histone H1, involved in alternative nonhomologous end-joining was not affected by NKX3.1 knockdown (28) (Figure 7D). A negative control for the ChIP experiments is shown in Figure 8E. Thus NKX3.1 influences both the efficiency and mechanism of repair at the ERG IV breakpoint site after AR-induced juxtaposition with TMPRSS2 and transcription-induced DNA oxidation.
Discussion
The most common genetic abnormality in human prostate cancer is loss of heterozygosity at 8p21 (29–31). This loss reduces the NKX3.1 copy number and accounts for one of several mechanisms by which NKX3.1 expression is decreased in both preneoplastic and early prostate cancer (1). NKX3.1 has diverse functions including the support of prostate epithelial terminal differentiation (32), suppression of cell proliferation (33), enhancing transcription of antioxidant and DNA repair genes (9, 10), and augmentation of DNA repair (11, 12). The role of NKX3.1 in DNA repair may be most relevant to the earliest phases of prostate carcinogenesis. The prostate is susceptible to inflammation from infectious causes, from “culture-negative” noninfectious prostatitis, and from inflammatory atrophy of aging (34, 35). Inflammation is accompanied by generation of reactive oxygen species that increases the chance of DNA damage. In regions of prostatic inflammation NKX3.1 protein is lost, precisely in the regions of the gland where it may be most needed to enhance the DNA damage response (5, 36). NKX3.1 protein loss would predispose to genetic rearrangements and DNA damage caused by reactive oxygen species generated by inflammation. Those genetic rearrangements are targeted at AR enhancer binding sites where AR and NKX3.1 colocalize and where NKX3.1 plays a critical role in faithful DNA repair and suppression of gene rearrangement (14).
TMPRSS2-ERG rearrangements in prostate cancer are pathogenic events initiated by action of the androgen receptor (6, 7, 20). Gene rearrangement juxtaposing a member of the ETS transcription factor family with the androgen-responsive TMPRSSS2 promoter is a hallmark of the majority of prostate cancer cases (2, 37). TMPRSS2-ERG gene rearrangement is mediated by the androgen receptor that juxtaposes promoter and enhancer regions in the two genes as a precursor to gene rearrangement. Since transcriptional activation by nuclear receptors involves creation of 8-oxo-G adducts and subsequent DNA single-strand opening for nucleotide excision repair, efficient DNA repair may be critical to avoid gene recombination between TMPRSS2 and ERG. Our data show that NKX3.1 is intimately involved with AR at an ERG gene breakage site and loss of NKX3.1 predisposes to an increased frequency of DNA rearrangement. Thus, NKX3.1 loss that is common in intraepithelial neoplasia is a likely precursor to TMPRSS2-ERG rearrangement that is much more common in invasive cancer than in PIN (38).
Although clinical prostate cancer occurs predominantly in the seventh through ninth decades of life, there are histologic changes of microscopic prostate cancer in younger men (39) but these changes are not predictive of future clinical prostate cancer. Thus there may be early mutational events that underlie latent prostate cancer, but over time clinical prostate cancer must be caused by gatekeeper inactivation that facilitates initiation of a pathogenic mutational cascade (31). Loss of NKX3.1, ubiquitously expressed in prostate epithelial cells (40), is the gatekeeper that both restricts prostate epithelial cell proliferation and mediates the DNA damage response. Consistent with this notion is genomic evidence that relatively few pathogenic events during prostate carcinogenesis result in mutations of many genes (31). One marker of genetic rearrangement, TMPRSS2-ERG fusion, is seen in nearly half of all prostate cancer, but in only 16–21% of high-grade intraepithelial neoplasia (HGPIN) and not at all in histologically normal cells (3). In fact, most TMPRSS2-ERG-positive cancer was found to have TMPRSS2-ERG-negative HGPIN (38, 41) and the presence of the rearrangement in PIN predicts for the development of invasive cancer, further suggesting that TMPRSS2-ERG rearrangement is an event that marks the transition from in situ to invasive prostate cancer (42).
NKX3.1 expression is decreased in the aging prostate via a variety of mechanisms that target DNA, RNA, and protein. DNA analysis of prostate cancers identified those with NKX3.1 allelic loss and placed that deletion upstream from TP53 loss (31). Interestingly, Nkx3.1 was found to stabilize p53 in a mouse model of prostate cancer driven by targeted loss of Pten (43). Thus the role of NKX3.1 in the DNA damage response may extend to affecting P53 as well. Other genes with known oncogenic potential were also placed downstream of NKX3.1 gene loss including chromosome 10 open ready frame 90, C10orf90 (FATS), that has been implicated as an activator of P53 expression (44, 45) and neuroblastoma break point family 1, NBPF1, that has been disrupted in human cancer (46).
Prostate cancer prognosis is predicted well by histology represented by Gleason grade, an index that is used universally (47). Our previous data suggested an inverse correlation between NKX3.1 expression levels and Gleason grade (1). We now have validated that finding in the 86 primary cancers examined for this report. Baca et al provided Gleason scoring information on 55 prostate cancer specimens among which it appeared that low scores were unlikely to correlate with the presence of TMPRSS2-ERG rearrangements (see Figure 1 in (31)). We found a statistically strong association between the presence of TMPRSS2-ERG rearrangement and higher Gleason grade. The association of TMPRSS2-ERG rearrangement of prostate cancer prognosis has been controversial with as many groups finding a correlation of the rearrangement and poor prognosis (48–51) as not (52–54). We have taken a more focused approach by analyzing histologic grade of the tumor region that we subject to FISH for TMPRSS2-ERG gene rearrangement. Perhaps because we did not rely on records from surgical pathology reports, we may have made a more accurate assessment of the relationship between histologic grade and TMPRSS2-ERG rearrangement. Our results show a clear correlation between the presence of TMPRSS2-ERG rearrangement and higher Gleason grade, suggesting that both NKX3.1 loss and the presence of TMPRSS2-ERG rearrangement reflect genetic instability that favors tumor progression. It is noteworthy that reduced expression of NKX3.1 may also lead to increased expression of the TMPRSS2-ERG chimeric mRNA, thus further underscoring the influence of NKX3.1 loss on the pathogenic effects of ERG gene expression (55).
Supplementary Material
Acknowledgements
This work was supported by Public Health Service grant CA154293 and by the Molecular Pathology Shared Resource and the Imaging Shared Resource of the Herbert Irving Comprehensive Cancer Center, CA013696-36.
Footnotes
The authors have no financial or employment conflicts.
Contributor Information
Cai Bowen, Email: bc2283@columbia.edu.
Tian Zheng, Email: tz33@columbia.edu.
Edward P. Gelmann, Email: gelmanne@columbia.edu.
Reference List
- 1.Asatiani E, Huang WX, Wang A, Rodriguez OE, Cavalli LR, Haddad BR, et al. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005;65:1164–1173. doi: 10.1158/0008-5472.CAN-04-2688. [DOI] [PubMed] [Google Scholar]
- 2.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 3.Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, et al. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15:4706–4711. doi: 10.1158/1078-0432.CCR-08-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowski MC, Bowen C, Gelmann EP. Inflammatory cytokines induce phosphorylation and ubiquitination of prostate suppressor protein NKX3.1. Cancer Res. 2008;68:6896–6901. doi: 10.1158/0008-5472.CAN-08-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, et al. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 2006;66:10683–10690. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]
- 6.Mani RS, Tomlins SA, Callahan K, Ghosh A, Nyati MK, Varambally S, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, et al. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 9.Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-Function of Nkx3.1 Promotes Increased Oxidative Damage in Prostate Carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- 10.Yang CC, Chung A, Ku CY, Brill LM, Williams R, Wolf DA. Systems analysis of the prostate tumor suppressor NKX3.1 supports roles in DNA repair and luminal cell differentiation. F1000Res. 2014;3:115. doi: 10.12688/f1000research.3818.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowen C, Gelmann EP. NKX3.1 activates cellular response to DNA damage. Cancer Res. 2010;70:3089–3097. doi: 10.1158/0008-5472.CAN-09-3138. [DOI] [PubMed] [Google Scholar]
- 12.Bowen C, Ju JH, Lee JH, Paull TT, Gelmann EP. Functional activation of ATM by the prostate cancer suppressor NKX3.1. Cell Rep. 2013;4:516–529. doi: 10.1016/j.celrep.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, et al. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 2007;67:455–464. doi: 10.1158/0008-5472.CAN-06-1591. [DOI] [PubMed] [Google Scholar]
- 14.Puc J, Kozbial P, Li W, Tan Y, Liu Z, Suter T, et al. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015;160:367–380. doi: 10.1016/j.cell.2014.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlbradt E, Asatiani E, Ortner E, Wang A, Gelmann EP. NKX3.1 activates expression of insulin-like growth factor binding protein-3 to mediate insulin-like growth factor-I signaling and cell proliferation. Cancer Res. 2009;69:2615–2622. doi: 10.1158/0008-5472.CAN-08-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 17.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 18.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Androgen-induced TOP2B–mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42:668–675. doi: 10.1038/ng.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 20.Bastus NC, Boyd LK, Mao X, Stankiewicz E, Kudahetti SC, Oliver RT, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res. 2010;70:9544–9548. doi: 10.1158/0008-5472.CAN-10-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E. Histone demethylation catalysed by LSD1 is a flavin-dependent oxidative process. FEBS Lett. 2005;579:2203–2207. doi: 10.1016/j.febslet.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 22.Vidal AE, Boiteux S, Hickson ID, Radicella JP. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo J. 2001;20:6530–6539. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, et al. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- 24.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276:42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 25.Meyer B, Voss KO, Tobias F, Jakob B, Durante M, Taucher-Scholz G. Clustered DNA damage induces pan-nuclear H2AX phosphorylation mediated by ATM and DNA-PK. Nucleic Acids Res. 2013;41:6109–6118. doi: 10.1093/nar/gkt304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci U S A. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosidi B, Wang M, Wu W, Sharma A, Wang H, Iliakis G. Histone H1 functions as a stimulatory factor in backup pathways of NHEJ. Nucleic Acids Res. 2008;36:1610–1623. doi: 10.1093/nar/gkn013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swalwell JI, Vocke CD, Yang Y, Walker JR, Grouse L, Myers SH, et al. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes Chromosomes Cancer. 2002;33:201–205. doi: 10.1002/gcc.10015. [DOI] [PubMed] [Google Scholar]
- 30.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Norton CR, et al. Roles for Nkx3.1 in prostate development and cancer. Genes and Development. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, et al. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002;62:2999–3004. [PubMed] [Google Scholar]
- 34.Nelson WG, De Marzo AM, DeWeese TL, Isaacs WB. The role of inflammation in the pathogenesis of prostate cancer. J Urol. 2004;172:S6–S11. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- 35.Platz EA, De Marzo AM. Epidemiology of inflammation and prostate cancer. J Urol. 2004;171:S36–S40. doi: 10.1097/01.ju.0000108131.43160.77. [DOI] [PubMed] [Google Scholar]
- 36.Khalili M, Mutton LN, Gurel B, Hicks JL, De Marzo AM, Bieberich CJ. Loss of Nkx3.1 expression in bacterial prostatitis: a potential link between inflammation and neoplasia. Am J Pathol. 2010;176:2259–2268. doi: 10.2353/ajpath.2010.080747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soller MJ, Isaksson M, Elfving P, Soller W, Lundgren R, Panagopoulos I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer. 2006;45:717–719. doi: 10.1002/gcc.20329. [DOI] [PubMed] [Google Scholar]
- 38.Cerveira N, Ribeiro FR, Peixoto A, Costa V, Henrique R, Jeronimo C, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–832. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanchez-Chapado M, Olmedilla G, Cabeza M, Donat E, Ruiz A. Prevalence of prostate cancer and prostatic intraepithelial neoplasia in Caucasian Mediterranean males: an autopsy study. Prostate. 2003;54:238–247. doi: 10.1002/pros.10177. [DOI] [PubMed] [Google Scholar]
- 40.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- 41.Mosquera JM, Perner S, Genega EM, Sanda M, Hofer MD, Mertz KD, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–3385. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park K, Dalton JT, Narayanan R, Barbieri CE, Hancock ML, Bostwick DG, et al. TMPRSS2:ERG gene fusion predicts subsequent detection of prostate cancer in patients with high-grade prostatic intraepithelial neoplasia. J Clin Oncol. 2014;32:206–211. doi: 10.1200/JCO.2013.49.8386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, et al. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Zhang Q, Zhang J, Qiu L, Yan SS, Feng J, et al. FATS is a transcriptional target of p53 and associated with antitumor activity. Mol Cancer. 2010;9:244. doi: 10.1186/1476-4598-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan S, Qiu L, Ma K, Zhang X, Zhao Y, Zhang J, et al. FATS is an E2-independent ubiquitin ligase that stabilizes p53 and promotes its activation in response to DNA damage. Oncogene. 2013 doi: 10.1038/onc.2013.494. [DOI] [PubMed] [Google Scholar]
- 46.Vandepoele K, Andries V, Van RN, Staes K, Vandesompele J, Laureys G, et al. A constitutional translocation t(1;17)(p36.2;q11.2) in a neuroblastoma patient disrupts the human NBPF1 and ACCN1 genes. PLoS One. 2008;3:e2207. doi: 10.1371/journal.pone.0002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Albertsen PC, Hanley JA, Penson DF, Barrows G, Fine J. 13-year outcomes following treatment for clinically localized prostate cancer in a population based cohort. J Urol. 2007;177:932–936. doi: 10.1016/j.juro.2006.10.051. [DOI] [PubMed] [Google Scholar]
- 48.Yoshimoto M, Joshua AM, Cunha IW, Coudry RA, Fonseca FP, Ludkovski O, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008;21:1451–1460. doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 49.Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rostad K, Hellwinkel OJ, Haukaas SA, Halvorsen OJ, Oyan AM, Haese A, et al. TMPRSS2:ERG fusion transcripts in urine from prostate cancer patients correlate with a less favorable prognosis. APMIS. 2009;117:575–582. doi: 10.1111/j.1600-0463.2009.02517.x. [DOI] [PubMed] [Google Scholar]
- 51.Leinonen KA, Tolonen TT, Bracken H, Stenman UH, Tammela TL, Saramaki OR, et al. Association of SPINK1 expression and TMPRSS2:ERG fusion with prognosis in endocrine-treated prostate cancer. Clin Cancer Res. 2010;16:2845–2851. doi: 10.1158/1078-0432.CCR-09-2505. [DOI] [PubMed] [Google Scholar]
- 52.Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–3400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 53.Hermans KG, Boormans JL, Gasi D, van Leenders GJ, Jenster G, Verhagen PC, et al. Overexpression of prostate-specific TMPRSS2(exon 0)-ERG fusion transcripts corresponds with favorable prognosis of prostate cancer. Clin Cancer Res. 2009;15:6398–6403. doi: 10.1158/1078-0432.CCR-09-1176. [DOI] [PubMed] [Google Scholar]
- 54.Boormans JL, Porkka K, Visakorpi T, Trapman J. Confirmation of the association of TMPRSS2(exon 0):ERG expression and a favorable prognosis of primary prostate cancer. Eur Urol. 2011;60:183–184. doi: 10.1016/j.eururo.2011.03.028. [DOI] [PubMed] [Google Scholar]
- 55.Thangapazham R, Saenz F, Katta S, Mohamed AA, Tan SH, Petrovics G, et al. Loss of the NKX3.1 tumorsuppressor promotes the TMPRSS2-ERG fusion gene expression in prostate cancer. BMC Cancer. 2014;14:16. doi: 10.1186/1471-2407-14-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.