Abstract
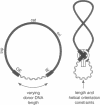
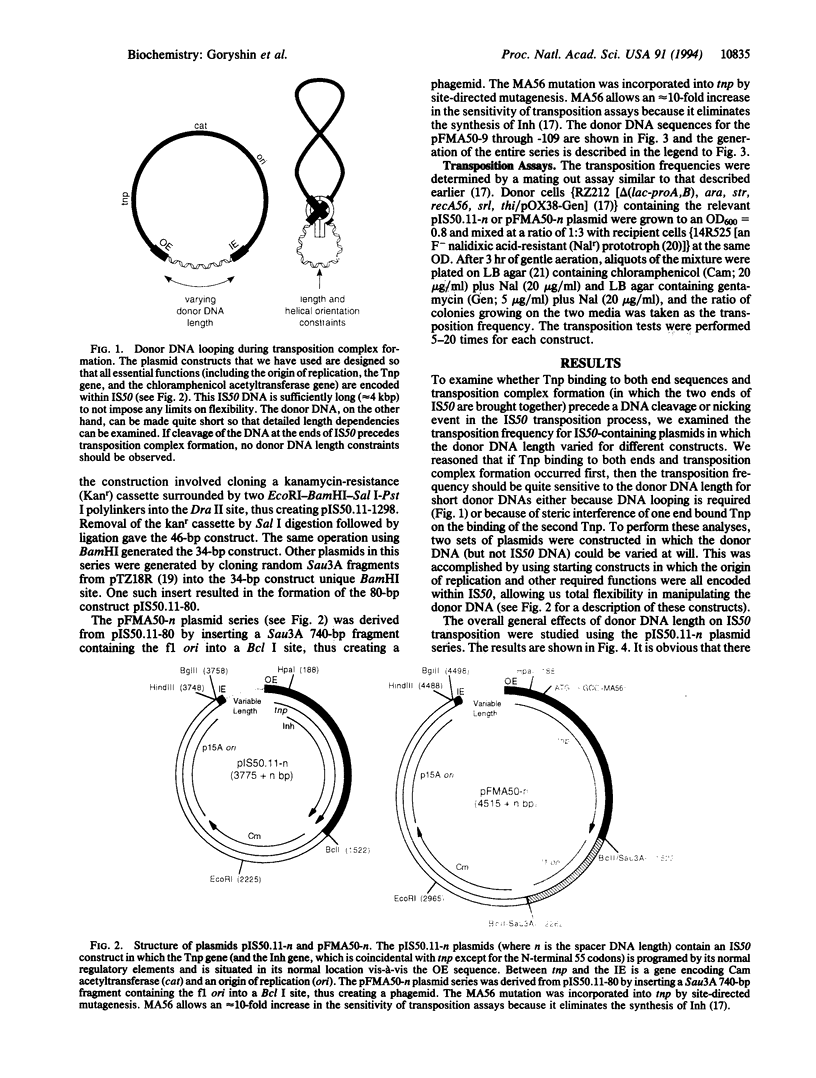
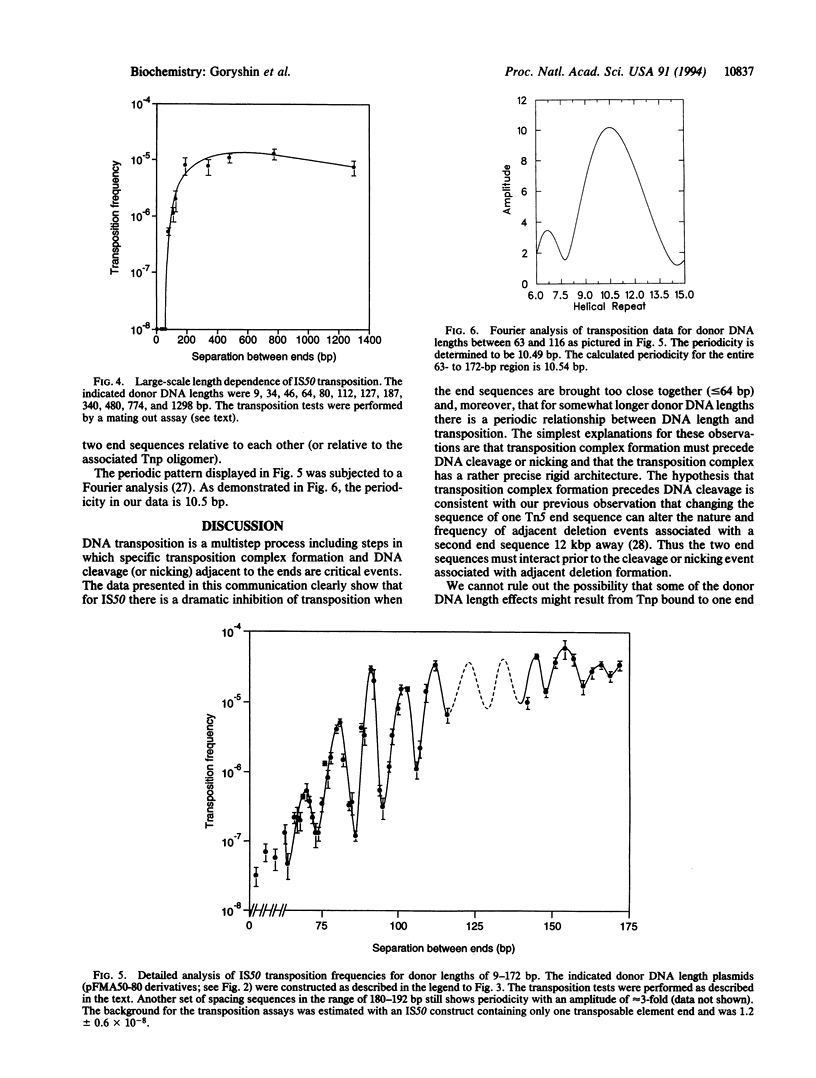
Transposition is a multistep process in which a transposable element DNA sequence moves from its original genetic location to a new site. Early steps in this process include the formation of a transposition complex in which the end sequences of the transposable element are brought together in a structurally precise fashion through the action of the element-encoded transposase protein and the cleavage of the element free from the adjoining DNA. If transposition complex formation must precede DNA cleavage (or nicking), then changing the length of the donor DNA between closely spaced ends should have dramatic effects on the frequency of the transposition. This question has been examined by studying the effects of altering donor DNA length on IS50 transposition. Donor DNA < or = 64 bp severely impaired transposition. Donor DNA > or = 200 bp demonstrated high transposition frequencies with only modest length dependencies. Constructs with donor DNA lengths between 66 and 174 bp demonstrated a dramatic periodic effect on transposition (periodicity approximately 10.5 bp).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. E., Bliska J. B., Cozzarelli N. R. Cre-lox recombination in Escherichia coli cells. Mechanistic differences from the in vitro reaction. J Mol Biol. 1992 Aug 5;226(3):661–673. doi: 10.1016/0022-2836(92)90623-r. [DOI] [PubMed] [Google Scholar]
- Bainton R., Gamas P., Craig N. L. Tn7 transposition in vitro proceeds through an excised transposon intermediate generated by staggered breaks in DNA. Cell. 1991 May 31;65(5):805–816. doi: 10.1016/0092-8674(91)90388-f. [DOI] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986 Jun 20;45(6):801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haykinson M. J., Johnson R. C. DNA looping and the helical repeat in vitro and in vivo: effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 1993 Jun;12(6):2503–2512. doi: 10.1002/j.1460-2075.1993.tb05905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschel B. J., Galas D. J., Berg D. E., Chandler M. Structure and stability of transposon 5-mediated cointegrates. J Mol Biol. 1982 Aug 25;159(4):557–580. doi: 10.1016/0022-2836(82)90101-2. [DOI] [PubMed] [Google Scholar]
- Hirschel B. J., Galas D. J., Chandler M. Cointegrate formation by Tn5, but not transposition, is dependent on recA. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4530–4534. doi: 10.1073/pnas.79.15.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isberg R. R., Lazaar A. L., Syvanen M. Regulation of Tn5 by the right-repeat proteins: control at the level of the transposition reaction? Cell. 1982 Oct;30(3):883–892. doi: 10.1016/0092-8674(82)90293-8. [DOI] [PubMed] [Google Scholar]
- Jilk R. A., Makris J. C., Borchardt L., Reznikoff W. S. Implications of Tn5-associated adjacent deletions. J Bacteriol. 1993 Mar;175(5):1264–1271. doi: 10.1128/jb.175.5.1264-1271.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. DNA sequences at the ends of transposon Tn5 required for transposition. Nature. 1983 Jul 21;304(5923):280–282. doi: 10.1038/304280a0. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Reznikoff W. S. Role of the IS50 R proteins in the promotion and control of Tn5 transposition. J Mol Biol. 1984 Aug 25;177(4):645–661. doi: 10.1016/0022-2836(84)90042-1. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Kil' Iu V., Goryshin I. Iu, Lantsov V. A. Rekombinatsionnyi mekhanizm tochnoi ékstsizii mobil'nogo élementa IS50 v kletkakh Escherichia coli K12. Mol Biol (Mosk) 1994 May-Jun;28(3):563–573. [PubMed] [Google Scholar]
- Krebs M. P., Reznikoff W. S. Transcriptional and translational initiation sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986 Dec 20;192(4):781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- Law S. M., Bellomy G. R., Schlax P. J., Record M. T., Jr In vivo thermodynamic analysis of repression with and without looping in lac constructs. Estimates of free and local lac repressor concentrations and of physical properties of a region of supercoiled plasmid DNA in vivo. J Mol Biol. 1993 Mar 5;230(1):161–173. doi: 10.1006/jmbi.1993.1133. [DOI] [PubMed] [Google Scholar]
- Lee D. H., Schleif R. F. In vivo DNA loops in araCBAD: size limits and helical repeat. Proc Natl Acad Sci U S A. 1989 Jan;86(2):476–480. doi: 10.1073/pnas.86.2.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Niederweis M., Lederer T., Hillen W. An accurate method for determining the helical repeat of DNA in solution reveals differences to the crystal structures of two B-DNA decamers. J Mol Biol. 1992 Nov 20;228(2):322–326. doi: 10.1016/0022-2836(92)90820-a. [DOI] [PubMed] [Google Scholar]
- Reznikoff W. S. The Tn5 transposon. Annu Rev Microbiol. 1993;47:945–963. doi: 10.1146/annurev.mi.47.100193.004501. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Helical periodicity of DNA determined by enzyme digestion. Nature. 1980 Aug 7;286(5773):573–578. doi: 10.1038/286573a0. [DOI] [PubMed] [Google Scholar]
- Sasakawa C., Carle G. F., Berg D. E. Sequences essential for transposition at the termini of IS50. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7293–7297. doi: 10.1073/pnas.80.23.7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992 Oct 20;227(4):1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Wiater L. A., Grindley N. D. Integration host factor increases the transpositional immunity conferred by gamma delta ends. J Bacteriol. 1990 Sep;172(9):4951–4958. doi: 10.1128/jb.172.9.4951-4958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand T. W., Reznikoff W. S. Characterization of two hypertransposing Tn5 mutants. J Bacteriol. 1992 Feb;174(4):1229–1239. doi: 10.1128/jb.174.4.1229-1239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J. C., Krebs M. P., Reznikoff W. S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988 Jan 5;199(1):35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]