Abstract
Background
Recently the Type VI secretion system (T6SS), which can play a significant role in bacterial survival and pathogenesis, was reported in Campylobacter spp., having the hcp gene as a key component.
Methods
Campylobacteriosis is associated with the consumption of infected chicken meat. Our study aimed to explore the presence of T6SS in C. jejuni (n = 59) and C. coli (n = 57) isolates, from retail raw chicken and to investigate their pathogenic potential. The hcp gene was used as an indicator for the T6SS presence.
Results
Using multiplex PCR we have identified a significantly higher prevalence of hcp in C. coli isolates (56.1%) than in C. jejuni (28.8%) and AFLP analysis of the isolates showed a high degree of genetic similarity between the isolates carrying the hcp gene. Genome sequencing data showed that 84.3% of the C. coli and 93.7% of the C. jejuni isolates had all 13 T6SS open reading frames. Moreover, the virulence characteristics of hcp + isolates, including motility and the ability to invade human intestinal epithelial cells in vitro, were significantly greater than in the control strain C. jejuni 12502; a human isolate which is hcp positive.
Conclusion
Overall, it was discovered that hcp+C. coli and C. jejuni isolated from retail chicken isolates posses genetic and phenotypic properties associated with enhanced virulence. However, since human infections with C. coli are significantly less frequent than those of C. jejuni, the relationship between virulence factors and pathogenesis requires further study.
Keywords: Campylobacter jejuni, Retail chicken, Type VI secretion system
Background
Campylobacter species are the most common foodborne pathogens in humans but also commensals in many farm animals, including cattle, swine and poultry. Campylobacters are Gram-negative, microaerophilic microorganisms possessing a corkscrew motility phenotype which has proven significance in achieving penetration of the human gut epithelium in order to establish infection in humans, and colonisation in poultry [1, 2].
The most frequent cause of campylobacteriosis in the UK is poultry meat [3] and up to 90% of poultry carcasses are contaminated with C. jejuni [4, 5] and C. coli [6]. The virulence properties of campylobacters are, however, yet to be fully defined and will be dependent on the genetic content of each individual isolate. Campylobacters can cause a range of illnesses; diarrhoea, reactive arthritis and in some cases infection can subsequently lead to serious neuromuscular disorders such as Guillain-Barré syndrome [7, 8]. One pathogenic property employed by bacteria and reported in Campylobacter spp., is the use of secretion systems to export toxins (proteins) into their environment, or directly through membranes into neighbouring eukaryotic [9] or prokaryotic [10] cells.
The Type VI secretion system (T6SS), has been reported in Campylobacter [11], as well as other Gram negative bacteria [12]. In C. jejuni this secretion system is potentially associated with more severe forms of disease as it can confer cytotoxicity toward red blood cells [11]. A significant component of the T6SS is the product of the hcp gene and the presence of this gene has been described as being indicative of a functional T6SS in C. jejuni [13]. Further, it has been reported as being more prevalent in strains isolated from patients experiencing bloody diarrhoea, than those having non-bloody diarrhoea [13]. Also, hcp+ strains of C. jejuni have shown increased abilities to adhere to, and invade, the host gastrointestinal epithelium in vivo [14].
This study has focused only on the hcp+ isolates as information on their virulence is lacking for this emerging virulence mechanism. This study aimed to analyze the C. jejuni and C. coli isolates obtained from retail packs of raw chicken produced in Northern Ireland for the presence of the T6SS and to test their pathogenic potential.
Methods
Microbiology
All media were supplied by Thermo Fisher Scientific Ltd, Basingstoke UK, and all reagents supplied by Sigma-Aldrich Ltd, Gillingham, UK unless otherwise stated. All incubations were performed microaerobically (85% N2, 10% CO2 and 5% O2, all v/v) in a Don Whitley MACS 500 workstation (Don Whitley Scientific, Shipley, UK) at 41.5°C unless otherwise stated.
Campylobacter isolation and identification
Samples of retail chicken were prepared as previously described [15]. Briefly, we have used a stomacher bag and buffer peptone water (225 ml) to emulsify 25 g of skin and flesh sample in a Seward 400 blender (Seward Ltd, Worthing, UK). The emulsified sample (25 ml) was transferred into a container with 225 ml of BBW (Bolton broth). The BS EN ISO10272-1:2006 [16] was followed as previously described [17]. The 225 ml BB were first incubated for 4 h at 37°C, followed by a second incubation step of 24 h at 41.5°C. The resulting culture was plated on modified charcoal cefoperazone deoxycholate agar (mCCDA) and incubated at 41.5°C until single colonies were countable. In order to confirm that the resulting colonies represent a typical Campylobacter colony the motility and oxidase tests were performed. DNA was extracted from each individual isolate using half of a 10 μl loopful in 1 ml of SET buffer (150 mmol l−1 NaCl, 15 mmol l−1 EDTA, 10 mmol l−1 Tris–HCl, pH 8.0). Long-term stocks (−80°C) were prepared in 1 ml of NB plus (nutrient broth plus) containing 10% (v/v) glycerol Overall 59 C. jejuni and 57 C. coli poultry isolates from Northern Ireland producers were selected from the culture collection yielded by the survey [15] and investigated in this study.
Genome sequencing, assembly and annotation
We have performed genome sequencing of all our 52 [18] C. jejuni and C. coli isolates as described previously by Ugarte Ruiz et al. [19] using Illumina MiSeq 2 × 150 bp paired-end sequencing. To analyze the data quality FastQC was used [20]. In order to evaluate the sequencing reads the Trimmomatic was used at the following parameters: (v0.32) ‘leading’ and ‘trailing’ setting of 3, a ‘slidingwindow’ setting of 4:20 and a ‘minlength’ of 36 nucleotides) [21]. BWA-MEM (v0.7.7-r441) was used to map the reads using the genome sequence of C. jejuni NCTC 11168 (AL111168) as Ref. [22]. Velvet Optimiser (v2.2.5) using n50 optimization [23, 24] was used to perform assembly. The reference strain C. jejuni NCTC 11168 (AL111168) was used to complete Contigs using ABACAS (v1.3.1) [25]. In order to finalize annotation of all the genomes we have used RATT [26] and the references species C. jejuni NCTC 11168 (AL111168), C. jejuni 414 (CM000855), C. jejuni RM1221 (CP000025), C. coli 76339 (HG326877), C. coli CVM N29710 (CP004066), C. concisus 13826 (CP000792), C. fetus 82-40 (CP000487), C. jejuni 81-176 (CP000538), C. jejuni M1 (CP001900) and C. lari RM2100 (CP000932). The Artemis and ACT software [27] were used to read the genomes. T6SS ORFs were identified using BLAST [28, 29].
Amplified fragment length polymorphism (AFLP)
The ABI 3100 Genetic Analyzer was used to perform automated amplified fragment length polymorphism (AFLP) as previously described [30, 31]. The resulting data was analysed using the BioNumerics V4.6.1 software and the Pearson product-moment correlation coefficient was required to normalize the profile similarities. Following normalization the unweighted pair group with mathematical average (UPGMA) method was used for clustering and dendrogram assembly. As controls DNA from standard C. jejuni and C. coli cultures were used in each run. In our experiments the similarities between standards were ≥90% having a position tolerance setting of 0.07%, and a profile size range of 50–500 bp.
PCR detection of hcp and gltA genes
To detect the hcp multiplex PCR analysis was used as previously described using the gltA as a control housekeeping gene [13]. For amplification of gltA the primers gltAF (gcccaaagcccatcaagcgga) and gltAR (gcgctttggggtcatgcaca) and for the amplification of the hcp gene primers hcpF (caagcggtgcatctactgaa) and hcpR (taagctttgccctctctcca) were used. C. jejuni NCTC 12502 served as the hcp+ control.
Motility assay
The motility of all hcp+ isolates (n = 51), plus one hcp− strain and NCTC 12502, was compared based on the method of Corcionivoschi et al. [32]. Briefly, 5 µl of culture [48 h, grown on blood agar and recovered in 1 ml brain heart infusion (BHI) broth] was inoculated into the centre of semi-solid BHI plates (0.4% agar). The radius of the disc of visible growth was measured after incubation (48 h). The experiment was carried out in triplicate.
Resistance to bile salts
The resistance of the isolates to bile salts was studied based on the method of Stef et al. [33]. Briefly, each isolate was grown confluently on blood agar (48 h) then 2.5 ml BHI broth added and mixed. This cell suspension (1 ml) was used to inoculate BHI broth (10 ml), and BHI broth containing 4.0% (wt/vol) bile salts (10 ml), dispensed into 30 ml bottles. The bottles were incubated (24 h) after which 20 μl was removed and added to 100 μl of water. Serial dilutions were made in maximum recovery diluent, plated onto mCCDA, incubated (48 h), and enumerated. The experiment was carried out in triplicate. Due to complete lack of resistance of some of the isolates we have scored the resistance as (+) resistant, (±) intermediate and (−) sensitive.
Gentamicin protection assay
In order to quantify the invasive capability of C. jejuni and C. coli isolates the gentamicin protection assay has been used as previously described [34]. Intestinal epithelial cells (HCT-8) were grown for up to 18 h until a confluence of 1 × 105 has been reached. The bacterial isolates were cultivated on blood agar plates for 2 days followed by serial dilution to reach an OD600 of 0.4. Once the HCT-8 cells reached the desired confluence they were washed with 1 ml of PBS followed by the addition and 2 ml of fresh tissue culture media. In our current study an MOI (multiplicity of infection) of 10 was used. One centrifugation step was necessary (250×g for 5 min) followed by incubation for 3 h at 37°C, in microaerophilic conditions (10% CO2). The infected cells, previously treated with 400 μg/ml, were washed three times in 1 ml PBS followed by exposure to 0.1% Triton X-100 at 37°C during a 2 h period. The lysate was diluted and spread on Mueller–Hinton agar plates and colonies counted after 2–3 days incubation or until colonies were visible on plates. The ratio between the total number of CFU and the initial inoculum was used to calculate the efficiency of invasion. The experiments were performed in triplicate.
Visualisation of capsular polysaccharide (CPS)
CPS was extracted from the isolates using the method of Hitchcock [35]. Briefly, plate grown bacteria were lysed for 5 min at 100°C in 31.25 mM Tris–HCl (pH 6.8), 4% sodium dodecyl sulphate, 0.025% bromophenol blue, and 20% glycerol. Proteinase K (20 mg ml−1 proteinase) was added to the lysate and incubated for 1 h at 50°C. The lysate, containing the polysaccharides, were separated in 10% Bis–Tris gels (NuPage Novex) for 1 h at 100 V (Invitrogen, Paisley, United Kingdom) and he gels were stained with Alcian blue [36].
Statistical analysis
Experiments were repeated three times in separate instances. Means of standard deviations (±) were used to represent the repeated experiments. Prism software was required for graph design and the unpaired Student t test to calculate the significance of data. The resulting P values were considered significant if their calculated values were <0.05.
Results
Prevalence of hcp and identification of T6SS open reading frames in C. coli and C. jejuni isolates
Using PCR, the presence of the hcp gene in the C. jejuni and C. coli isolates obtained packed retail chicken produced in Northern Ireland was determined, Figure 1. A higher prevalence was found in C. coli isolates (56.1%) than in C. jejuni (28.8%), Table 1. The gltA housekeeping gene in the multiplex PCR served as a positive control, and confirmed the isolates were all Campylobacter spp. C. jejuni strain 108 (JX436460) was used as reference when analysing the genomes of the chicken isolates in order to identify the presence of the T6SS ORFs. The hcp+ isolates were next investigated for the T6SS ORFs integrity. The results showed that 84.3% of the C. coli and 93.7% of the C. jejuni isolates, detected positive for hcp, possess all 13 T6SS ORFs, Table 2.
Figure 1.
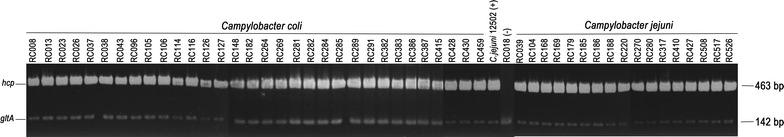
PCR detection of hcp in C. jejuni (n = 17) and C. coli (n = 32) isolates. Only positive isolates are shown, and one negative isolate (RC018). The positive control for hcp was C. jejuni 12502 and gltA served as the negative control for PCR reaction. Photoshop software was used to achieve the desired resolution.
Table 1.
Prevalence of hcp gene in Campylobacter spp. isolated from raw, retail chicken
| Species | Total number of isolates | Number of hcp positive strains (%) |
|---|---|---|
| Campylobacter coli | 57 | 32 (56.1) |
| Campylobacter jejuni | 59 | 17 (28.8) |
Table 2.
Presence of T6SS components in C. coli and C. jejuni chicken isolates in comparison with the amino acid sequence matches of C. jejuni strain 108
| TssA | TssB | TssC. | TssD | TssE | TssF | TssG | TssH | TssI | TssJ | TssK | TssL | TssM | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amino acids | 415 | 161 | 484 | 171 | 130 | 573 | 302 | 299 | 838 | 148 | 465 | 257 | 1175 | ||
| REFERENCE | TssA | TssB | TssC | TssD | TssE | TssF | TssG | TssH | TssI | TssJ | TssK | TssL | TssM | PCR results | Spp. |
| RC008 | + | C.colli | |||||||||||||
| RC013 | + | C.colli | |||||||||||||
| RC023 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 568 (99.1%) | 302 (100%) | 294 (98.3%) | 758 (90.5%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1171 (99.7%) | + | C.colli |
| RC026 | + | C.colli | |||||||||||||
| RC037 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC043 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC096 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 791 (94.4%) | 147 (99.3%) | 436 (93.8%) | 257 (100%) | 1151 (97.8%) | + | C.colli |
| RC105 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RG106 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC116 | 311 (74.9%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC126 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC127 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC148 | 389 (93.7%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 465 (100%) | 254(98.8%) | 1170 (99.6%) | + | C.colli |
| RC182 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 432 (92.9%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC264 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 465 (100%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC269 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC281 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC282 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC284 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC285 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC289 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1168(99.4%) | + | C.colli |
| RC291 | + | C.colli | |||||||||||||
| RC382 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC383 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC386 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC387 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC430 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 459 (98.7%) | 226 (87.9%) | 1170 (99.6%) | + | C.colli |
| RC415 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC428 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.colli |
| RC459 | 126 (97.0%) | 567 (99.0%) | 146 (48.3%) | 295(98.7%) | 599 (71.5%) | 1170 (99.6%) | + | C.colli | |||||||
| 12502 | 413 (99.5%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 127 (97.7%) | 569 (99.3%) | 301 (99.7%) | 294 (98.3%) | 825 (98.4%) | 147 (99.3%) | 464 (99.8%) | 254 (98.8%) | 1171 (99.7%) | + | C.jejuni |
| RC018 | 344 (71.1%) | 127 (97.7%) | 569 (99.3%) | 302 (100%) | 821 (98.0%) | 332 (28.3%) | + | C.colli | |||||||
| RC039 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 169 (98.8%) | 126 (97.0%) | 541 (94.4%) | 302 (100%) | 294 (98.3%) | 821 (98.0%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC104 | 340 (81.9%) | 155 (96.3%) | 477 (99.4%) | 156 (91.2%) | 115 (88.5%) | 567 (99.0%) | 272 (90.1%) | 296 (99.0%) | 782 (93.3%) | 146(98.6%) | 465 (100%) | 255 (99.2%) | 1158 (98.6%) | + | C.jejuni |
| RC168 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 526 (91.8%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC169 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC179 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC186 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC185 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC220 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 416 (89.5%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC270 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC280 | 367 (88.4%) | 156 (96.9%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 226 (87.9%) | 1170 (99.6%) | + | C.jejuni |
| RC317 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC410 | + | C.jejuni | |||||||||||||
| RC427 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 449 (96.6%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC508 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 294 (98.3%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | 1170 (99.6%) | + | C.jejuni |
| RC517 | 412 (99.3%) | 160 (99.4%) | 481 (99.4%) | 171 (100%) | 126 (97.0%) | 567 (99.0%) | 302 (100%) | 820 (97.9%) | 147 (99.3%) | 464 (99.8%) | 257 (100%) | + | C.jejuni | ||
| RC526 | 422 (87.2%) | 126 (97.0%) | 147 (49.2%) | 493 (58.8%) | + | C.jejuni |
Relative motility of isolates
Motility represents and important virulence factor in campylobacters increasing their ability to invade and colonise the gut epithelium. We have next investigated the motility of the hcp positive C. jejuni and C. coli isolates in order to differentiate and select them for further pathogenicity investigations, using motility as an exclusion factor. Our initial results show that none of the hcp+C. jejuni and C. coli isolates displayed motility significantly less than that of the control strain C. jejuni 12502 (data not shown), p < 0.05 and five isolates from each species were selected for further study (Figure 2), based on their high levels of motility.
Figure 2.
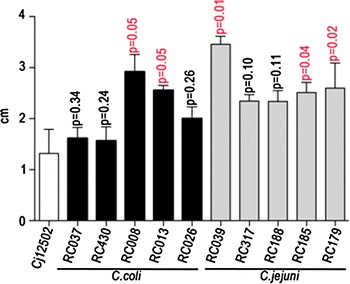
Relative motility assay of hcp + C. jejuni and C. coli isolates. The motility of five hcp positive C. jejuni and C. coli isolates was compared to hcp + control strain, C. jejuni 12502 in 0.4% BHI agar. Results are the mean of three separate experiments. The statistical significance was determined using the Student t test.
AFLP analysis of a subset of C. jejuni and C. coli strains
AFLP was used to determine the genetic difference between the five strains of C. coli and of C. jejuni selected for further study. The C. coli isolates; RC037, RC038 and RC013, 026 produced two clusters with 95.8% genetic similarity and 97.8% respectively. The degree of genetic similarity between these two clusters and C. coli isolates RC430 and RC008 was over 80%. When the hcp+ isolates were compared with the negative control, RC018, the degree of similarity was also very high, >70%, (Figure 3a). The C. jejuni 12502 control has been included in the AFLP diagram to specifically emphasize the genetic differences between C. jejuni and C. coli isolates and similarly for C. coli RC018 for the C. jejuni clustering.
Figure 3.
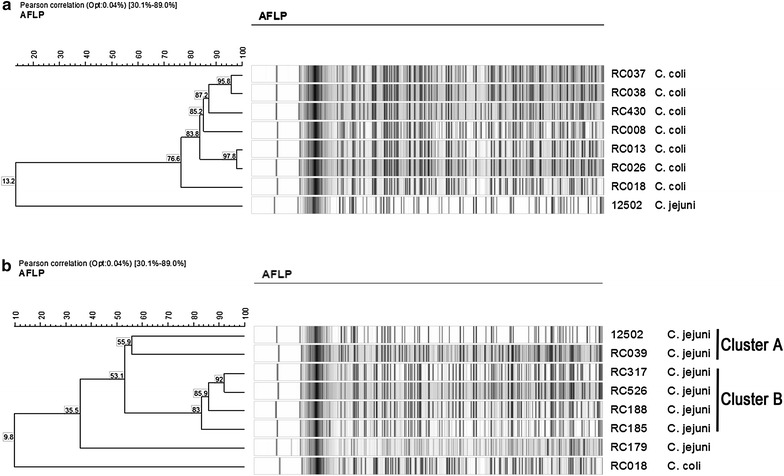
AFLP analysis of C. coli and C. jejuni hcp positive chicken isolates. a shows the AFLP fingerprints of the five most motile C. coli isolates and b the C. jejuni isolates. The percentage of amino acid identity between banding patterns is indicated. For comparison we have used the hcp positive C. jejuni 12502 and the negative isolate RC018.
C. jejuni isolates showing higher motility were also investigated by AFLP (panel b). Isolate RC039 formed a separate cluster with the control strain C. jejuni 12502 at a degree of similarity of over 50% (cluster A) and isolates RC317, RC526, RC188 and RC185 clustered together at a similarity >80% (cluster B). When cluster A was compared to cluster B the degree of similarity was approximately 50%. Isolate RC179 showed a low degree of similarity, approximately 35%, when compared to cluster A and B.
In vitro cell invasion abilities of hcp+ isolates
The ability of hcp+C. coli and C. jejuni isolates to invade HCT-8 cells was studied using C. jejuni 12502 as a reference strain (Figure 4). Using gentamicin, higher invasiveness rates were determined for C. coli isolates RC037 and RC130 when compared to C. jejuni invasiveness (p < 0.05). For Approximately 4% of the inoculums was internalised with C. coli isolates RC008, RC013 and RC026, and C. jejuni isolates RC317, RC188, RC185 and RC179: significantly more the level of less than 2% seen for the control strain (p < 0.01). C. jejuni isolate RC039 was most invasive, with approximately 6% of the inoculum being internalised (p < 0.001). A positive correlation between motility and invasiveness was seen for of all the hcp+C. coli and C. jejuni isolates. The hcp+ negative isolate (RC018) displayed invasiveness of less than 0.01% of the inoculum (data not shown).
Figure 4.
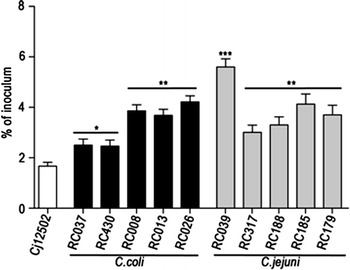
Invasive ability of C. coli and C. jejuni isolates. Invasion of HCT-8 cells of C. coli (RC037, 430, 008, 013 and 026) and C. jejuni (RC039, 317, 188, 185 and 179) isolates. Statistical significance (Student t test) relative to the level of C. jejuni 12502 is indicated. The experiments were done in triplicate and on three separate occasions. The error bars represent standard deviations.
Capsule polysaccharide profiles of C. coli isolates
Five isolates were chosen for study based on the high levels recorded in the motility determinations noted above. C. coli isolates RC037 and RC038 showed low amounts of CPS with most of the polysaccharides having very high molecular weights, similar to the control strain C. jejuni 12502. Isolates RC008, RC013 and RC026 had CPS molecular weights similar to the negative control, isolate RC018. The C. jejuni positive isolates had very different CPS profiles, with only the isolates RC526 and RC018 having profiles similar to each other, but dissimilar to C. jejuni 12502. Lower amounts of LOS were also detected, following staining with Alcian blue, for isolates RC039, RC526 and RC179, Figure 5. Due to the diversity of the capsule profiles obtained, between the different isolates, a capsule deficient mutant has not been used for comparison. We will focus in the future on the most virulent isolate and a mutant will be created for control purposes.
Figure 5.
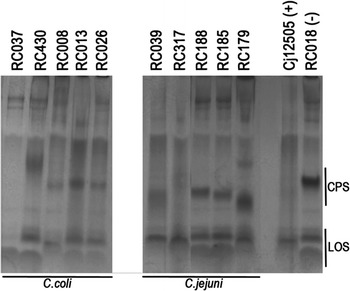
Capsule polysaccharides profiles of hcp positive isolates. Capsular polysaccharides were separated in 10% SDS-PAGE gels and stained with Alcian blue. The profiles of C. jejuni 12502 and C. coli RC018 served as controls for lipo-oligosaccharide (LOS).
Bile salts resistance
The ability of hcp+ chicken isolates to survive in the presence of bile salts (Table 3). Most of the C. coli isolates showed no resistance to bile salts (RC037, RC430, RC013 and RC026) with only isolate RC008 displaying intermediate resistance (i.e. lower counts compared to the control strain C. jejuni 12502). A similar pattern was also found with C. jejuni, with isolates RC039, RC317, RC188 and RC185 showing no resistance to bile salts in vitro.
Table 3.
Resistance to bile salts
| Isolate | Control | Campylobacter coli | Campylobacter jejuni | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12502 | RC037 | RC430 | RC008 | RC013 | RC026 | RC039 | RC317 | RC188 | RC185 | RC179 | |
| Bile salt resistance | + | – | – | ± | – | – | – | – | – | – | ± |
(+) resistance, (±) intermediate, (−) sensitive.
Discussion
Campylobacters are the main cause of in human food poisoning in Europe and North America, with chicken meat identified as a major source of campylobacters, which grow profusely in the gut and crop of broiler chickens. The European Food Safety Authority (EFSA) stated that reducing the numbers of Campylobacter in the intestines at slaughter 1,000-fold would reduce the public health risk by at least 90%. Previous studies have shown that the virulence abilities, of Campylobacter chicken isolates, are increased compared to human isolates [13, 37]. The Type VI secretion system novel protein translocation system has been described as a potentially new virulence factor active in vitro [11] and in vivo [14]. However, the role of T6SS in acute human disease remains to be determined, as the biological mechanisms involved are not yet fully understood. This study investigated the prevalence of T6SS in C. coli and C. jejuni chicken isolates and to characterise the virulence potential of these isolates in vitro.
The hcp gene is currently used as the main indicator for the presence of T6SS C. jejuni [11]. The incidence of hcp has been reported as being relatively higher in C. jejuni isolates from chickens in Asia, whilst and only a small proportion of chicken isolates in the UK were identified as positive for T6SS [13]. Given the pathogenic potential of hcp+ positive campylobacters, this study investigated, by multiplex PCR, the presence of the hcp gene in both C. coli and C. jejuni isolates obtained from retail chicken produced in Northern Ireland. It has been reported that the prevalence of C. coli isolates in retail chicken is much lower than that of C. jejuni [18] but this study found that the incidence of hcp was higher in C. coli, than in C. jejuni, hence these strains could potentially display increased virulence abilities. However, epidemiological studies have found that only 7% of reported cases of campylobacteriosis are caused by C. coli and 93% by C. jejuni, with most of the C. coli cases being recorded in older patients [38]. No epidemiological connection to the presence of hcp has been reported in C. coli. Our results show that the majority of the C. coli and C. jejuni isolates contained the complete set of ORFs required for a fully active T6SS. The characterization of the T6SS ORF was undertaken as it has been suggested [13] that the detection of the hcp gene alone might not actually indicate the presence of a full T6SS locus. This was demonstrated in Spanish isolates where it was shown that only 14% of the strains (n = 9) had a complete T6SS ORF [19].
In this study, analysis of the genetic similarities between isolates revealed similar results to those previously reported, i.e. suggesting that C. coli isolates were less diverse, based on the AFLP profiles, that C. jejuni isolates [39], probably due to C. coli showing more host specificity than C. jejuni [40].
Campylobacter species have emerged as gastrointestinal pathogens in humans and commensals in birds [41] with motility as a phenotypic feature required to achieve both colonization [42] and internalization in surface intestinal epithelial layer [43]. The results reported above show that all of the hcp+ chicken isolates had greater motility than an hcp+ human isolate (C. jejuni 12502), but no direct link between the presence of the hcp gene and motility in Campylobacter has yet been reported. However, the observations reported above are supported by studies with other Gram-negative bacteria such as Aeromonas hydrophila. A. hydrophila produces a cytotoxic enterotoxin, and associated with this toxin is a type II secretion system. However, it has also been reported that A. hydrophila possess a T6SS and that hcp increases not only motility but also protease and biofilm formation, thus significantly influencing the pathogenicity and survival of this microorganism [44].
In Campylobacter, motility is recognised as an important virulence factor [1] and the chicken isolates in this study were shown, not only to be highly motile, but also very invasive when in vitro gentamicin protection assays were performed with human intestinal cells [33]. In general hcp− strains were poorly invasive, e.g. C. jejuni 11168, with less than 0.1% of the inoculum invading the epithelial cells [34] but with hcp+ strains over 1% invades [14]. All of the hcp+ chicken isolates investigated in this study demonstrated greater invasiveness in the gentamicin protection assay [14], when compared to the control. The involvement of hcp in C. coli in vitro virulence is still under debate since our genomic sequencing results show that even the isolates with an incomplete ORF are not significantly different in virulence compared to strains with complete ORFs. This suggests that in these isolates other virulence factors might also be involved.
To act as a haemolysin, hcp requires to be synthesised and subsequently injected into the host cell, using the fully functional T6SS. This process will require modifications of the bacterial surface structures, especially its polysaccharide capsule, to allow the T6SS to act effectively. The decrease in capsule production has been reported as necessary for effective T6SS mediated toxicity and virulence [11]. Accordingly, the capsule polysaccharides of hcp+ isolates were studied to detect any differences in the molecular weights, or amounts, between these isolates. With isolate RC039, less CPS appeared to coincide with greater virulence characteristics, however this was not the case with the other isolates. However, in Campylobacter de-capsulation can occur in the presence of epithelial cells [34], hence the CPS profiles may not reflect their state during pathogenesis.
In order to successfully colonise humans, Campylobacter will have to be resistant to the potentially lethal conditions present in the intestinal tract, and possess resistance to bile salts [45]. It has been suggested that deoxycholic acid, at physiological levels found in the human gut, can inhibit cell growth in C. jejuni, however when the concentration falls to those found in the proximal colon [46], the T6SS system is expressed and this contributes to colonic inflammation in humans [14]. In this study the resistance of the isolates to bile salts were compared with the human isolate C. jejuni 12502, revealing that most isolates had no resistance to levels of bile salts previously described as minimally inhibitory concentrations [45].
Conclusion
Taken together, all these results show that the hcp gene and the complete T6SS are prevalent in C. coli as well as in C. jejuni isolates obtained from retail chicken at a very high rate. Overall the results suggest that C. jejuni and C. coli chicken isolates containing the complete T6SS ORFs potentially have greater virulence in vitro, than do reference strains, but the consequences of these factors, in vivo, has yet to be determined.
Availability of supporting data
The data set supporting the results of this article are available in the European Nucleotide Archive under the project accession number PRJEB9722.
Authors’ contributions
NC, RM, OG participated in the design of the study. NC, RM wrote the manuscript. LS, AC carried out the infection studies. OG performed the genome sequencing experiments. CK, LM, PS carried out motility, bile salts resistance and AFLP analysis, BW, ND revised the manuscript critically and interpreted the data. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Agri-Food and Biosciences Institute, Belfast, Northern Ireland and by a grant from the Romanian Authority for Scientific Research, CNS-UEFISCDI, project number PN-II-RU-TE-2012-3-0092.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Contributor Information
Nicolae Corcionivoschi, Email: Nicolae.corcionivoschi@afbini.gov.uk.
Ozan Gundogdu, Email: Ozan.Gundogdu@lshtm.ac.uk.
Lynn Moran, Email: Lynn.Moran@afbini.gov.uk.
Carmel Kelly, Email: Carmel.Kelly@afbini.gov.uk.
Pam Scates, Email: Pam.Scates@afbini.gov.uk.
Lavinia Stef, Email: lavi_stef@animalsci-tm.ro.
Ada Cean, Email: teleaada@gmail.com.
Brendan Wren, Email: Brendan.wren@lshtm.ac.uk.
Nick Dorrell, Email: Nick.dorrell@lshtm.ac.uk.
Robert H Madden, Email: Bob.madden@afbini.gov.uk.
References
- 1.van Vliet AH, Ketley JM. Pathogenesis of enteric Campylobacter infection. Symp Ser Soc Appl Microbiol. 2001;90:45S–56S. doi: 10.1046/j.1365-2672.2001.01353.x. [DOI] [PubMed] [Google Scholar]
- 2.Yao R, Burr DH, Doig P, Trust TJ, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- 3.Strachan NJ, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, et al. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis. 2009;199:1205–1208. doi: 10.1086/597417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin O, Morishita TY, Zhang Q. Campylobacter colonization in poultry: sources of infection and modes of transmission. Anim Health Res Rev. 2002;3:95–105. doi: 10.1079/AHRR200244. [DOI] [PubMed] [Google Scholar]
- 5.Wagenaar JA, Mevius DJ, Havelaar AH. Campylobacter in primary animal production and control strategies to reduce the burden of human campylobacteriosis. Rev Sci Tech. 2006;25:581–594. [PubMed] [Google Scholar]
- 6.Behringer M, Miller WG, Oyarzabal OA. Typing of Campylobacter jejuni and Campylobacter coli isolated from live broilers and retail broiler meat by flaA-RFLP, MLST, PFGE and REP-PCR. J Microbiol Methods. 2011;84:194–201. doi: 10.1016/j.mimet.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Crushell E, Harty S, Sharif F, Bourke B. Enteric campylobacter: purging its secrets? Pediatr Res. 2004;55:3–12. doi: 10.1203/01.PDR.0000099794.06260.71. [DOI] [PubMed] [Google Scholar]
- 8.Humphreys H, Bourke S, Dooley C, McKenna D, Power B, Keane CT, et al. Effect of treatment on Campylobacter pylori in peptic disease: a randomised prospective trial. Gut. 1988;29:279–283. doi: 10.1136/gut.29.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz S, Hood RD, Mougous JD. What is type VI secretion doing in all those bugs? Trends Microbiol. 2010;18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe. 2010;8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleumink-Pluym NM, van Alphen LB, Bouwman LI, Wosten MM, van Putten JP. Identification of a functional type VI secretion system in Campylobacter jejuni conferring capsule polysaccharide sensitive cytotoxicity. PLoS Pathog. 2013;9:e1003393. doi: 10.1371/journal.ppat.1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bingle LE, Bailey CM, Pallen MJ. Type VI secretion: a beginner’s guide. Curr Opin Microbiol. 2008;11:3–8. doi: 10.1016/j.mib.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Harrison JW, Dung TT, Siddiqui F, Korbrisate S, Bukhari H, Tra MP, et al. Identification of possible virulence marker from Campylobacter jejuni isolates. Emerg Infect Dis. 2014;20:1026–1029. doi: 10.3201/eid2006.130635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lertpiriyapong K, Gamazon ER, Feng Y, Park DS, Pang J, Botka G, et al. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One. 2012;7:e42842. doi: 10.1371/journal.pone.0042842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madden RH, Moran L, Scates P, McBride J, Kelly C. Prevalence of Campylobacter and Salmonella in raw chicken on retail sale in the republic of Ireland. J Food Prot. 2011;74:1912–1916. doi: 10.4315/0362-028X.JFP-11-104. [DOI] [PubMed] [Google Scholar]
- 16.1 IBEI . Microbiology of food and animal feeding stuffs-horizontal method for detection and enumeration of Campylobacter spp. London: BSI; 2006. [Google Scholar]
- 17.Moran L, Kelly C, Madden RH. Factors affecting the recovery of Campylobacter spp. from retail packs of raw, fresh chicken using ISO 10272-1:2006. Lett Appl Microbiol. 2009;48:628–632. doi: 10.1111/j.1472-765X.2009.02581.x. [DOI] [PubMed] [Google Scholar]
- 18.Moran L, Scates P, Madden RH. Prevalence of Campylobacter spp. in raw retail poultry on sale in Northern Ireland. J Food Prot. 2009;72:1830–1835. doi: 10.4315/0362-028x-72.9.1830. [DOI] [PubMed] [Google Scholar]
- 19.Ugarte-Ruiz M, Stabler RA, Dominguez L, Porrero MC, Wren BW, Packard CA, et al. Prevalence of Type VI secretion system in Spanish Campylobacter jejuni isolates. Zoonoses Public Health. 2014 doi: 10.1111/zph.12176. [DOI] [PubMed] [Google Scholar]
- 20.Andrews S (2010) FastQC, a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 25 May 2014
- 21.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gladman S ST (2012) VelvetOptimiser, v2.2.5
- 24.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25:1968–1969. doi: 10.1093/bioinformatics/btp347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: rapid annotation transfer tool. Nucleic Acids Res. 2011;39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 30.Harrington CS, Thomson-Carter FM, Carter PE. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;35:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokotovic B, On SL. High-resolution genomic fingerprinting of Campylobacter jejuni and Campylobacter coli by analysis of amplified fragment length polymorphisms. FEMS Microbiol Lett. 1999;173:77–84. doi: 10.1111/j.1574-6968.1999.tb13487.x. [DOI] [PubMed] [Google Scholar]
- 32.Corcionivoschi N, Alvarez LA, Sharp TH, Strengert M, Alemka A, Mantell J, et al. Mucosal reactive oxygen species decrease virulence by disrupting Campylobacter jejuni phosphotyrosine signaling. Cell Host Microbe. 2012;12:47–59. doi: 10.1016/j.chom.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stef L, Cean A, Vasile A, Julean C, Drinceanu D, Corcionivoschi N. Virulence characteristics of five new Campylobacter jejuni chicken isolates. Gut Pathog. 2013;5:41. doi: 10.1186/1757-4749-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corcionivoschi N, Clyne M, Lyons A, Elmi A, Gundogdu O, Wren BW, et al. Campylobacter jejuni cocultured with epithelial cells reduces surface capsular polysaccharide expression. Infect Immun. 2009;77:1959–1967. doi: 10.1128/IAI.01239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hitchcock PJ. Aberrant migration of lipopolysaccharide in sodium dodecyl sulfate/polyacrylamide gel electrophoresis. Eur J Biochem. 1983;133:685–688. doi: 10.1111/j.1432-1033.1983.tb07517.x. [DOI] [PubMed] [Google Scholar]
- 36.Karlyshev AV, Wren BW. Detection and initial characterization of novel capsular polysaccharide among diverse Campylobacter jejuni strains using alcian blue dye. J Clin Microbiol. 2001;39:279–284. doi: 10.1128/JCM.39.1.279-284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cean A, Stef L, Simiz E, Julean C, Dumitrescu G, Vasile A, et al. Effect of human isolated probiotic bacteria on preventing Campylobacter jejuni colonization of poultry. Foodborne Pathog Dis. 2015;12:122–130. doi: 10.1089/fpd.2014.1849. [DOI] [PubMed] [Google Scholar]
- 38.Gillespie IA, O’Brien SJ, Frost JA, Adak GK, Horby P, Swan AV, et al. A case-case comparison of Campylobacter coli and Campylobacter jejuni infection: a tool for generating hypotheses. Emerg Infect Dis. 2002;8:937–942. doi: 10.3201/eid0809.010817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kittl S, Korczak BM, Niederer L, Baumgartner A, Buettner S, Overesch G, et al. Comparison of genotypes and antibiotic resistances of Campylobacter jejuni and Campylobacter coli on chicken retail meat and at slaughter. Appl Environ Microbiol. 2013;79:3875–3878. doi: 10.1128/AEM.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopkins KL, Desai M, Frost JA, Stanley J, Logan JM. Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping, and phage typing. J Clin Microbiol. 2004;42:229–235. doi: 10.1128/JCM.42.1.229-235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wassenaar TM, van der Zeijst BA, Ayling R, Newell DG. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J Gen Microbiol. 1993;139(Pt 6):1171–1175. doi: 10.1099/00221287-139-6-1171. [DOI] [PubMed] [Google Scholar]
- 42.Lee A, O’Rourke JL, Barrington PJ, Trust TJ. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect Immun. 1986;51:536–546. doi: 10.1128/iai.51.2.536-546.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szymanski CM, King M, Haardt M, Armstrong GD. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect Immun. 1995;63:4295–4300. doi: 10.1128/iai.63.11.4295-4300.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sha J, Rosenzweig JA, Kozlova EV, Wang S, Erova TE, Kirtley ML, et al. Evaluation of the roles played by Hcp and VgrG type 6 secretion system effectors in Aeromonas hydrophila SSU pathogenesis. Microbiology. 2013;159:1120–1135. doi: 10.1099/mic.0.063495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mavri A, Smole Mozina S. Resistance to bile salts and sodium deoxycholate in macrolide- and fluoroquinolone-susceptible and resistant Campylobacter jejuni and Campylobacter coli strains. Microb Drug Resist. 2013;19:168–174. doi: 10.1089/mdr.2012.0217. [DOI] [PubMed] [Google Scholar]
- 46.Hamilton JP, Xie G, Raufman JP, Hogan S, Griffin TL, Packard CA, et al. Human cecal bile acids: concentration and spectrum. Am J Physiol Gastrointest Liver Physiol. 2007;293:G256–G263. doi: 10.1152/ajpgi.00027.2007. [DOI] [PubMed] [Google Scholar]


