Abstract
Cell-to-cell variability and functional heterogeneity are integral features of multicellular organisms. Chemical classification of cells into cell type is important for understanding cellular specialization as well as organismal function and organization. Assays to elucidate these chemical variations are best performed with single cell samples because tissue homogenates average the biochemical composition of many different cells and oftentimes include extracellular components. Several single cell microanalysis techniques have been developed but tend to be low throughput or require preselection of molecular probes that limit the information obtained. Mass spectrometry (MS) is an untargeted, multiplexed, and sensitive analytical method that is well-suited for studying chemically complex individual cells that have low analyte content. In this work, populations of cells from the rat pituitary, the rat pancreatic islets of Langerhans, and from the Aplysia californica nervous system, are classified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI) MS by their peptide content. Cells were dispersed onto a microscope slide to generate a sample where hundreds to thousands of cells were separately located. Optical imaging was used to determine the cell coordinates on the slide, and these locations were used to automate the MS measurements to targeted cells. Principal component analysis was used to classify cellular subpopulations. The method was modified to focus on the signals described by the lower principal components to explore rare cells having a unique peptide content. This approach efficiently uncovers and classifies cellular subtypes as well as discovers rare cells from large cellular populations.
Cell-to-cell chemical variability and heterogeneity are fundamental features of multicellular organisms. Cells have historically been classified by their morphology and localization within an organism. However, a cell’s chemical content can also suggest cellular function and specialization. Further, even within supposedly homogeneous cell populations, chemical heterogeneities can be observed due to a variety of endogenous and exogenous factors. Although chemical analyses of cells are often conducted on tissue homogenates, these assays may be less useful for cell classification because homogenization typically mixes many cell types as well as extracellular materials. Signals from rare cells can also be missed because their unique chemical content is diluted during homogenization. Single cell chemical analysis is therefore important for categorizing individual cells based on their chemical content. As a recent example, single cell transcriptomics uncovered molecularly distinct cellular classes in the cortex and the hippocampus, demonstrating the value of single cell analysis for molecular cellular classification.1
Beyond the transcriptome, there also have been many advances in single cell metabolomics and peptidomics analyses, often using mass spectrometry (MS) and different separation methods.2−4 The nontargeted and multiplexed nature of mass spectrometric methods makes them useful for single cell characterization but many are serial approaches. Consequently, the required separation times and sampling processes have restricted investigations to relatively few cells,3,5−7 thereby limiting capabilities for categorizing populations of cells. Higher throughput methods have been developed. Mass cytometry, for example, enables classification of immune cell types based on a panel of markers,8 but the reliance on molecular probes requires a priori knowledge of the cellular chemical content and restricts the number of analytical channels available per analysis. Another high throughput approach, microarray MS, uses arrays of hydrophilic wells surrounded by an omniphobic material, depositing one to a few cells into each well,4 and has been used to study metabolites from single cell organisms like algae and yeast.9,10 Mass spectrometry imaging (MSI) is another option that can obtain thousands of spectra from tissues,11−14 although MSI has yet to be demonstrated for high-throughput single cell profiling.
In this work, we scale up single cell matrix-assisted laser desorption/ionization (MALDI) MS to enable label-free mass spectrometric categorization of cells in endocrine systems based on their peptide profiles. We analyzed a variety of endocrine and nervous system cell types, including cells from the rat pituitary and pancreatic islets of Langerhans, and the Aplysia californica central nervous system. These systems were chosen because there is detailed information on the peptide content of these cells, and we have extensive experience working with these cell types,3,5,7 important factors in allowing the efficacy of our approach to be evaluated. The analysis begins by spreading a population of fluorescently labeled, intact cells onto a microscope slide so that the cells are randomly distributed. The population is optically imaged, and the cell coordinates are determined. The coordinates are then used to automate the MALDI-TOF MS analysis to target the individual cell or cells of interest. This approach is a refinement of the stretched sample method, in which MSI, or profiling, is conducted on tissue samples that are placed on an array of beads embedded on a Parafilm substrate and analyzed via MALDI MS.15−18 A similar approach has also been used for laser ablation electrospray ionization MSI.19 Instead of analyzing tissues or tissues on beads, here we focused on determining distinct subpopulations of cells based on their peptide profiles. Although a cell population prepared in this way can also be analyzed via traditional MSI, this targeted approach greatly reduces data size and complexity, and improves the quality of the data as MS acquisitions are only from the cells of interest (and not from cellular debris or other features).
Along with optimizing the data collection process, we also worked on effective data mining. A challenge in analyzing single cell data sets involves finding both the major and minor patterns that characterize cell populations. We conducted principal component analysis (PCA) and PCA-based outlier detection, enabling identification of subpopulations and rare cells. Using this data collection and analysis method, we profiled the peptide content of populations of hundreds to thousands of cells, classifying multiple cell types within the pituitary and pancreas, as well as revealing several rare cells having a unique cellular content.
Experimental Section
Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), unless noted otherwise.
Sample Preparation
Details for the Aplysia neuronal samples are provided in the Supporting Information. The pituitary and islets of Langerhans cellular populations were extracted from male Sprague–Dawley outbred rats (Rattus norvegicus, 2.5–3 months-old; Harlan Laboratories, Indianapolis, IN). Animals were housed on a 12 h light cycle and fed ad libitum. Euthanasia was performed in accordance with the protocols established by the University of Illinois Institutional Animal Care and Use Committee, and in accordance with all state and federal regulations for the humane care and treatment of animals.
For pituitary isolation, rats were sacrificed by decapitation using a guillotine. The pituitaries were immediately surgically removed and placed into ∼2 mL of ice cold Modified Gey’s balanced salt solution (mGBSS) containing 1.5 mM CaCl2, 4.9 mM KCl, 0.2 mM KH2PO4, 11 mM MgCl2, 0.3 mM MgSO4, 138 mM NaCl, 27.7 mM NaHCO3, 0.8 mM NaH2PO4, and 25 mM HEPES dissolved in Milli-Q water (Millipore, Billerica, MA), with the pH adjusted to 7.2 using NaOH in Milli-Q water. Enzymatic treatment was done in an mGBSS solution containing 1% trypsin and 0.2% collagenase at 37 °C for 20 min, followed by 5 min incubation in mGBSS containing 1% trypsin, and another 5 min in 0.2% collagenase and 5 mg/mL of DNase I (Boehringer-Mannheim, Mannheim, Germany) dissolved in mGBSS. Finally, the sample was washed with and kept for 30–180 min in a mixture containing 33% glycerol and 67% mGBSS. The cell nucleus was stained by adding 10 μL of 1 mg/mL of Hoechst 33342. Gentle trituration with a wide-bore plastic Pasteur pipet was used to form the cell suspension, which was deposited onto indium tin oxide (ITO) slides. To remove excess extracellular glycerol, the samples were rinsed with 150 mM ammonium acetate buffer (pH 10), which minimizes damage to the cells,20 and which was pH balanced to reduce removal of endogenous peptides. A total of four sample slides containing cells from four animals were used for analysis.
Islets of Langerhans isolation was performed according to a previously described protocol with minor modifications.21 Briefly, the pancreas was injected with 2 mL of 1.4 mg/mL collagenase P solution from Clostridium histolyticum (Clostridiopeptidase A, EC 3.4. 24. 3; cat. no. 11 213 857 001, Roche Diagnostics, Indianapolis, IN) dissolved in mGBSS. The injected pancreas was surgically dissected and placed in a 10 mL glass vial containing 3 mL of the collagenase P solution and then incubated in a recirculating water bath for 20–30 min at 37 °C. The resulting suspension was washed twice with mGBSS for 3 min at 300g, resuspended in 10 mL of mGBSS, and sieved through a 1 × 1 mm mesh into a Petri dish. Manual collection of islets of Langerhans into 2 mL of mGBSS in a 35 mm Petri dish was performed with a micropipette under an inverted light microscope. To stain the nuclei, 2 μL of 1 mg/mL Hoechst 33342 was added to the dish, followed by incubation for 15 min at 15 °C. Islets were then treated with 0.25% trypsin-EDTA at 37 °C for 20 min. Islets were triturated into single cells and transferred to an ITO-slide, stabilized in a 40% glycerol, 60% mGBSS solution, and followed by washing with 150 mM ammonium acetate.
Optical Imaging
Each dispersed cell population was imaged using an AxioVert 200 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) with an X-CITE 120 mercury lamp (Lumen Dynamics, Mississauga, Canada) and a 31000v2 DAPI filter set (Chroma Technology, Irvine, CA). A 10× objective was used to obtain a mosaic image with 10% overlap between neighboring images. Images were taken using an AxioCam HRC color camera (Carl Zeiss) set to a resolution of 4164 × 3120.
Cell Localization
All mosaic optical images were stitched in ImageJ to output a list of relative offsets between images.22 Individual images were batch processed with the “color threshold” and “analyze particles” features in ImageJ to find the cell coordinates in each image. Cell coordinates from each image were added to the corresponding image offset to obtain the cell coordinates on the sample slide.
Geometry File Creation
Before fluorescence imaging, a silver sharpie was used to draw an ∼5 mm thick line along two opposite sides of the cell population. An ultrafleXtreme mass spectrometer (Bruker Daltonics, Billerica, MA) was used to generate multiple fiducial markers into each line by laser ablation.
After microscopy imaging and cell localization, 3000 rat pituitary cells or ∼600 islets of Langerhans cells were randomly chosen from their respective cellular populations. The determined cell coordinates were used to create a custom geometry file for automatic single cell MALDI-TOF MS analysis. The geometry file was created using the Java JDK 1.6 Applet available at http://neuroproteomics.scs.illinois.edu/imaging.html, as described previously.15 All fiducial markers were used when calculating the scaling and rotation parameters, but only four markers were input into the applet.
Matrix Application
A solution of the MALDI matrix 2,5-dihydroxybenzoic acid (50 mg/mL) in 1:1 acetone:water plus 0.5% trifluoroacetic acid (TFA) was sprayed onto the samples using an artist’s airbrush (Paasche Airbrush Company, Chicago, IL) propelled with nitrogen at 40 psi. The airbrush was held ∼60 cm from the sample, and the matrix coating was built up by alternatively spraying 1 and 2 mL of the matrix solution. The slide was rotated after each spray cycle and dried completely. Coating thickness was estimated by weighing the sample slide before and after matrix application and measuring the coated area. A MALDI matrix coating of 0.2–0.4 mg/cm2 was applied.
MALDI-TOF MS
MALDI-TOF MS analysis was conducted using the Bruker ultrafleXtreme mass spectrometer with a frequency tripled Nd:YAG solid state laser. The laser was set to the “small” footprint setting at an ∼30 μm diameter. Acquisition was automated to each cell location using the autoXecute feature of the instrument and the custom geometry file generated earlier. For pituitary cells, signals from 250 laser shots fired at 1000 Hz were summed for each cell. Islets of Langerhans cells were analyzed with 1000 laser shots fired at 1000 Hz. Signals were summed from one spot on the cell for both types of cells. Details for analyzing Aplysia neurons are included in the Supporting Information. To confirm and identify the markers for pituitary subpopulations, tandem MS (MS/MS) was conducted on the pituitary extracts using the LIFT mode of the mass spectrometer with argon as the collision gas and an isolation window of 10 Da for the precursor ions.
Identifying the Peptides in Pituitary Extracts
For the pituitary samples, we also identified the peptides present using larger samples, extracting the peptides and characterizing them via liquid chromatography (LC)–MS. Details of the peptide extraction, capLC–MALDI MS, and capLC–electrospray ionization (ESI) MS/MS are provided in the Supporting Information.
Statistical Analysis
For all cell types, the raw mass spectra were first processed using two R packages, MALDIquant and MALDIquantForeign,23 in order to identify those without a signal. Mass spectra were baseline subtracted using the Top Hat algorithm with a half-window size of 50, smoothed using the Savitzky-Golay algorithm with a half window size of 4, and peak picked with a minimum signal-to-noise ratio of 3. After peak picking, the intensity of the strongest peak was used to sort the mass spectral data set in increasing intensity. Spectra sorted in this manner were manually examined to exclude those without signal.
The remaining mass spectra were processed and visualized using a custom Python script (see the Supporting Information). The spectra were down-binned to unit m/z resolution, baseline subtracted with a window of 10, and normalized to the total ion count before further analysis. PCA was used to categorize major cell subpopulations by projecting the data onto the heavily weighted principal vectors. Principal component (PC)-based outlier detection was performed to find rare cells.24 The same program also generated false-color peptide distribution maps of the population. Signal intensity is illustrated using a rainbow color scheme. To further enhance contrast, colors associated with lower intensities (blue and green) were also made more transparent using the GNU Image Manipulation Program (GIMP; see http://www.gimp.org).
Results and Discussion
The goal of our approach is to classify populations of neurons and endocrine cells based on their peptide profiles in order to determine categories of cells (cell types) in a complex sample, as well as to identify unusual peptide profiles among the studied cells. MALDI appears well-suited for this application because of its ability to profile the hormone and neuropeptide content of individual cells.7,25−28 The cell population is dispersed over a microscope slide so that the distance between the cells is large enough to enable MALDI MS to assess individually isolated cells; next, each cell location is profiled via MALDI-TOF MS. To validate the protocol and our ability to target and assay individual cells, a preliminary experiment was conducted on a population of A. californica neurons as they are larger and easier to visualize. Our results confirm that the method enables targeting of cells randomly localized on a slide (see the Supporting Information). Rather than further characterize our protocol using the 30–50 μm diameter Aplysia neurons, we then moved to smaller mammalian neuroendocrine cells.
High-Throughput MALDI-TOF MS Analysis of Pituitary Cells
We demonstrate the capability of the approach by analyzing populations of rat pituitary cells. The pituitary was chosen because it contains different morphological and functional regions, with many cell types expressing high levels of well-characterized cell-to-cell signaling peptides.7,29−31 Challenges faced when working with these samples include their relatively small cell size (10–20 μm diameter) and cell optical transparency (Figure S3 of the Supporting Information). Therefore, live pituitary cells were labeled with the nuclear stain Hoechst 33342 and stabilized with glycerol before being dispersed onto the slides. We have previously used a similar glycerol stabilization protocol to conduct quantitative single cell MALDI MS peptide measurements26 and in another study showed that glycerol stabilization helps to maintain cell integrity during sample preparation and decreases CE−MS measurement variability.5 We have also obtained pituitary peptide profiles using this protocol that are similar to those reported by others using different approaches.30 Nuclear staining enables cell coordinates to be determined from fluorescent images based on nucleus size and brightness. The stain did not produce interfering MS signals in the peptide mass range (Figure S4 of the Supporting Information).
To speed up the MALDI MS analysis while still demonstrating high-throughput capabilities, 3000 cells were randomly chosen from each population and deposited on the four microscope slides. These cells were typically distributed over a roughly 2.5 × 4 cm2 area. If we used traditional MSI to analyze these cell populations, ∼10000000 pixels would have been needed to image the area at a 10 μm lateral resolution. This high a resolution would be necessary for spatially resolving single cells when using a raster pattern in imaging mode. Assuming the same MALDI MS conditions as used in this study, each pixel would need at least 0.25 s to be acquired (summing signals from 250 laser shots fired at 1000 Hz). These conditions would lead to an overall data collection time of several hundred hours. With the targeted profiling technique presented here, the MS analysis can be done in about an hour.
Each cell was profiled with a single MALDI MS acquisition. The laser spot was set at a diameter of ∼30 μm, which is larger than typical pituitary cells, ensuring that the entire cell was sampled. This reduces signal variations that may result from only partially sampling a cell. Even from a single cell, several pituitary peptides were detected, including arginine vasopressin (AVP), oxytocin, and alpha-melanocyte stimulating hormone (α-MSH) (Figure 1), which are markers of the posterior and intermediate pituitary. The identity of these peptides was confirmed via MALDI MS/MS (Figure S5 of the Supporting Information). Melanotrophs in the intermediate pituitary express high amounts of peptides derived from the pro-opiomelanocortin (POMC) prohormone, such as α-MSH, joining peptide and β-endorphin.32 As is typical for melanotrophs, mass spectra with intense α-MSH signals also contain other POMC peptides (Figure 2). The detection of AVP and oxytocin in cells selected by nuclear stain and cell morphology is surprising. According to the classical view, AVP and oxytocin are expressed in the soma of distantly located hypothalamic neurons and then transported to their release sites in the posterior pituitary.33 Thus, no AVP and oxytocin cell bodies were expected. The posterior pituitary is primarily composed of these hypothalamic-originating terminals and neurites rather than cells, so it is intriguing to find pituitary cells with AVP and oxytocin signals. It is possible that hypothalamic neuron terminals were colocalized with some pituitary cells, which is consistent with the weak-to-moderate coappearance of POMC peptides with AVP in many spectra (Figure S6 of the Supporting Information). However, several reports have demonstrated AVP-related immunostaining34 and vasopressin RNA35 in some pituicytes,36 and AVP may be internalized into some cells,37,38 and so either is a possibility. The current data do not resolve the source of these peptides.
Figure 1.
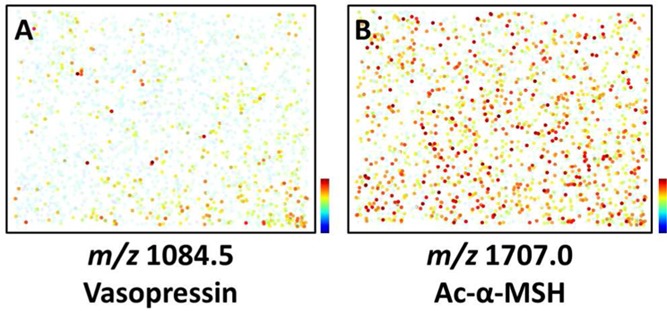
Spatial distribution maps for peptides detected in a dispersed population of pituitary cells. Distribution of (A) AVP-containing cells and (B) ac-α-MSH-containing melanotrophs. Signal intensity is color coded and increases from blue to red, with blue indicating noise level. The intensity of the color scale is distinct for each ion.
Figure 2.

Mass spectrum acquired from a melanotroph. Several peptides from the POMC prohormone are detected, including α-MSH, joining peptide (J-peptide), corticotropin-like intermediate peptide (CLIP), and β-endorphin.
Determination of Major Pituitary Cell Subpopulations using PCA
We also worked on effective data mining. Compared to traditional MSI, the data set collected with this approach is simplified and minimized because it contains only spectra from cells rather than a mix of spectra from cells, debris, media, and empty spaces. PCA was conducted to classify cell types (Figure 3). Although more manual statistical methods (e.g., examining biaxial plots) can also be used, multivariate methods take into account the entire data set. Our nonsupervised analysis facilitates the detection of features because it reveals those that are responsible for major patterns in the data. For the described data set, the chemical profiles obtained via MALDI MS revealed multiple pituitary cellular populations. The loading spectra have a strong contribution from ac-α-MSH in PC 1, lipidlike signals for PC 2, and AVP for PC 3 (Figure 3B). This pattern is consistent with examining peptide distribution maps, with PC 1 highlighting melanotrophs and PC 3 showing cells with strong AVP signal. PC 2 may be composed of cells from the anterior pituitary, where lipids may be the dominant chemical species observed in the targeted mass range. Another possibility may be partially lysed cells that still retain the stained nucleus. Although the pituitary contains distinct cell types with well-characterized content, the PCA shows a high degree of heterogeneity, with data points spreading out along the PCs to show a continuum of signal instead of an on–off behavior (Figure 3A). We have taken care to reduce measurement variations by validating our ability to target cells using an Aplysia neuronal population. Our previous work with the stretched sample method also corroborates our targeting accuracy.15−18 Together, these factors support the notion that the observed variations may be biological in origin rather than a measurement artifact.
Figure 3.

PCA of the MS data set acquired from the pituitary cell population. (A) Score plot projected onto the PC 1 by PC 2 plane, with a rainbow color scale indicating scores on PC 3 (warmer color = higher PC 3 score). PC1: 49.1% variance, PC2: 10.7% variance, and PC3: 6.0% variance. (B) Loading spectra for PCs 1, 2, and 3. Blue: PC 1; black: PC 2; and red: PC 3.
Determination of Chemically Distinct Rare Cells in the Pituitary
Examining the first few PCs is useful for finding major patterns; however, minor variations, such as chemically rare cells, may be overlooked because they are not captured in the first few PCs. Mass spectra corresponding to rare cells may not strongly influence the largest principal vectors but may be identified by projecting the data onto the smallest principal vectors. In practice, with high-dimensional data sets, it is often more computationally efficient to project onto the largest principal vectors then back-project into the original data space and, finally, subtract the resulting spectra from the original. The difference reveals the parts of the data that were not captured by typical PCA.24 The number of PCs used for back-projection determines the sensitivity for detecting rare signals, with more PCs leading to the observation of more unique signals.
This PCA-outlier detection approach was applied using the first 23 PCs (∼90% of the total variance) for back-projection. Figure 4 shows the generated difference mass spectrum. Although some major pituitary peptides are still present, many uncommon signals are also visible. Several signals are tentatively labeled as contaminants (and not from cells) by examining the relevant mass spectra and the ion distribution pattern on the slide. Table 1 summarizes the peptide-containing cell types found using traditional and extended PCA. Other than melanotrophs, each cell type is labeled with their distinguishing peptide marker. The characteristic ions for each population are listed in Table S1 of the Supporting Information. Cells identified as melanotrophs, or those containing AVP and oxytocin signals, are classified as major cell populations. AVP and oxytocin-exhibiting cells are separated into two groups because some differences were seen in their spatial distribution. A characteristic ion at m/z 2264.2 was detected in a large portion of melanotrophs and used to classify a subpopulation of melanotrophs. For rare cells, it is not surprising that most of the apparent biomarkers detected were not identified. One exception is the signal at m/z 1622.8, which matches by mass to unmodified α-MSH. Most α-MSH in the intermediate pituitary is acetylated, but the nonacetylated form is also present39 and has been observed in prior MSI studies of the pituitary.31 Revealing these rare cells in conjunction with larger subpopulations demonstrates the usefulness of our PCA-based statistical workflow to highlight major and minor patterns.
Figure 4.
Difference mass spectrum for the studied pituitary cell population after accounting for 90% of the total variance (23 PCs were used for back-projection). Known pituitary peptides are labeled; other unidentified ions are labeled with their m/z ratios. Possible contaminant signals are marked by asterisks.
Table 1. Major Cell Populations, A Subpopulation of Melanotrophs, And Rare Cells Found in the Rat Pituitary Using Peptide Biomarkers Revealed via Traditional PCA and the Extended PCA Approacha.
| major cell population biomarkers | subpopulation |
|---|---|
| POMC peptides (melanotrophs) | POMC peptides and m/z 2264.2 (melanotrophs) |
| AVP | – |
| oxytocin | – |
| m/z 2105.3 | – |
| rare cells | |
|---|---|
| m/z 944.5 | m/z 3175.1 |
| m/z 1125.6 | m/z 3455.3 |
| m/z 1622.8 (unmodified α-MSH) | m/z 5882.6 |
| m/z 2491.4 | |
Each presented biomarker classifies a cell population, subpopulation, or a rare cell type. Only cells with peptide signals are classified.
Another interesting signal at m/z 2105.3 was detected in many cells and does not appear to have originated from the intermediate or posterior pituitary. Using LC–MALDI MS/MS, we identified this ion as a putative somatotropin-related peptide. However, only a portion of the sequence is supported by genomic data (Figure S7 of the Supporting Information). This assignment suggests the detection of somatotrophs from the anterior pituitary. LC–ESI MS/MS measurements on the same sample uncovered a number of other pituitary peptides, including known somatotropin-related peptides (Table S2 of the Supporting Information). Peptidomic analysis revealed peptides from the prolactin prohormone, suggesting the presence of lactotrophs in the sample. Both somatotrophs and lactotrophs are acidophiles, while other major cell types in the anterior pituitary more readily take up basophilic stains.40 Peptides from prohormones known to be expressed in basophiles (corticotropin, thyrotropin, lutropin, and follicle-stimulating hormone) were not detected in our peptidomic experiments. It is possible that the predominant detection of peptides from acidophilic cell types may have been caused by the acidic extraction and separation conditions.
Determination of Islets of Langerhans Cell Populations
In order to explore whether the approach will work with smaller endocrine cells, we studied the rat pancreatic islet of Langerhans. As the islet cells are smaller than cells from pituitary, being between 3–6 μm in diameter, these represent a more difficult challenge for accurate cell targeting. In these cases, we dispersed cells from individual islets for each experiment.
PCA was performed on a MALDI MS data set with spectra from ∼600 single cells. PC 1, 2, and 3 distinguished cell types based on known pancreatic prohormones (Figure 5, Table 2).41,42 In the loading spectrum for PC 1, peptides characteristic for beta cells (insulin 1 and 2 prohormones) separated cells containing peptides characteristic for alpha cells (the glucagon prohormone), with the peptides having opposite loading signs. Beta cells also have higher scores on PC 3 than alpha cells. In PC 2, the somatostatin-14 peptide, characteristic of delta cells, appeared with an opposite sign to the loading for insulin peptide from beta cells, hence separating these cell types as well. Although these three cell types are separated via PCA, gamma cells, which are characterized by the pancreatic hormone, appear convoluted in the same direction as alpha cells in the score plot. The loading for pancreatic hormone in PC 1 has the same sign as the glucagon prohormone peptide, and the loading in PC 2 is close to zero. Gamma cells are much less abundant than other cell types in islets of Langerhans throughout a majority of the rat pancreas.41 We expect that the low count of these observations did not provide enough data variability to separate this cell type from others, and explains why alpha and gamma cells are grouped together in the score plot. Our results show successful categorization of most cell types from the rat pancreatic islets of Langerhans, further demonstrating the method’s capability to conduct population-level single cell analysis, and to classify cells based on peptide biomarkers.
Figure 5.

PCA of the MS data set acquired from the cell population of an islet of Langerhans. (A) Score plot projected onto PC 1 by the PC 2 plane, with a rainbow color scale indicating scores on PC 3 (warmer color = higher PC 3 score). PC1: 27.4% variance; PC2: 19.8% variance; and PC3: 15.2% variance. (B) Loading spectra for PCs. Blue: PC 1; black: PC 2; and red: PC 3. Peak annotations are 1: somatostatin-14; 2: insulin 1 A+B [M + 2H]2+; 3: insulin 2 C-peptide; 4: insulin 1 C-peptide; 5: glicentin-related polypeptide; 6: glucagon; 7: pancreatic hormone; 8: insulin 1 and 2 A+B chains.
Table 2. Cell Populations in the Rat Pancreatic Islet of Langerhans Classified Using Peptide Biomarkersa.
| cell type | biomarkers |
|---|---|
| alpha | glucagon |
| glicentin-related polypeptide | |
| beta | insulin 1 A+B chain |
| insulin 2 A+B chain | |
| insulin 1 C-peptide | |
| insulin 2 C-peptide | |
| gamma | pancreatic hormone |
| delta | somatostatin-14 |
Peptide signals characteristic of all four cell types were detected.
Conclusions
We have developed a novel approach that combines in vitro live cell labeling, optical microscopy, image processing, high throughput single cell mass spectrometry, and multivariate statistical analysis, to enable cellular classification via multiplexed analysis of cell peptide content. Cells in a randomly distributed population were individually examined by finding the cell coordinates using optical microscopy and performing automated single cell MALDI-TOF MS analysis. The entire cell is profiled via MALDI MS, and the MS data obtained from hundreds to thousands of single cells allows cellular subpopulations and rare cells to be revealed. We demonstrate this method on multiple cell populations from the rat pituitary and pancreatic islets of Langerhans, and the A. californica nervous system.
The multiplexed chemical data that is generated, which includes spatial information, has parallels to data obtained using MSI. However, the approach described here saves time and improves specificity by examining only the cells of interest. One could use more specific stains than the nuclear stains used here. For example, stains that differentiate glia and neurons can be used that enable populations of cells to be classified based on the localization stain, and then their chemical differences compared. In addition, although we have categorized cells here based only on their peptide profiles, morphological information generated during optical cell finding may contribute another dimension for cell classification. Beyond the endocrine cell populations covered here, this approach may be used to conduct chemical classification of many cell types across multiple species and can easily be extended to lipids and other molecular classes that are detected via MALDI MS.
Acknowledgments
This work was supported by award numbers P30 DA018310 from the National Institute on Drug Abuse, R01 NS031609 from the National Institute of Neurological Disease and Stroke, and the R21 MH100704 from the National Institutes of Mental Health. In addition, a Springborn Postdoctoral Fellowship was awarded to D.J.K., and the Aplysia were from the National Resource for Aplysia (Miami, FL), partially funded by PHS Grant P40 OD010952. T.J.C. acknowledges funding from the Training Program at Chemistry-Interface with Biology (NIH T32 GM070421). The authors acknowledge the help of Xiying Wang with Aplysia and Stephanie Baker with manuscript preparation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the awarding agencies.
Supporting Information Available
Supporting experimental details, figures (Figures S1–S7), and tables (Tables S1–S2) as noted in the text. The custom Python script is provided as a .txt file (see instructions in the PDF). The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b01557.
The authors declare no competing financial interest.
Supplementary Material
References
- Zeisel A.; Muñoz-Manchado A. B.; Codeluppi S.; Lönnerberg P.; La Manno G.; Juréus A.; Marques S.; Munguba H.; He L.; Betsholtz C.; Rolny C.; Castelo-Branco G.; Hjerling-Leffler J.; Linnarsson S. Science 2015, 347, 1138–1142. [DOI] [PubMed] [Google Scholar]
- Trouillon R.; Passarelli M. K.; Wang J.; Kurczy M. E.; Ewing A. G. Anal. Chem. 2013, 85, 522–542. [DOI] [PubMed] [Google Scholar]
- Nemes P.; Knolhoff A. M.; Rubakhin S. S.; Sweedler J. V. Anal. Chem. 2011, 83, 6810–6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban P. L.; Jefimovs K.; Amantonico A.; Fagerer S. R.; Schmid T.; Madler S.; Puigmarti-Luis J.; Goedecke N.; Zenobi R. Lab Chip 2010, 10, 3206–3209. [DOI] [PubMed] [Google Scholar]
- Nemes P.; Knolhoff A. M.; Rubakhin S. S.; Sweedler J. V. ACS Chem. Neurosci. 2012, 3, 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts J. T.; Louis K. R.; Crandall S. R.; Govindaiah G.; Cox C. L.; Sweedler J. V. Anal. Chem. 2014, 86, 3203–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin S. S.; Churchill J. D.; Greenough W. T.; Sweedler J. V. Anal. Chem. 2006, 78, 7267–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornatsky O.; Bandura D.; Baranov V.; Nitz M.; Winnik M. A.; Tanner S. J. Immunol. Methods 2010, 361, 1–20. [DOI] [PubMed] [Google Scholar]
- Fagerer S. R.; Schmid T.; Ibanez A. J.; Pabst M.; Steinhoff R.; Jefimovs K.; Urban P. L.; Zenobi R. Analyst 2013, 138, 6732–6736. [DOI] [PubMed] [Google Scholar]
- Ibáñez A. J.; Fagerer S. R.; Schmidt A. M.; Urban P. L.; Jefimovs K.; Geiger P.; Dechant R.; Heinemann M.; Zenobi R. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 8790–8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietz C. B.; Gemperline E.; Li L. Adv. Drug Delivery Rev. 2013, 65, 1074–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris J. L.; Caprioli R. M. Chem. Rev. 2013, 113, 2309–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni E. J.; Rubakhin S. S.; Sweedler J. V. J. Proteomics 2012, 75, 5036–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai K.; Heeren R. M. A. Chem. Rev. 2010, 110, 3237–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman T. A.; Rubakhin S. S.; Romanova E. V.; Tucker K. R.; Sweedler J. V. Anal. Chem. 2009, 81, 9402–9409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker K. R.; Serebryannyy L. A.; Zimmerman T. A.; Rubakhin S. S.; Sweedler J. V. Chem. Sci. 2011, 2, 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe E. B.; Jurchen J. C.; Koszczuk B. A.; Losh J. L.; Rubakhin S. S.; Sweedler J. V. Anal. Chem. 2006, 78, 6826–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman T. A.; Monroe E. B.; Sweedler J. V. Proteomics 2008, 8, 3809–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.; Smith B.; Shrestha B.; Márk L.; Vertes A. In Mass Spectrometry Imaging of Small Molecules, He L., Ed.; Springer: New York, 2015; pp 117–127. [Google Scholar]
- Berman E. S. F.; Fortson S. L.; Checchi K. D.; Wu L.; Felton J. S.; Wu K. J. J.; Kulp K. S. J. Am. Soc. Mass Spectrom. 2008, 19, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Carter J. D.; Dula S. B.; Corbin K. L.; Wu R.; Nunemaker C. S. Biol. Proced. Online 2009, 11, 3–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preibisch S.; Saalfeld S.; Tomancak P. Bioinformatics 2009, 25, 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb S.; Strimmer K. Bioinformatics 2012, 28, 2270–2271. [DOI] [PubMed] [Google Scholar]
- Gnanadesikan R.; Kettenring J. R. Biometrics 1972, 28, 81–124. [Google Scholar]
- Romanova E. V.; Aerts J. T.; Croushore C. A.; Sweedler J. V. Neuropsychopharmacology 2014, 39, 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin S. S.; Sweedler J. V. Anal. Chem. 2008, 80, 7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova E. V.; Sasaki K.; Alexeeva V.; Vilim F. S.; Jing J.; Richmond T. A.; Weiss K. R.; Sweedler J. V. PLoS One 2012, 7, e48764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann A.; Annangudi S. P.; Richmond T. A.; Ament S. A.; Xie F.; Southey B. R.; Rodriguez-Zas S. R.; Robinson G. E.; Sweedler J. V. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova E. V.; Rubakhin S. S.; Sweedler J. V. Anal. Chem. 2008, 80, 3379–3386. [DOI] [PubMed] [Google Scholar]
- Che F. Y.; Lim J.; Pan H.; Biswas R.; Fricker L. D. Mol. Cell. Proteomics 2005, 4, 1391–1405. [DOI] [PubMed] [Google Scholar]
- Guenther S.; Rompp A.; Kummer W.; Spengler B. Int. J. Mass Spectrom. 2011, 305, 228–237. [Google Scholar]
- Raffin-Sanson M. L.; de Keyzer Y.; Bertagna X. Eur. J. Endocrinol. 2003, 149, 79–90. [DOI] [PubMed] [Google Scholar]
- Strand F. L.Neuropeptides: Regulators of Physiological Processes; MIT Press: Cambridge, MA, 1999. [Google Scholar]
- Boersma C. J.; Sonnemans M. A.; Van Leeuwen F. W. Brain Res. 1993, 611, 117–129. [DOI] [PubMed] [Google Scholar]
- Pu L. P.; Van Leeuwen F. W.; Tracer H. L.; Sonnemans M. A.; Loh Y. P. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 10653–10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochman K. J. Anim. Feed Sci. 2013, 22, 283–294. [Google Scholar]
- Lutz W.; Sanders M.; Salisbury J.; Kumar R. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 6507–6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso L.; Peteri-Brunback B.; Mienville J. M. J. Neuroendocrinol. 2004, 16, 313–318. [DOI] [PubMed] [Google Scholar]
- Rudman D.; Hollins B. M.; Kutner M. H.; Moffitt S. D.; Lynn M. J. Am. J. Physiol. 1983, 245, E47–54. [DOI] [PubMed] [Google Scholar]
- Yeung C. M.; Chan C. B.; Leung P. S.; Cheng C. H. Int. J. Biochem. Cell Biol. 2006, 38, 1441–1449. [DOI] [PubMed] [Google Scholar]
- Elayat A. A.; el-Naggar M. M.; Tahir M. J. Anat. 1995, 186, 629–637. [PMC free article] [PubMed] [Google Scholar]
- Stewart K. W.; Phillips A. R.; Whiting L.; Jullig M.; Middleditch M. J.; Cooper G. J. Rapid Commun. Mass Spectrom. 2011, 25, 3387–3395. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



